Abstract
The Asian spongy moth, Lymantria dispar asiatica, is one of the most devastating forestry defoliators. The absence of a high-quality genome limited the understanding of its adaptive evolution. Here, we conducted the first chromosome-level genome assembly of L. dispar asiatica using PacBio HIFI long reads, Hi-C sequencing reads and transcriptomic data. The total assembly size is 997.59 Mb, containing 32 chromosomes with a GC content of 38.91% and a scaffold N50 length of 35.42 Mb. The BUSCO assessment indicated a completeness estimate of 99.4% for this assembly. A total of 19,532 protein-coding genes was predicted. Our study provides a valuable genomics resource for studying the mechanisms of adaptive evolution and facilitate an efficient control of L. dispar asiatica.
Subject terms: Genome, Molecular biology, Evolutionary genetics
Background & Summary
The spongy moth, Lymantria dispar, is one of the most important forestry pests. It is widely distributed across the temperate forests of the northern hemisphere, such as Europe, China and North America1,2. The larvae are destructive polyphagous folivores, and they consume more than 600 plant species ranging from oaks to conifers3. They completely defoliate entire trees, resulting in significant ecological and economic losses4,5. The spongy moth is divided into Asian (L. dispar asiatica) and European (L. dispar dispar) species based on origin6. Introduced to North America in 1869, the European variant has spread widely over 150 years7. The Asia spongy moth poses a greater threat due to its robust reproductive capacity and flight abilities8. Females are particularly drawn to lights in ports, often laying their egg masses on cargo and the superstructure of ships9. At present, how to effectively control the invasion and spread of the spongy moth has become a global research hotspot10.
Chemical control is the primary control method to combat the spongy moth11. Since the last century, a variety of insecticides have been used for spongy moth control12. Unfortunately, the frequent and extensive use of pesticides not only adversely affects biodiversity but also hastens the development of insecticide resistance. Xenobiotic detoxification is a crucial mechanism enabling insects to resist toxic phytochemicals or pesticides. It depends on the constitutive quantitative changes in the expression and activity of multiple detoxification enzymes, including cytochrome P450s (CYP450s), UDP-glucuronosyltransferase (UGTs), glutathione S-transferases (GSTs) and ATP-binding cassette transporters (ABC) family13. Currently, the development of RNAi insecticides targeting these detoxification genes is the focus of the pest control field. However, the lack of genomic information significantly constrains the identification of effective targets in the spongy moth. Additionally, this deficit impedes the understanding of insecticide resistance mechanisms in the spongy moth from the genomic diversity and evolution perspective.
Here, we constructed the first high-quality chromosome-level reference genome of L. dispar using PacBio long-read sequencing and Hi-C sequencing technologies. The final genome size was 997.59 Mb with N50 sizes of 35.42 Mb, and 991.35 Mb genome sequences were further clustered and ordered into 32 chromosomes. A total of 19,532 protein-coding genes was predicted in the genome of L. dispar asiatica. This chromosome-level genome assembly of L. dispar asiatica provides a valuable genomics resource for investigating its evolutionary dynamics and aiding in the control of L. dispar asiatica.
Methods
Insect rare and sample collection
The egg masses of L. dispar asiatica were obtained from a poplar filed in Harbin, Heilongjiang Province and maintained at 4 °C before hatching. Hatched larvae were fed with an artificial diet at 25 ± 1 °C, 14:10 (L:D) photoperiod and 65 ± 5% relative humidity referring to our previous studies14,15. The 2nd, 3rd, 4th, 5th instar larvae, pupae, and adult moth were collected separately. The samples were frozen in liquid nitrogen and then stored at −80 °C.
Genome sequencing and assembly
Genomic DNA was isolated from a fresh female pupa of Lymantria dispar asiatica using the sodium dodecyl sulfate (SDS) extraction method16. For PacBio long-read sequencing, 8 µg DNA was sheared into fragments of 15–20 kb in length by g-TUBE (Covaris USA) and then purified with AMPure PB Beads. High-fidelity (HiFi) libraries were constructed with SMRTbell Express Template Prep Kit 2.0 and sequenced on Pacbio Sequel IIe platform (Pacifc Biosciences, Menlo Park, USA). A total of 33.02 Gb HiFi reads with N50 sizes of 20,583 bp were obtained using Circular Consensus Sequencing (CCS) mode (Table 1). The PacBio HiFi reads of L. dispar asiatica were de novo assembled by using Hifiasm software v0.19.517,18 with default parameters. The draft genome had a total size of 997.53 Mb containing 102 contigs with N50 sizes of 32.048 Mb (Table 2).
Table 1.
Statistics of the HIFI sequence data used for genome assembly.
| sample | Read_base | Read_Number | Read_length(max) | Read_length(mean) | Read_length(N50) |
|---|---|---|---|---|---|
| pupa | 33,02,05,01,680 | 16,12,392 | 49,587 | 20,479 | 20,583 |
Table 2.
Summary statistics of the Lymantria dispar asiatica genome assembly.
| Contig level | |
|---|---|
| Assembly length (bp) | 99,75,33,159 |
| Longest contig (bp) | 4,42,20,918 |
| Number of contigs | 102 |
| GC (%) | 38.91 |
| Contig N50 (bp) | 3,20,48,912 |
| BUSCO | C:99.3%[S:97.8%,D:1.5%],F:0.3%,M:0.4%,n:1367 |
| Chromosome level | |
| Number of chromosomes | 32 |
| Chromosome length (bp) | 99,75,90,907 |
| Scafold N50 (bp) | 3,54,22,674 |
| BUSCO | C:99.4%[S:97.9%,D:1.5%],F:0.2%,M:0.4%,n:1367 |
Hi-C scaffolding
To construct Hi-C libraries, the 5th instar female larva of L. dispar asiatica was used as inputs following previously described standard protocols19. In detail, the larva was cut into small pieces and pulverized in liquid nitrogen. The tissues were cross-linked by 4% formaldehyde solution for 30 mins. After quenching the crosslinking reaction with 2.5 M glycine, tissue sample was centrifuged at 2500 rpm at 4°C for 10 mins. The pellet was washed with 500 μl PBS and then centrifuged for 5 min (2500 rpm). Subsequently, the pellet was resuspended in 20 ul of lysis buffer, followed by twice washing with 100 μl ice cold 1x NEB buffer. The nuclei were collected by centrifuging at 5000 rpm for 5 min, resuspended with 100 μl NEB buffer, and solubilized with dilute SDS. After quenching the SDS with Triton X-100, the samples were digested overnight at 37 °C with a 4-cutter restriction enzyme MboI (400 units). The linked DNA was labelled with biotin-14-dCTP and then ligated by T4 DNA polymerase. The ligated DNA was sheared fragments by sonication (200–600 base pairs) and sequenced on Illumina HiSeq-2500 platform (PE 125 bp) with the paired-end module. About 110.96 Gb of raw data were obtained from L. dispar asiatica (Table 3).
Table 3.
Statistics of the Hic sequence data used for genome assembly.
| Sample | Raw paired reads | Raw Base(bp) | Duplication rate(%) | Effective Rate(%) | Error Rate(%) | Q20(%) | Q30(%) | GC Content(%) |
|---|---|---|---|---|---|---|---|---|
| L. dispar asiatica larvae | 36,98,62,582 | 1,10,95,87,74,600 | 20.31% | 78.97 | 0.03 | 97.47 | 93.23 | 40.02 |
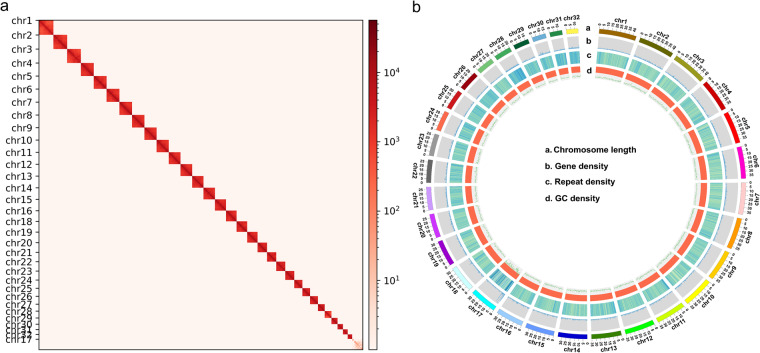
The high-quality sequencing reads were filtered by fastp v0.23.420. The cleaned Hi-C reads were then mapped to the draft genome using Juicer v1.621. The unique high-quality paired-end reads were taken as input for 3D-DNA v190716 pipeline22 with parameters “-r 0”. Chromosome interaction matrix was manually adjusted by using JuicerBox v1.11.0821. The Hi-C heatmap was drawn with HiCExplorer v3.7.223. Finally, a total of 32 chromosomes was obtained, which contained 99.38% of the assembled contigs (Fig. 1).
Fig. 1.
The genome features of Lymantria dispar asiatica. (a) genome-wide Hi-C heatmap of chromatin interaction counts. (b) Circos plot of the 32 chromosomes of Lymantria dispar asiatica. From the outermost layer to the innermost layer, the chromosome length, gene density, repeat density, and GC density are sequentially displayed.
After Hi-C scaffolding, the genome integrity was evaluated using Benchmarking Universal Single-Copy Orthologs (BUSCO v5.4.3)24. This analysis revealed that L. dispar asiatica chromosome level assembly contained C:99.4% [S:97.9%, D:1.5%], F:0.2%, M:0.4%, n:1367 (Table 4). The results indicated that the genome assembly of L. dispar asiatica were of considerable contiguity, completeness and accuracy.
Table 4.
Statistics of repetitive elements in the Lymantria disapr asiatica genome.
| Elements | Repeat type | Number of elements | length (bp) | percentage in genome |
|---|---|---|---|---|
| SINEs | Retroelements | 331135 | 22932667 | 2.30% |
| Penelope | Retroelements | 0 | 0 | 0.00% |
| LINEs | Retroelements | 1188736 | 242743224 | 24.33% |
| CRE/SLACS | Retroelements | 640 | 105470 | 0.01% |
| L2/CR1/Rex | Retroelements | 98136 | 21226467 | 2.13% |
| R1/LOA/Jockey | Retroelements | 138973 | 73208592 | 7.34% |
| R2/R4/NeSL | Retroelements | 14042 | 2586546 | 0.26% |
| RTE/Bov-B | Retroelements | 632322 | 90599808 | 9.08% |
| L1/CIN4 | Retroelements | 227 | 17956 | 0.00% |
| BEL/Pao | Retroelements | 11923 | 8500706 | 0.85% |
| Ty1/Copia | Retroelements | 7017 | 1982373 | 0.20% |
| Gypsy/DIRS1 | Retroelements | 28427 | 10317472 | 1.03% |
| Retroviral | Retroelements | 3967 | 281397 | 0.03% |
| hobo-Activator | DNA transposons | 140277 | 18453863 | 1.85% |
| Tc1-IS630-Pogo | DNA transposons | 91034 | 26313765 | 2.64% |
| En-Spm | DNA transposons | 0 | 0 | 0.00% |
| MULE-MuDR | DNA transposons | 994 | 78938 | 0.01% |
| PiggyBac | DNA transposons | 7452 | 1244764 | 0.12% |
| Tourist/Harbinger | DNA transposons | 5026 | 808317 | 0.08% |
| Other (Mirage, P-element, Transib) | DNA transposons | 2115 | 256393 | 0.03% |
| Rolling-circles | Rolling-circles | 564710 | 78046078 | 7.82% |
| Satellites | Small RNA | 5385 | 524461 | 0.05% |
| Simple repeats | Small RNA | 188889 | 9526362 | 0.95% |
| Low complexity | Small RNA | 23215 | 1097031 | 0.11% |
| Unclassified: | 792468 | 88703522 | 8.89% |
RNA sequencing
Larvae in the 2nd, 3rd, 4th, and 5th instars, along with adult females, were collected for RNA extraction. Total RNA was extracted from each tissue respectively using TRIzol reagent. Sequencing libraries were generated by NEBNext Ultra RNA Library Prep Kit (NEB, USA). The transcriptomes were sequenced on the Illumina Hiseq 4000 platform with PE150 strategy, and a total of 33.99 Gb short-read RNA-seq raw data were obtained (Table 5).
Table 5.
Statistics of RNA sequcing data of Lymantria disapr asiatica.
| Sample | Raw Reads | Clean Reads | Raw Base(G) | Clean Base(G) | Effective(%) | Error(%) | Q20(%) | Q30(%) | GC(%) |
|---|---|---|---|---|---|---|---|---|---|
| 2 instar | 49084562 | 43903568 | 7.36 | 6.59 | 89.44 | 0.03 | 97.74 | 93.6 | 45.02 |
| 3 instar | 50192094 | 46752784 | 7.53 | 7.01 | 93.15 | 0.03 | 97.6 | 93.27 | 43.41 |
| 4 instar | 44017684 | 41351054 | 6.6 | 6.2 | 93.94 | 0.03 | 97.46 | 92.96 | 43.37 |
| 5 instar | 44119946 | 40964012 | 6.62 | 6.14 | 92.85 | 0.03 | 97.79 | 93.73 | 42.71 |
| Adult female | 39174758 | 36613146 | 5.88 | 5.49 | 93.46 | 0.03 | 97.67 | 93.55 | 45.17 |
Genome annotation
For insect genome annotation, we mainly referred to a genome annotation pipeline developed by the Institute of Insect Sciences, Zhejiang University25. In detail, a de novo repeat library of insect specialization was firstly constructed using RepeatModeler v2.0.426 and RepeatMasker v4.1.527 for repeat sequence annotation. Then the genome was masked by RepeatMasker v4.1.5 with “-xsmall” parameters. In the genomic sequences, a total of 527.67 Mb (52.89%) repetitive elements were identified, mainly including 28.98% retroelements, 5.94% DNA transposons, 7.82% rolling-circles, and 8.89% unclassified repeat sequence (Table 2). To predict protein-coding genes, homology proteins were obtained from other insect species (downloaded from InsectBase 2.025). Transcriptome data was aligned to the genome using HISAT v2.2.128 and the open reading frame (ORF) was predicted using StringTie v2.2.129 combined with TransDecoder v5.7.030. Both homology proteins and transcriptome-based evidence were as inputs to BRAKER v3.0.331, which containing ab initio gene prediction generated by Augustus v3.4.032 and Genemark-ETP mode v1.033. A total of 19,532 protein-coding genes was predicted in the genome of L. dispar asiatica. Functional annotation of protein-coding genes was evaluated based on eggNOG-mapper v2 (http://eggnog-mapper.embl.de/)34.
Data Records
Illumina, PacBio and Hi-C raw data for L. dispar asiatica genome sequencing have been deposited in the NCBI Sequence Read Archive with accession number SRR2605746935, SRR2603651136 and SRR260463037. Illumina transcriptome data for larvae and adult have been deposited in the NCBI Sequence Read Archive with accession number SRP45959738. The final assembled L. dispar asiatica genome has been submitted to the GenBank database of NCBI with accession number GCA_032191425.139. The annotation file is available in figshare40.
Technical Validation
The completeness of L. dispar asiatica genome assembly was evaluated using the BUSCO (in the insects_odb10 database), and the completeness was 99.40% (97.9% single-copied genes and 1.5% duplicated genes), 0.2% fragmented, and 0.4% missing genes. The Hi-C heatmap revealed a well-structured interaction pattern in and around the chromosome inversion regions, with the notable exception of chromosome 17. This chromosome showed a lower probability of contact compared to others, leading to speculation that it may be associated with the W sex chromosome, which is specific to females. Besides, the mapping rates of short-reads sequencing data exceeds 90%. All evidence strongly supported that the completeness and accuracy of L. dispar asiatica genome assembly.
Acknowledgements
This work was financially supported by National Key Research and Development Program (2022YFD1401001), Natural Science Foundation of China (32201559, 32071773, 32271882), Postdoctoral Grant Program of Heilongjiang Province (LBH-Z22049).
Author contributions
Zhe Xu and Jianyang Bai conceived the project and performed the experiments; Yue Zhang, Jingyu Cao and Lu Li performed the bioinformatic analyses; Yue Zhang, Mengru Min and Jingxin Cao wrote the manuscript; Fei Li, Yanchun Xu and Ling Ma evaluated the results; All authors read and approved the final manuscript.
Code availability
This study did not employ a custom script; data processing was conducted following the protocols and manuals of the relevant bioinformatics software mentioned in Methods section.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Zhe Xu, Jianyang Bai, Yue Zhang.
References
- 1.Boukouvala, M. C. et al. Lymantria dispar (L.) (Lepidoptera: Erebidae): Current Status of Biology, Ecology, and Management in Europe with Notes from North America. Insects13 (2022). [DOI] [PMC free article] [PubMed]
- 2.Keena M. A., Richards, J. Y. Comparison of Survival and Development of Gypsy Moth Lymantria dispar L. (Lepidoptera: Erebidae) Populations from Different Geographic Areas on North American Conifers. Insects11 (2020). [DOI] [PMC free article] [PubMed]
- 3.Srivastava V, Griess VC, Keena MA. Assessing the Potential Distribution of Asian Gypsy Moth in Canada: A Comparison of Two Methodological Approaches. Sci Rep. 2020;10:22. doi: 10.1038/s41598-019-57020-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakajima T, Haruki S. Defoliation by gypsy moths negatively affects the production of acorns by two Japanese oak species. Trees. 2015;29:1559–1566. doi: 10.1007/s00468-015-1237-9. [DOI] [Google Scholar]
- 5.Bigsby KM, Ambrose MJ, Tobin PC, Sills EO. The cost of gypsy moth sex in the city. Urban For Urban Gree. 2014;13:459–468. doi: 10.1016/j.ufug.2014.05.003. [DOI] [Google Scholar]
- 6.Gray D, Keena M. A Phenology Model for Asian Gypsy Moth Egg Hatch. Environ Entomol. 2019;48:903–910. doi: 10.1093/ee/nvz051. [DOI] [PubMed] [Google Scholar]
- 7.Friedline CJ, et al. Evolutionary genomics of gypsy moth populations sampled along a latitudinal gradient. Mol Ecol. 2019;28:2206–2223. doi: 10.1111/mec.15069. [DOI] [PubMed] [Google Scholar]
- 8.Chen F, et al. DNA Barcoding of Gypsy Moths From China (Lepidoptera: Erebidae) Reveals New Haplotypes and Divergence Patterns Within Gypsy Moth Subspecies. J Econ Entomol. 2016;109:366–374. doi: 10.1093/jee/tov258. [DOI] [PubMed] [Google Scholar]
- 9.Zhao J, Wu Y, Kurenshchikov DK, Ilyinykh AV, Shi J. Underestimated mitochondrial diversity in gypsy moth Lymantria dispar from Asia. Agr Forest Entomol. 2019;21:235–242. doi: 10.1111/afe.12325. [DOI] [Google Scholar]
- 10.Tobin PC, Bai BB, Eggen DA, Leonard DS. The ecology, geopolitics, and economics of managing Lymantria dispar in the United States. Int J Pest Manage. 2012;58:195–210. doi: 10.1080/09670874.2011.647836. [DOI] [Google Scholar]
- 11.Rongrong, Wen Q, Zhao B, Wang Y, Ma L. Molecular characterization and functional analysis of USP-1 by RNA interference in the Asian gypsy moth Lymantria dispar. J Forestry Res. 2020;v.31:449–457. [Google Scholar]
- 12.Liebhold & McManus. The evolving use of insecticides in - Gypsy moth management. J Forest, 20–23 (1999).
- 13.Kalsi M, Palli SR. Cap n collar transcription factor regulates multiple genes coding for proteins involved in insecticide detoxification in the red flour beetle, Tribolium castaneum. Insect Biochem Mol Biol. 2017;90:43–52. doi: 10.1016/j.ibmb.2017.09.009. [DOI] [PubMed] [Google Scholar]
- 14.Xu ZB, et al. concentration of emamectin benzoate inhibits the growth of gypsy moth by inducing digestive dysfunction and nutrient metabolism disorder. Pest Manag Sci. 2021;77:4073–4083. doi: 10.1002/ps.6432. [DOI] [PubMed] [Google Scholar]
- 15.Bai J, et al. Temporospatial modulation of Lymantria dispar immune system against an entomopathogenic fungal infection. Pest Manag Sci. 2020;76:3982–3989. doi: 10.1002/ps.5947. [DOI] [PubMed] [Google Scholar]
- 16.Zhou J, Bruns MA, Tiedje JM. DNA recovery from soils of diverse composition. Appl Environ Microbiol. 1996;62:316–322. doi: 10.1128/aem.62.2.316-322.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cheng H, Concepcion GT, Feng X, Zhang H, Li H. Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods. 2021;18:170–175. doi: 10.1038/s41592-020-01056-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cheng H, et al. Haplotype-resolved assembly of diploid genomes without parental data. Nat Biotechnol. 2022;40:1332–1335. doi: 10.1038/s41587-022-01261-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Belton JM, et al. Hi-C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58:268–276. doi: 10.1016/j.ymeth.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34:i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Durand NC, et al. Juicer Provides a One-Click System for Analyzing Loop-Resolution Hi-C Experiments. Cell Syst. 2016;3:95–98. doi: 10.1016/j.cels.2016.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dudchenko O, et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017;356:92–95. doi: 10.1126/science.aal3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramírez F, et al. High-resolution TADs reveal DNA sequences underlying genome organization in flies. Nat Commun. 2018;9:189. doi: 10.1038/s41467-017-02525-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 25.Mei Y, et al. InsectBase 2.0: a comprehensive gene resource for insects. Nucleic Acids Res. 2022;50:D1040–d1045. doi: 10.1093/nar/gkab1090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Flynn JM, et al. RepeatModeler2 for automated genomic discovery of transposable element families. Proc Natl Acad Sci. 2020;117:9451–9457. doi: 10.1073/pnas.1921046117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tempel S. Using and understanding RepeatMasker. Methods Mol Biol. 2012;859:29–51. doi: 10.1007/978-1-61779-603-6_2. [DOI] [PubMed] [Google Scholar]
- 28.Kim D, Paggi JM, Park C, Bennett C, Salzberg SLJNPG. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37:907–915. doi: 10.1038/s41587-019-0201-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pertea M, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haas BJ, et al. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat Protoc. 2013;8:1494–1512. doi: 10.1038/nprot.2013.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoff KJ, Lomsadze A, Borodovsky M, Stanke M. Whole-Genome Annotation with BRAKER. Methods Mol Biol. 2019;1962:65–95. doi: 10.1007/978-1-4939-9173-0_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Stanke M, Diekhans M, Baertsch R, Haussler D. Using native and syntenically mapped cDNA alignments to improve de novo gene finding. Bioinformatics. 2008;24:637–644. doi: 10.1093/bioinformatics/btn013. [DOI] [PubMed] [Google Scholar]
- 33.Stanke M, Schöffmann O, Morgenstern B, Waack S. Gene prediction in eukaryotes with a generalized hidden Markov model that uses hints from external sources. BMC Bioinformatics. 2006;7:62. doi: 10.1186/1471-2105-7-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cantalapiedra CP, Hernández-Plaza A, Letunic I, Bork P, Huerta-Cepas J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol Biol Evol. 2021;38:5825–5829. doi: 10.1093/molbev/msab293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.2023. NCBI Sequence Read Archive. SRR26057469
- 36.2023. NCBI Sequence Read Archive. SRR26036511
- 37.2023. NCBI Sequence Read Archive. SRR26046303
- 38.2023. NCBI Sequence Read Archive. SRP459597
- 39.Xu Z, Bai J, Ma L. 2023. whole genome shotgun sequencing project. GenBank. GCA_032191425.1
- 40.Xu Z. 2023. Annotations of Lymantira dispar asiatica. figshare. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- 2023. NCBI Sequence Read Archive. SRR26057469
- 2023. NCBI Sequence Read Archive. SRR26036511
- 2023. NCBI Sequence Read Archive. SRR26046303
- 2023. NCBI Sequence Read Archive. SRP459597
- Xu Z, Bai J, Ma L. 2023. whole genome shotgun sequencing project. GenBank. GCA_032191425.1
- Xu Z. 2023. Annotations of Lymantira dispar asiatica. figshare. [DOI]
Data Availability Statement
This study did not employ a custom script; data processing was conducted following the protocols and manuals of the relevant bioinformatics software mentioned in Methods section.