Abstract
In Acinetobacter sp. strain ADP1, benzoate degradation requires the ben genes for converting benzoate to catechol and the cat genes for degrading catechol. Here we describe a novel transcriptional activator, BenM, that regulates the chromosomal ben and cat genes. BenM is homologous to CatM, a LysR-type transcriptional activator of the cat genes. Unusual regulatory features of this system include the abilities of both BenM and CatM to recognize the same inducer, cis,cis-muconate, and to regulate some of the same genes, such as catA and catB. Unlike CatM, BenM responded to benzoate. Benzoate together with cis,cis-muconate increased the BenM-dependent expression of the benABCDE operon synergistically. CatM was not required for this synergism, nor did CatM regulate the expression of a chromosomal benA::lacZ transcriptional fusion. BenM-mediated regulation differs significantly from that of the TOL plasmid-encoded conversion of benzoate to catechol in pseudomonads. The benM gene is immediately upstream of, and divergently transcribed from, benA, and a possible DNA binding site for BenM was identified between the two coding regions. Two mutations in the predicted operator/promoter region rendered ben gene expression either constitutive or inducible by cis,cis-muconate but not benzoate. Mutants lacking BenM, CatM, or both of these regulators degraded aromatic compounds at different rates, and the levels of intermediary metabolites that accumulated depended on the genetic background. These studies indicated that BenM is necessary for ben gene expression but not for expression of the cat genes, which can be regulated by CatM. In a catM-disrupted strain, BenM was able to induce higher levels of catA expression than catB expression.
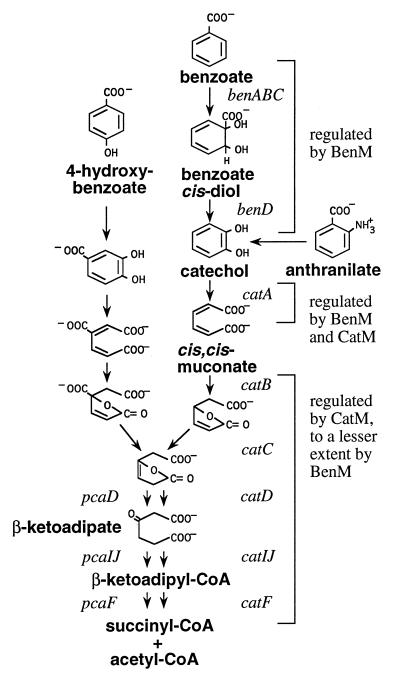
In the soil bacterium Acinetobacter sp. strain ADP1 (formerly Acinetobacter calcoaceticus), the enzymes needed for the conversion of benzoate to tricarboxylic acid cycle intermediates via the β-ketoadipate pathway are encoded by the ben and cat genes (Fig. 1 and 2) (11). Although cat gene expression has been characterized, little is known about the regulation of the adjacent ben genes. The ben-encoded enzymes convert benzoate to catechol, a compound subsequently degraded by cat-encoded enzymes (26). During growth with benzoate, tight regulation of metabolic flow prevents the accumulation of most intermediary compounds (7). One metabolite whose transient production from benzoate can be detected, however, is cis,cis-muconate, the product of catechol ring cleavage and the inducer of all of the cat-encoded enzymes.
FIG. 1.
The β-ketoadipate pathway of Acinetobacter sp. strain ADP1. Compounds and genes relevant to these studies are indicated by boldfaced and italicized text, respectively.
FIG. 2.
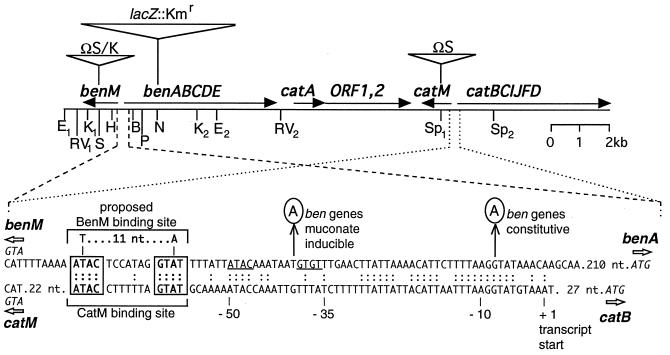
Restriction map of the chromosomal ben-cat region. The locations of genes, and their transcriptional directions, are shown relative to some of the known restriction endonuclease sites: EcoRI (E), EcoRV (RV), KpnI (K), SalI (S), HindIII (H), BsaAI (B), PvuII (P), NsiI (N), and SphI (Sp). Triangles mark the insertion sites of ΩS (29), ΩK (6), and the lacZ::Kmr cassette (18) of strains listed in Table 1. Below the map, the DNA sequence of the benM-benA intergenic region was aligned with the corresponding catM-catB intergenic region, with identical nucleotides indicated (:). The known CatM binding site (30) was used to predict a possible BenM binding site that has the consensus sequence (T-11 nt-A) and dyad symmetry (ATAC, GTAT) involved in binding LysR-type activators. An adjacent region with similar sequences is underlined. Circled nucleotides indicate mutations identified in strains ACN147 and ACN149 (G→A) and in strain ACN146 (T→A).
cis,cis-Muconate regulates its own formation and degradation. Acting on the LysR-type transcriptional regulator CatM, this compound activates transcription of catA and the catBCIJFD genes (30). Moreover, cis,cis-muconate derived from benzoate can prevent the degradation of an alternative carbon source, p-hydroxybenzoate, by an as yet undefined regulatory mechanism (7). The accumulation of cis,cis-muconate during growth with benzoate most likely reflects differential expression of catA and the catBCIJFD genes, which are physically separated on the ADP1 chromosome (Fig. 2). Unlike the other cat genes, catA is expressed in response to benzoate as well as cis,cis-muconate (23). Benzoate in the absence of its metabolism, however, yields only partial induction of CatA. In a similar fashion, partial induction of the ben-encoded enzymes can be mediated by cis,cis-muconate (24). Full induction of these enzymes, however, requires growth with benzoate.
The ability of both cis,cis-muconate and benzoate to induce CatA and the Ben enzymes, albeit to different levels, suggests interconnected regulation of the corresponding genes. The goal of the current investigation was to determine the mechanisms and inducers of ben gene expression and to clarify the basis of benzoate-mediated expression of catA. The first step in this process was to characterize a genetic region adjacent to benA that appeared to control cat gene expression in a catM mutant strain (30). As described in this report, the region contains a gene immediately upstream of and divergently transcribed from benA, designated benM, that encodes a LysR-type transcriptional regulator with striking similarity to CatM. The ability of BenM to regulate the ben and cat genes in response to different inducers was tested. In addition, the effect of benM disruption on metabolic flow was examined by using nuclear magnetic resonance (NMR) techniques to monitor benzoate degradation (7).
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
Acinetobacter strains and plasmids are described in Table 1 and Fig. 2. All Acinetobacter strains were derived from BD413 (14), also designated ADP1. Confusion about the taxonomic classification of ADP1 has prompted the use of Acinetobacter sp. until further characterization is complete (9). Escherichia coli DH5α (Gibco BRL) and S17-1 (35) were used as plasmid hosts. Bacteria were cultured in Luria-Bertani (LB) broth and minimal medium (MM) at 37°C as previously described (32, 34). Carbon sources added to MM, at the final concentrations specified, were 10 mM succinate, 3 mM benzoate, 3 mM cis,cis-muconate, 2.5 mM anthranilate, and 2 mM catechol. Antibiotics were added as needed at the following final concentrations: tetracycline, 6 μg/ml; kanamycin, 25 μg/ml; streptomycin, 25 μg/ml; spectinomycin, 25 μg/ml; and ampicillin, 150 μg/ml for Acinetobacter strains and 50 μg/ml for E. coli.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid(s) | Relevant characteristic(s)a | Reference or source |
|---|---|---|
| Acinetobacter strains | ||
| ADP1 | Wild type (BD413) | 14 |
| ADP205 | Δ(catM-catB3205) | 20 |
| ISA13 | catM::ΩS4013 | 30 |
| ISA25 | Δ(catBCIJF4025) | 7 |
| ISA29 | Δ(benE-catA4029) | 7 |
| ISA35 | Δ(benMABC4025)catM::ΩS4013 | 30 |
| ISA36 | benM::ΩS4036 | This study |
| ACN9 | benM::ΩK5008 catM::ΩS4013 | This study |
| ACN32 | benA::lacZ-Kmr5032 | This study |
| ACN39 | benA::lacZ-Kmr5032 Δ(catBCIJF4025) | This study |
| ACN41 | benA::lacZ-Kmr5032 Δ(benE-catA4029) | This study |
| ACN42 | benA::lacZ-Kmr5032 benM::ΩS4036 Δ(catM-catB3205) | This study |
| ACN47 | benA::lacZ-Kmr5032 benM::ΩS4036 | This study |
| ACN146 | benM::ΩS4036, point mutation T→A (Fig. 2) | This study |
| ACN147 | benM::ΩS4036, point mutation G→A (Fig. 2) | This study |
| ACN149 | benM::ΩS4036, point mutation G→A (Fig. 2) | This study |
| ACN157 | benA::lacZ-Kmr5032 benM::ΩS4036, point mutation T→A (Fig. 2) | This study |
| ACN158 | benA::lacZ-Kmr5032 benM::ΩS4036, point mutation G→A (Fig. 2) | This study |
| ACN160 | benA::lacZ-Kmr5032 benM::ΩS4036, point mutation G→A (Fig. 2) | This study |
| Plasmids | ||
| pUC19 | Apr, cloning vector | 37 |
| pRK415 | Tcr, cloning vector | 16 |
| pKOK6 | Source of promoterless lacZ-Kmr cassette | 18 |
| pHP45 | Source of ΩS | 29 |
| pUI1637 | Source of ΩK | 6 |
| pIB1351, pIB1352 | benM to -D (E1 to E2; Fig. 2) in pUC19 | 24 |
| pIB1354 | benA to -D (H to E2; Fig. 2) in pUC19 | 24 |
| pIGG5 | Part of benM (E1 to S; Fig. 2) in pUC19 | This study |
| pIGG16 | Part of benK, benCDE (E1 to K1, K2 to RV2; Fig. 2) in pRK415 | This study |
| pIGG23 | Part of benM (E1 to K1; Fig. 2) in pUC19 | This study |
| pIGG24 | Part of benM (E1 to H; Fig. 2) in pUC19 | This study |
| pIGG26 | Part of benM (S to E2; Fig. 2) in pUC19 | This study |
| pIGG27 | ΩS in benM SalI site (S; Fig. 2) of pIGG24 | This study |
| pBAC12 | ΩK in benM SalI site (S; Fig. 2) of pIGG24 | This study |
| pBAC14 | All of benM (E1 to P; Fig. 2) in pRK415 | This study |
| pBAC54 | lacZ-Kmr in benA NsiI site (N; Fig. 2) of pIB1354 | This study |
| pBAC64 | Part of benM (RV1 to K1; Fig. 2) in pUC19 | This study |
Kmr, kanamycin resistant; Apr, ampicillin resistant; Tcr, tetracycline resistant.
DNA manipulations and plasmid constructions.
Standard methods were used for chromosomal and plasmid DNA purifications, restriction enzyme digestions, electrophoresis, ligations, and E. coli transformations (32). Plasmids carrying DNA fragments upstream of benA were generated from pIB1351 and pIB1352 (24). The sole SalI ADP1 DNA fragment of the pIB1351 or pIB1352 was deleted to form pIGG5 or pIGG26, respectively. The two KpnI ADP1 DNA fragments of pIB1351 were deleted to form pIGG23. The sole HindIII ADP1 DNA fragment of pIB1351 was deleted to form pIGG24. The DNA fragment between the EcoRV and KpnI sites of pIGG23 was ligated to pUC19 between the vector’s HincII and KpnI sites to form pBAC64.
Plasmids pIGG27 and pBAC12, used for interposon mutagenesis, were made by inserting an omega (Ω) cassette conferring resistance to spectinomycin (Spr) and streptomycin (Smr) (ΩS) or kanamycin (Kmr) (ΩK), respectively, in the sole SalI site of pIGG24. Plasmids pHP45 (29) and pUI1637 (6) were the sources of the ΩS and ΩK fragments, respectively. Each of these fragments carries transcriptional and translational stop signals following the drug resistance determinant. To generate the benA::lacZ fusion of pBAC54, the promoterless lacZ-Kmr cartridge of pKOK6 (18) was isolated as a PstI fragment and inserted into the compatible NsiI site in benA of plasmid pIB1354 (24). To generate pBAC14, carrying benM, we made an intermediate plasmid in which the EcoRI-PvuII fragment (E1 to P in Fig. 2) was ligated into pUC19 digested with EcoRI and HincII. The benM fragment was excised from this plasmid by digestion with EcoRI and PstI, and it was ligated into similarly digested pRK415 (16) to form pBAC14.
Strain construction by interposon mutagenesis and allelic replacement.
A linear DNA fragment carrying either the benM::ΩS4036 or the benM::ΩK5008 allele was generated by restriction endonuclease digestion of plasmid pIGG27 with PvuII or plasmid pBAC12 with BglI, respectively. Each DNA fragment with an Ω cassette was electrophoretically separated and purified from an agarose gel with the GeneClean purification kit (Bio101). As described previously (20), the DNA fragments were used to transform and to replace the corresponding chromosomal regions of recipient Acinetobacter strains ADP1 and ISA13, generating ISA36 and ACN9, respectively. Similarly, a linear DNA fragment carrying the benA::lacZ fusion was generated by digestion of pBAC54 with restriction endonuclease Asp718 and used to transform ADP1, thereby generating strain ACN32.
A crude cell lysate was made from strain ACN32 and used as previously described (20) to introduce the benA::lacZ fusion into the chromosomes of strains with different genetic backgrounds. In this way, new strains ACN39, ACN41, and ACN47 were generated from recipient strains ISA25, ISA29, and ISA36, respectively. DNA in a crude cell lysate made from strain ACN47 was used to transform ADP205 and generate strain ACN42. As previously described, the chromosomal configurations of all strains were confirmed by Southern hybridization analyses, and use of probes made from vector sequences confirmed that allelic replacement rather than plasmid integration had introduced modified DNA into the chromosomes of the recipient strains (9).
Isolation of plasmids with the benA operator/promoter regions of mutants.
The gap repair method of Gregg-Jolly and Ornston (9a) was used to isolate the chromosomal DNA of selected mutants corresponding to the region between K1 and K2 in Fig. 2. Plasmid pIGG16 was constructed by ligating ADP1 DNA fragments E1 to K1 and K2 to RV2 (Fig. 2) into the vector pRK415 (16). Recipient strains were transformed with pIGG16, linearized at the sole KpnI site, and homologous recombination in vivo generated plasmids carrying the benM-benA intergenic regions of the mutants. The intergenic regions between benM and benA were sequenced as described below from plasmids isolated by the gap repair method, using two oligonucleotide primers, 5′-CAAGATTTTGAATTTGTCGGC-3′ and 5′-GCTAGTATTAATGACGGGAAT-3′, purchased from National Biosciences Inc.
DNA sequence analysis.
The DNA sequence of plasmids pIGG5, -23, -24, and -26 and pBAC64 were determined with double-stranded templates and sequencing primers (Promega) which recognize the pUC19 cloning vector. An automated DNA sequencer (ABI373A; Applied Biosystems, Inc.) was used in the University of Georgia Molecular Genetics Instruments Facility. In addition, two oligonucleotide primers, 5′-GCTCTACTGGAATTGGTT-3′ and 5′-GCTCAGTAAAACCTTCAT-3′ (Genosys Biotechnologies), were used with the pIGG5 DNA template. DNA sequences were analyzed with the Wisconsin Genetics Computer Group programs (5).
CatA and β-galactosidase (LacZ) assays.
Catechol 1,2-dioxygenase (CatA) activity was measured in cell extracts of cultures grown in LB medium with or without 2 mM benzoate or 2 mM cis,cis-muconate as described previously (30). CatA activity was assayed spectrophotometrically by the increase in A260, indicative of cis,cis-muconate formation (25). For LacZ assays, cultures were grown overnight in 5 ml of LB with or without 3 mM benzoate, 3 mM cis,cis-muconate, 2.5 mM anthranilate, or 2 mM catechol. Cells were lysed with chloroform and sodium dodecyl sulfate, and β-galactosidase activities were determined as described by Miller (19).
Metabolite monitoring by HPLC.
To monitor benzoate metabolism, 1-ml culture samples were centrifuged to pellet cells. Any cells remaining in the supernatant fractions were removed by passage through a low-protein-binding, 0.22-μm-pore-size syringe filter (MSI). A 10-μl sample of the filtrate was analyzed on a C18 reversed-phase high-performance liquid chromatography (HPLC) column (Bio-Rad Laboratories). Elution at a rate of 0.8 ml/min was carried out with 30% acetonitrile and 0.1% phosphoric acid, and the eluant was monitored by UV detection at 254 nm to detect anthranilate, benzoate, and cis,cis-muconate and at 282 nm to detect catechol. Under these conditions, the retention times for benzoate, anthranilate, catechol, and cis,cis-muconate were 7.7, 6.0, 4.6, and 3.5 min, respectively. Peak areas corresponding to standards and experimental samples were integrated by using the ValuChrom software package (Bio-Rad). To assess the metabolism of benzoate by mutants blocked in its degradation, cultures were grown in LB broth with 2.5 to 3 mM benzoate added. Culture samples were analyzed by HPLC before and after 24 h of growth to determine the benzoate concentration. The concentration determinations of duplicate samples varied no more than 100 μM.
Metabolic observation.
Metabolic observation-NMR (MO-NMR) was carried out as described previously (7). Briefly, Acinetobacter strains were deuterated by overnight growth in a complex 2H-IG medium prepared (by Isogenetics, Inc.) from algae grown photoautotrophically in deuterated water (7). The Acinetobacter cells were used to inoculate a tube containing 6 ml of a defined deuterated minimal medium, 2H-MM (7), and 600 μl of 2H-IG medium. The addition of benzoate or a mixture of benzoate and p-hydroxybenzoate, dissolved in 2H2O, marked the start of each experiment (time zero). The culture tubes were incubated at 34°C with aeration. Culture samples were taken periodically and analyzed by NMR spectroscopy. Concentrations of compounds were determined by integrating the appropriate spectral peaks.
Northern hybridization analysis.
Total RNA was isolated from 20-ml Acinetobacter cultures by using a Snap-O-Sol kit (Biotecx Laboratories). RNA species were electrophoretically separated on agarose-formaldehyde gels with 2 to 8 μg of RNA per lane (32). A rapid downward transfer system was used to transfer RNA to Nytran nylon membranes (TurboBlotter; Schleicher & Schuell). RNA was cross-linked to the membranes by exposure to a total dose of UV light (254 nm) of 120 mJ cm−2. Sizes of the mRNAs were estimated by using RNA standards of sizes 9.4, 6.2, 3.9, 2.8, 1.9, 0.9, 0.6, and 0.4 kb (Promega) that were stained with methylene blue dye following transfer to the nylon membranes (32).
Nonradioactive probes, made by random-primed labeling of gel-purified DNA fragments with digoxigenin, were used in hybridizations as directed by the Genius system (Boehringer Mannheim Corp.). In the description of fragments, each purified and used in labeling reactions, the numbers in parentheses correspond to positions in the 15,923-nucleotide (nt) ben-cat region sequence in GenBank, accession no. M76991. The benA probe was derived from DNA between BsaAI (351) and NsiI (1446) sites. A benBC probe was derived from DNA between ClaI sites (2143 and 2988). DNA between EcoRI sites (3348 and 3803) was used for a benD probe. A benE probe was made from DNA between the ATG start (4173) and TAA termination (5357) codons.
Nucleotide sequence accession number.
The nucleotide sequence of benM was deposited in the GenBank database (accession no. AF009224).
RESULTS
Identification of the benM gene.
In previous studies, a deletion encompassing the benABC genes and a region upstream of benA was shown to eliminate inducible catA and catB expression in a catM mutant (30). As presented here, the DNA sequence upstream of benA revealed a divergently transcribed 912-bp open reading frame, designated benM. The deduced 304-amino-acid sequence of BenM is homologous to sequences of LysR-type transcriptional activators including CatM, to which it is 59% identical and 75% similar. Similarities are greatest within a subset of this family containing proteins that regulate the degradation of catechol, such as CatM and CatR, or that regulate the degradation of chlorocatechols, such as ClcR, TcbR, and TfdR (Fig. 3). In pairwise sequence comparisons of BenM with Pseudomonas regulators CatR (12, 31), ClcR (3), and TcbR (36) or TfdR from Ralstonia eutropha (15), identity between aligned residues ranges from 29 to 41% and similarity ranges from 50 to 61%.
FIG. 3.
Protein sequence alignment of homologous LysR-type transcriptional activators: ClcR (3), TcbR (36), TfdR (15), BenM, CatM (30), and CatR (31). Aligned residues identical or similar to those of BenM are in black or gray boxes, respectively. The underlined region, BenM residues 109 to 162, may be involved in inducer recognition.
Disruption of benM and isolation of suppressor mutations.
Strain ISA36 was created by interposon mutagenesis (29) of benM with the ΩS cassette (Fig. 2 and Table 1). The benM disruption did not prevent the catabolism of cis,cis-muconate, catechol, or anthranilate, and with each of these as the sole carbon source, ADP1 and ISA36 grew at comparable rates (data not shown). The degradation of these substrates requires the gene products of catA and catBCIJFD but not those of benABCD. In contrast, the benM disruption prevented benzoate degradation. Growth of ISA36 on catechol but not benzoate suggested that BenM regulates ben gene expression.
After 2 to 3 days of incubation on solid medium with benzoate as the sole carbon source, spontaneous mutants of ISA36 appeared at a rate of approximately 10−5. Crude DNA lysates made from six independently isolated mutants transformed an ISA36 recipient to be able to grow rapidly on benzoate, indicating that mutations distinct from the ΩS disruption conferred growth with benzoate. These transformants remained Smr Spr, consistent with their retention of the ΩS insertion in benM. When wild-type DNA was used to transform ISA36, benzoate-grown transformants were sensitive to streptomycin (Sms) and spectinomycin (Sps), as would be expected if the wild-type allele had replaced the ΩS-disrupted benM.
DNA upstream of benA was similar in sequence to the characterized catB operator/promoter (Fig. 2) (30). Because mutations in the ben gene operator/promoter region might allow ben gene expression in the absence of BenM, a chromosomal region including benA and benM was isolated from the six ISA36-derived mutants by gap repair methods (9a). DNA sequencing revealed that two of the independently isolated strains, ACN147 and ACN149, had a G-to-A transition in the same position upstream of the benA coding region (Fig. 2). Another strain, ACN146, had a T-to-A transversion (Fig. 2). No mutations were identified in the benA-benM intergenic region of the remaining three strains. To determine how the identified mutations affect gene expression, a better understanding of wild-type ben gene expression was needed.
Regulation of benA expression.
A benA::lacZ transcriptional fusion, on plasmid pBAC54 (Table 1), was introduced into the wild-type chromosome by allelic replacement, forming ACN32. In this strain, the promoterless lacZ cassette (18) lies 1.1 kbp downstream of the ATG initiation codon of benA, and β-galactosidase (LacZ) activity reflects benA expression. HPLC confirmed that the benA disruption had blocked further degradation of benzoate (data not shown). The addition of benzoate to the growth medium increased β-galactosidase activity in ACN32 approximately 10-fold compared to additive-free medium (Fig. 4), establishing benzoate as an inducer of benA expression.
FIG. 4.
β-Galactosidase (LacZ) activity resulting from expression of a chromosomal benA::lacZ fusion in strains ACN32, ACN39, ACN41, ACN47, ACN47(pBAC14), and ACN42(pBAC14) (Table 1). Genotypes are noted above bars. Strains were cultured in LB with the inducers indicated. Activities are averages of at least three independent repetitions, and the corresponding standard deviations were <20% of the average value.
Since a role for cis,cis-muconate in regulating benzoate degradation had previously been suggested (4, 24), the effect of this compound on expression of the benA::lacZ fusion was tested. The addition of cis,cis-muconate led to even higher β-galactosidase levels than did benzoate, approximately 25-fold greater than without an added inducer (Fig. 4). To ensure that the true inducer was cis,cis-muconate and not a subsequent metabolite, strain ACN39 was constructed to carry the chromosomal benA::lacZ fusion (Table 1) and a chromosomal deletion of genes catB to catF which prevents cis,cis-muconate degradation. In ACN39, the pattern of β-galactosidase induction was similar to that of ACN32 (Fig. 4).
The ability of cis,cis-muconate to induce higher ben gene expression than benzoate was surprising, since benzoate-grown cells oxidize benzoate faster and have higher Ben enzyme activities than cis,cis-muconate-grown cells (1a, 4, 24). One possible cause for higher ben gene expression during growth on benzoate than on cis,cis-muconate is the ability of both inducers together to cause higher levels of ben gene expression than either alone. Consistent with this possibility, the addition of both cis,cis-muconate and benzoate to all strains with a benA::lacZ fusion induced the highest β-galactosidase levels in these studies, up to 80-fold higher than in uninduced cells (Fig. 4). Catechol, another metabolite of benzoate, also induced benA expression. In strain ACN41 (Table 1), which carries the chromosomal benA::lacZ fusion, the catA gene was deleted from the chromosome to block catechol degradation. In this strain, induction by benzoate and/or cis,cis-muconate was similar to that in ACN32. Catechol-induced expression was comparable to that of benzoate, with β-galactosidase activities sevenfold higher than in uninduced cells (Fig. 4).
benM is required for benA expression.
ACN47 carries a chromosomal benM disruption as well as the chromosomal benA::lacZ fusion (Table 1). In ACN47, expression of the benA::lacZ fusion was not inducible (Fig. 4). The wild-type pattern of induction was restored by benM in trans on pBAC14, although the induced levels of β-galactosidase were all approximately 40% lower than in ACN32 (Fig. 4). This same pattern of expression was observed with benM in trans in strain ACN42. Since ACN42 additionally carries a chromosomal deletion of the catMBC genes (Fig. 4), CatM is not required for the synergistic effect of benzoate and cis,cis-muconate on benA expression.
Introduction of the chromosomal benA::lacZ fusion into the benM-disrupted ACN146, ACN147, and ACN149 (Table 1) generated ACN157, ACN158, and ACN160, respectively. These strains each carry a mutation upstream of benA allowing benzoate degradation (selected as described above). Grown in the presence or absence of benzoate, ACN158 and ACN160, each with the same mutation (depicted in Fig. 2), constitutively expressed the benA::lacZ fusion, yielding LacZ levels of approximately 3,000 Miller units. In contrast, ACN157 grown with or without benzoate had low LacZ levels of 150 Miller units. The presence of cis,cis-muconate or both cis,cis-muconate and benzoate in the growth medium increased this activity in ACN157 to approximately 3,000 Miller units. The ACN157 mutation (depicted in Fig. 2) caused expression of the benA::lacZ fusion to be inducible by cis,cis-muconate but not benzoate.
BenM-mediated regulation of catA expression.
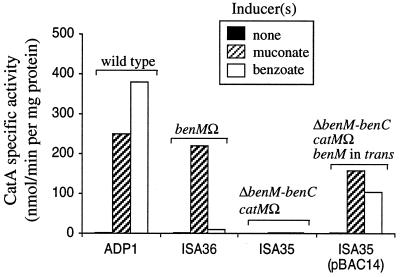
CatM has been shown to activate catA expression in response to cis,cis-muconate but not benzoate (30). Since benzoate induces catechol 1,2-dioxygenase (CatA) independently of cis,cis-muconate (23) and CatM, the possibility that BenM regulates catA expression was tested. CatA activity was measured in the wild-type and benM mutants grown on LB to which no inducer, cis,cis-muconate, or benzoate was added. In the benM mutant ISA36, cis,cis-muconate, but not benzoate, induced CatA to wild-type levels (Fig. 5). These results, consistent with CatM regulating catA expression in ISA36, suggested that the loss of benzoate induction was due to the absence of BenM-mediated catA expression.
FIG. 5.
CatA (catechol 1,2-dioxygenase) activity in strains ADP1, ISA36, ISA35, and ISA35(pBAC14) (Table 1). Genotypes are noted above bars. Strains were cultured in LB with the inducers indicated. Activities are the averages of at least three independent repetitions, and the corresponding standard deviations were <20% of the average value. In ADP1, but not the other strains, benzoate can be metabolized to cis,cis-muconate.
The ability of benM in trans to regulate catA expression was tested in ISA35 (Table 1), a strain lacking functional chromosomal catM and benM alleles. The roles of benzoate and cis,cis-muconate as effectors in ISA35 can be distinguished, since a chromosomal deletion encompassing the benABC genes prevents the conversion of benzoate to cis,cis-muconate. HPLC monitoring confirmed that no degradation of benzoate was detected during a 24-h period. Whereas ISA35 had no benzoate- or cis,cis-muconate-induced CatA activity, benM in trans restored induction in response to either inducer (Fig. 5). The response to benzoate was lower in ISA35(pBAC14) than that of the wild type (Fig. 5). In the latter case, however, benzoate can be degraded so that CatA levels stem from induction by both benzoate and cis,cis-muconate. CatA activity in ISA35 resulting from the presence of benM in trans was comparable to the level of benzoate-mediated CatA induction in a mutant unable to degrade benzoate (23). These results indicate that BenM is required for benzoate-mediated expression of catA whereas either CatM or BenM can regulate catA in response to cis,cis-muconate.
Coexpression of the benABCDE genes.
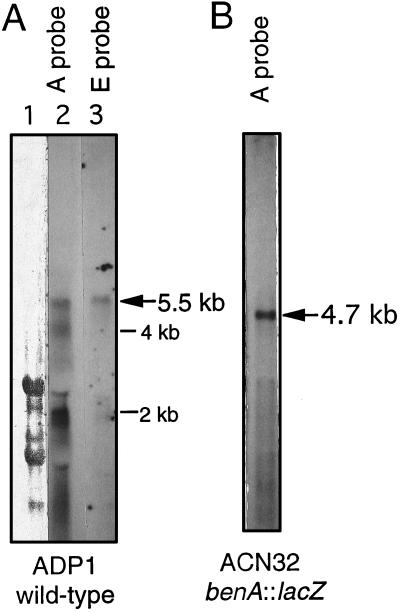
Northern hybridization studies assessed coexpression of the benABCDE genes, which form an approximately 5-kbp region with no more than 50 nt separating each coding region (GenBank accession no. M76990; Fig. 2). Labeled probes were hybridized to RNA from cells grown with or without benzoate. Probe A, from the 5′ region of benA, between BsaAI and NsiI restriction sites (Fig. 2), detected a 5.5-kb transcript in RNA from the wild type grown with benzoate (Fig. 6A, lane 2) but not in those grown without benzoate (data not shown). Hybridization signals, of variable relative intensities, corresponding to RNA sizes of approximately 4 and 2 kb were typically observed (Fig. 6A, lane 2).
FIG. 6.
Northern hybridization analysis of ben gene transcripts. Regions of benA and benE were labeled to make probes A and E, respectively, as described in Materials and Methods. Arrows indicate transcripts detected in RNA from the wild-type ADP1 grown with benzoate (A) or from ACN32, with a transcriptional terminator following the benA::lacZ fusion, grown with benzoate and cis,cis-muconate (B). Total wild-type RNA in one sample (A, lane 1) was stained with methylene blue to demonstrate the positions of the rRNA species.
The same benA probe detected a single transcript, 4.7 kb in size, in RNA from ACN32 grown with benzoate and cis,cis-muconate (Fig. 6B) but not from cells grown without inducers (data not shown). In ACN32, the known position of a transcriptional terminator that follows the lacZ-Kmr cassette in benA (Fig. 2) (18) enables the size of the transcript to predict the position of a single initiation site upstream of the benA coding region (discussed later). Additional ben region probes (described in Materials and Methods) detected a 5.5-kb transcript in wild-type RNA from benzoate-grown cells but not in RNA from cells grown without benzoate (results shown only for the benE probe [Fig. 6A, lane 3]). The benE gene encodes a membrane protein of unknown function.
Metabolic flow in regulatory mutants.
To assess the physiological significance of the individual CatM and BenM activators in ben and cat gene expression, metabolite flow was analyzed in a strain lacking a functional benM (ISA36) and a strain lacking both benM and catM (ACN9) (Table 1). Comparisons could then be made with results of previous studies in which identical methods were used to analyze the wild type and a catM mutant (ISA13) (7). Using MO-NMR techniques (7), either 3 mM [1H]benzoate or a mixture of 3 mM [1H]benzoate and 3 mM [1H]p-hydroxybenzoate was provided to deuterated bacterial cultures. 1H compounds were detected by NMR analyses of samples taken during growth of the batch culture.
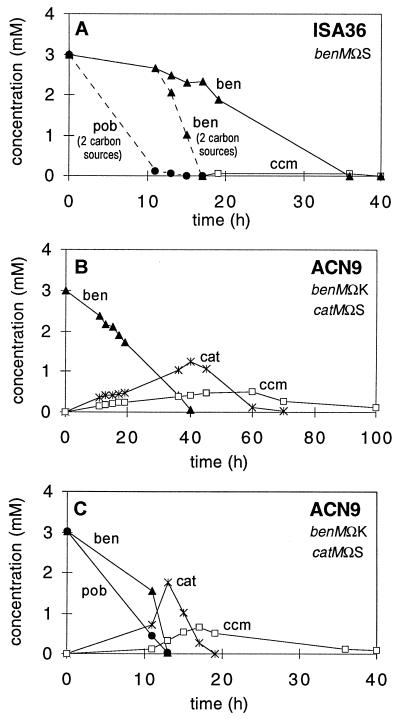
Although ISA36 cannot use benzoate as the sole carbon source, MO-NMR experimental conditions allowed this benM::Ω strain to degrade benzoate completely within 36 h of the addition of only this compound to the culture (Fig. 7A). During this time, ISA36 cells were most likely sustained by the small amount of the algae hydrolysate in the 2H-medium. Consistent with the inability of CatM to substitute for BenM in ben gene expression, the complete consumption of benzoate by ISA36 was slower than that by the wild type or the catM-disrupted ISA13 under the same conditions, 8 and 18 h, respectively (7). The accumulation of cis,cis-muconate by ISA36, approximately 100 μM (Fig. 7A), was similar to that of the wild type (7). The benM disruption did not cause catechol to accumulate by lowering catA expression. In the absence of BenM, CatM-regulated catA expression allows rapid catechol degradation.
FIG. 7.
Metabolites detected by MO-NMR following the addition, at time zero, of 3 mM benzoate (A and B) or 3 mM benzoate and 3 mM p-hydroxybenzoate (A and C) to deuterated cultures of ISA36 or ACN9. Concentrations of benzoate (ben), p-hydroxybenzoate (pob), catechol (cat), and cis,cis-muconate (ccm) were calculated by integration of NMR spectral peaks.
In contrast, catechol accumulated during the metabolism of benzoate by the catM- and benM-disrupted strain ACN9 (Fig. 7B). Although ACN9 did not grow with benzoate, cis,cis-muconate, anthranilate, or catechol as the sole carbon source, the growth medium used in the MO-NMR studies sustained the cells and allowed the very slow catabolism of benzoate, requiring more than 100 h for completion, to be monitored. The background levels of transcription occurring in the absence of regulated activation probably account for ben and cat gene expression. The high level of catechol accumulation by ACN9, as much as 1.3 mM, does not occur in either ISA36, ISA13, or ADP1 (7), suggesting that either CatM or BenM is sufficient to activate catA expression and allow the rapid degradation of catechol. In ISA13 (7), compared to ACN9, the relative decrease in catechol accumulation and increase in cis,cis-muconate accumulation from benzoate suggest that BenM activates catA expression to a greater extent than it does catB, since ISA13 differs from ACN9 only in having the wild-type benM allele.
The patterns of metabolites accumulating from benzoate were similar when ACN9 and ISA36 were provided with both [1H]benzoate and [1H]p-hydroxybenzoate (Fig. 7A and C). In each case, however, the time needed for complete benzoate consumption was much shorter in the presence of p-hydroxybenzoate (compare Fig. 7C with Fig. 7B). In the absence of a carbon source, such as p-hydroxybenzoate, low rates of benzoate catabolism may reflect carbon-limited growth, consistent with the inability of benM-disrupted strains to grow at the expense of benzoate as the sole carbon source.
DISCUSSION
BenM activates ben gene expression.
The dependence on benM for inducible expression of a benA::lacZ fusion identified BenM as a transcriptional activator of the benABCDE operon needed for the conversion of benzoate to catechol (Fig. 4). This metabolic conversion has been studied primarily in pseudomonads, where transcriptional control can be mediated by the XylS regulator encoded on the TOL plasmid pWWO. XylS, an AraC-type (8) rather than a LysR-type (33) activator, controls expression of the xylXYZ genes encoding broad-substrate-specific homologs of the BenABCD enzymes (10, 28). The benR locus of Pseudomonas putida has been suggested to encode a XylS-like transcriptional regulator of chromosomal ben genes (13). The lack of benR-xylS DNA hybridization, however, and differences between BenR- and XylS-mediated regulation (17) raise the possibility that BenR is not a XylS homolog and may be a BenM homolog.
The homology of BenM to LysR-type activators is not surprising, since several members of this large family regulate aromatic compound degradation (Fig. 3). The surprising observation was the relative effectiveness of benzoate and cis,cis-muconate in modulating benM-dependent transcription. Benzoate, or an intermediate in its conversion to cis,cis-muconate, had been the expected inducer of ben gene expression, since the initial rate of [14C]benzoate uptake and metabolism by cis,cis-muconate-grown ADP1 is only 65% of that of benzoate-grown cells (4). Here we found that cis,cis-muconate was more than twice as effective as benzoate in increasing benA expression (Fig. 4). Nevertheless, benzoate itself did increase BenM-regulated gene expression. Although HPLC analyses could not preclude the possibility that small amounts of benzoate were metabolized by strains with ben gene mutations, such mutations prevented the metabolism of benzoate from causing the expression of a cis,cis-muconate-regulated transcriptional fusion (4). Similarly, catechol itself, and not a subsequent metabolite, most likely causes BenM to activate benA expression in the catA-deleted strain ACN41 (Fig. 4).
Although cis,cis-muconate was more effective than benzoate as an individual inducer, both compounds together worked synergistically to induce higher levels of benA expression than either alone (Fig. 4). CatM is not essential for this synergism, since it occurred in the catM-deleted strain ACN42(pBAC14) (Fig. 4). In this strain, the relatively low response to all inducers may result from the expression of benM in trans, since a low response was also observed in a catM+ strain with benM in trans, ACN47(pBAC14) (Fig. 4). Low levels might be caused by gene copy number effects or differences in expression of benM from the plasmid and the chromosome. Moreover, the chromosomal benM-Ω insertion could have cis-acting effects on benA expression.
Interaction of BenM with effectors.
The ability of BenM to respond to multiple effectors raises questions about ligand binding. BenM probably binds cis,cis-muconate since its putative inducer binding region is approximately 60% identical in sequence to that of CatR and CatM, activators that also respond to this compound (Fig. 3, positions 109 to 162) (2, 33). The inducer binding regions of LysR-type proteins that recognize dissimilar ligands usually differ significantly from each other (33). For example, identity in sequence alignments of this region is only 15% between BenM and the N-acetylserine-responsive CysB, the only LysR-type protein for which the structure of the effector binding domain is known (35a). Therefore, the approximately 30% sequence identity in pairwise comparisons of this region of BenM with those of the chloromuconate-responsive ClcR, TcbR, and TfdR is significant. As expected, the central regions of ClcR, TcbR, and TfdR, are more similar to each other than each is to BenM.
If BenM binds to benzoate, it is not clear whether this occurs in the same region as that proposed for recognition of the structurally dissimilar cis,cis-muconate. NahR is a LysR-type transcriptional regulator that recognizes 2-hydroxybenzoate (salicylate), a compound similar in structure to benzoate (2). The effector binding region of NahR, however, is only 19% identical to the corresponding region of BenM. Mutations that allow NahR to recognize benzoate have been isolated (2) but are not sufficient to suggest amino acids in BenM specific for benzoate binding.
Transcription initiation of the ben operon.
Amino acids in BenM likely to be involved in DNA recognition are nearly identical to those in CatM, in the helix-turn-helix regions of the N-terminal domains (20). Therefore, CatM and BenM operator sequences would be expected to be similar, leading to the predication of a BenM binding site (Fig. 2) (30). Although protein binding to the proposed BenM operator depicted in Fig. 2 remains to be demonstrated, the ATACN7GTAT sequence is not only identical to that involved in CatM recognition and binding but also identical or nearly so to sequences that bind its most closely related homologs (discussed in reference 30). Binding of BenM to this site could have a negative autoregulatory function characteristic of LysR-type regulators (33). The spacing of this BenM binding site relative to a region in which two mutations affected benA expression corresponds to that between the operator and promoter regions upstream of catB (Fig. 2). The mutation causing constitutive benA expression, a G-to-A transition, may affect RNA polymerase interactions in the −10 promoter region, although confirmation requires mapping the transcription initiation site.
The mutation depicted in Fig. 2 that renders ben gene expression inducible by cis,cis-muconate but not benzoate might allow CatM to activate benA expression. In the mutant, CatM bound to the wild-type BenM operator could interact with RNA polymerase whose binding was altered by a promoter mutation. Alternatively, the mutation might create or alter an operator site to which CatM binds. As shown by the underlined sequences in Fig. 2, the mutation is adjacent to a motif that resembles, but does not exactly match, the dyad elements of the CatM operator. The inability of CatM to regulate benA expression in the wild type most likely does not reflect its inability to recognize benzoate, since effector binding to LysR-type regulators usually has a minimal effect on the affinity of the activators for their target DNA sequences (27). The ISA36 suppressor mutants that had wild-type ben region sequence might carry catM mutations.
The location of the benA regulatory region shown in Fig. 2 is consistent with the transcription initiation site predicted by the 4.7-kb truncated ben transcript in ACN32 (Fig. 6B), approximately 200 nt upstream of the ATG start codon in benA. The large size of the transcript, however, limits the accuracy of this prediction. The detection of only a single ACN32 transcript with the benA probe suggests that all signals detected by this probe in ADP1 RNA originated from the same transcriptional start site (Fig. 6A, lane 2). The small bands in ADP1 RNA, all variable in size and relative intensity, were detected with the benA but not the benE probe (Fig. 6A). These signals may result from different termination sites, degradation, and/or processing. The aberrant banding and exclusion patterns in regions corresponding in size to rRNA are not unusual.
Benzoate catabolism by benM-disrupted mutants.
As would be expected from the coexpression of benD with benABC that was indicated by Northern hybridization studies (Fig. 6), benzoate cis-diol was never detected by MO-NMR (Fig. 7). In strains lacking BenM, it was most likely basal levels of ben gene expression that enabled the slow degradation of benzoate (Fig. 7). These basal expression levels, as much as 80-fold lower than in fully induced cultures, were not negligible: LacZ levels were approximately 200 to 400 Miller units in strains with a benA::lacZ transcriptional fusion that lacked BenM (Fig. 4). Similarly, in the absence of BenM and CatM, basal levels of cat gene expression probably enabled catechol degradation. A comparison of metabolite accumulations in strains with different regulatory mutations helped to assess the individual roles of BenM and CatM in ben and cat gene expression in vivo.
Whereas a critical role for BenM in ben gene expression was indicated by the inability of the benM-disrupted mutant ISA36 to utilize benzoate as the sole carbon source, the relative contribution of BenM and CatM to cat gene expression was less clear. CatM with its effector cis,cis-muconate is known to activate expression of catA and the catBCIJFD genes (30). In this report, we demonstrated that BenM is responsible for the previously identified benzoate-specific induction of CatA (23). The benzoate inducibility is lost in strain ISA35 (lacking CatM and BenM) and restored by benM in trans (Fig. 5) but not by catM in trans (30). Although this benM-mediated restoration results in low CatA activity relative to that of the wild type induced with benzoate (Fig. 5), it is comparable to that of strains unable to convert benzoate to cis,cis-muconate (23). With catA expression, as well as with benA expression, cis,cis-muconate was more effective than benzoate in eliciting a BenM-mediated response (Fig. 5). Collectively, these results confirm that both CatM and BenM activate catA expression.
The MO-NMR experiments indicated that in vivo, either CatM or BenM alone allows the rapid degradation of catechol, presumably by activating catA expression (7) (Fig. 7). Strains in which either catM or benM was disrupted did not accumulate catechol, and it was only in the absence of both functional alleles in ACN9 that catA expression decreased to levels at which catechol was formed more rapidly than it was degraded. In contrast, the individual disruption of catM and benM had different effects on cis,cis-muconate accumulation, presumably reflecting differences in the ability of CatM and BenM to activate catB expression. In comparison to ACN9, the presence of a functional catM in ISA36 or a functional benM in ISA13 decreased or increased, respectively, the accumulation of cis, cis-muconate (7) (Fig. 7). These results indicate that in vivo, in a catM-disrupted background, BenM is better able to activate the transcription of catA than catB. Previous studies suggest that BenM is responsible for the CatM-independent regulation of the catBCIJFD genes (30), and recent gel shift mobility assays demonstrated the binding of BenM to the catB operator region in vitro (1).
BenM may also play a role in regulating p-hydroxybenzoate degradation. In ADP1, benzoate prevents p-hydroxybenzoate degradation, even in mutants unable to derive energy from benzoate degradation (7). The benM mutation in ISA36 and ACN9, however, alleviated this inhibition of p-hydroxybenzoate utilization, allowing p-hydroxybenzoate to be rapidly metabolized in the presence of benzoate (Fig. 7A and C). Since the inhibition appears to depend on the formation of cis,cis-muconate from benzoate (7), BenM may alleviate the repression either by delaying cis,cis-muconate formation or by a more direct mechanism.
ACKNOWLEDGMENTS
This research was supported by National Science Foundation grant MCB-9507393 (to E.L.N.) and by NIH SBIR phase I grant 1R43 ESO 6304-01 (to G.L.G.). Additional support was provided (to E.L.N.) from the University of Georgia Research Foundation, Inc.
We gratefully acknowledge the Wheaton College Chemistry Department for interesting discussions and the use of their 300-MHz NMR spectrometer. We also thank Alan Campbell for Northern hybridization data, Heidi Holmes and Molly McLendon for assaying β-galactosidase, and Anne Summers and Timothy Hoover for useful suggestions.
REFERENCES
- 1.Bundy, B. M., and E. L. Neidle. Unpublished observation.
- 1a.Canovas J L, Stanier R Y. Regulation of the enzymes of the β-ketoadipate pathway in Moraxella calcoaceticus. Eur J Biochem. 1967;1:289–300. doi: 10.1007/978-3-662-25813-2_40. [DOI] [PubMed] [Google Scholar]
- 2.Cebolla A, Sousa C, de Lorenzo V. Effector specificity mutants of the transcriptional activator NahR of naphthalene degrading Pseudomonas define protein sites involved in binding of aromatic inducers. J Biol Chem. 1997;272:3986–3992. doi: 10.1074/jbc.272.7.3986. [DOI] [PubMed] [Google Scholar]
- 3.Coco W M, Rothmel R K, Henikoff S, Chakrabarty A M. Nucleotide sequence and initial functional characterization of the clcR gene encoding a LysR family activator of the clcABD chlorocatechol operon in Pseudomonas putida. J Bacteriol. 1993;175:417–427. doi: 10.1128/jb.175.2.417-427.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collier L S, Nichols N N, Neidle E L. benK encodes a hydrophobic permease-like protein involved in benzoate degradation by Acinetobacter sp. strain ADP1. J Bacteriol. 1997;179:5943–5946. doi: 10.1128/jb.179.18.5943-5946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Devereaux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Eraso J M, Kaplan S. prrA, a putative response regulator involved in oxygen regulation of photosynthetic gene expression in Rhodobacter sphaeroides. J Bacteriol. 1994;176:32–43. doi: 10.1128/jb.176.1.32-43.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaines G L, III, Smith L, Neidle E L. Novel nuclear magnetic resonance spectroscopy methods demonstrate preferential carbon source utilization by Acinetobacter calcoaceticus. J Bacteriol. 1996;178:6833–6841. doi: 10.1128/jb.178.23.6833-6841.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gallegos M-T, Michan C, Ramos J L. The XylS/AraC family of regulators. Nucleic Acids Res. 1993;21:807–810. doi: 10.1093/nar/21.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gralton E M, Campbell A L, Neidle E L. Directed introduction of DNA cleavage sites to produce a high-resolution genetic and physical map of the Acinetobacter sp. strain ADP1 (BD413UE) chromosome. Microbiology. 1997;143:1345–1357. doi: 10.1099/00221287-143-4-1345. [DOI] [PubMed] [Google Scholar]
- 9a.Gregg-Jolly L A, Ornston L N. Recovery of DNA from the Acinetobacter calcoaceticus chromosome by gap repair. J Bacteriol. 1990;172:6169–6172. doi: 10.1128/jb.172.10.6169-6172.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harayama S, Rekik M, Bairoch A, Neidle E L, Ornston L N. Potential DNA slippage structures acquired during evolutionary divergence of Acinetobacter calcoaceticus chromosomal benABC and Pseudomonas putida TOL pWWO plasmid xylXYZ, genes encoding benzoate dioxygenases. J Bacteriol. 1991;173:7540–7548. doi: 10.1128/jb.173.23.7540-7548.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harwood C S, Parales R E. The β-ketoadipate pathway and the biology of self-identity. Annu Rev Microbiol. 1996;50:553–590. doi: 10.1146/annurev.micro.50.1.553. [DOI] [PubMed] [Google Scholar]
- 12.Houghton J E, Brown T M, Appel A J, Hughes E J, Ornston L N. Discontinuities in the evolution of Pseudomonas putida cat genes. J Bacteriol. 1995;177:401–412. doi: 10.1128/jb.177.2.401-412.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeffrey W H, Cuskey S M, Chapman P J, Resnick S, Olsen R H. Characterization of Pseudomonas putida mutants unable to catabolize benzoate: cloning and characterization of Pseudomonas genes involved in benzoate catabolism and isolation of a chromosomal DNA fragment able to substitute for xylS activation of TOL lower-pathway promoter. J Bacteriol. 1992;174:4986–4996. doi: 10.1128/jb.174.15.4986-4996.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Juni E, Janik A. Transformation of Acinetobacter calco-aceticus (Bacterium anitratum) J Bacteriol. 1969;98:281–288. doi: 10.1128/jb.98.1.281-288.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaphammer B, Kukor J J, Olsen R H. Regulation of tfdCDEF by tfdR of the 2,4-dichlorophenoxyacetic acid degradation plasmid pJP4. J Bacteriol. 1990;172:2280–2286. doi: 10.1128/jb.172.5.2280-2286.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keen T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 17.Kessler B, Marques S, Kohler T, Ramos J L, Timmis K T, de Lorenzo V. Cross talk between catabolic pathways in Pseudomonas putida: XylS-dependent and -independent activation of the TOL meta operon requires the same cis-acting sequences within the Pm promoter. J Bacteriol. 1994;176:5578–5582. doi: 10.1128/jb.176.17.5578-5582.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kokotek W, Lotz W. Construction of a lacZ-kanamycin-resistance cassette, useful for site-directed mutagenesis and as a promoter probe. Gene. 1989;84:467–471. doi: 10.1016/0378-1119(89)90522-2. [DOI] [PubMed] [Google Scholar]
- 19.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 20.Neidle E L, Hartnett C, Ornston L N. Characterization of Acinetobacter calcoaceticus catM, a repressor gene homologous in sequence to transcriptional activator genes. J Bacteriol. 1989;171:5410–5421. doi: 10.1128/jb.171.10.5410-5421.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neidle E L, Hartnett C, Ornston L N, Bairoch A, Rekik M, Harayama S. Nucleotide sequences of the Acinetobacter calcoaceticus benABC genes for benzoate 1,2-dioxygenase reveal evolutionary relationships among multicomponent oxygenases. J Bacteriol. 1991;173:5385–5395. doi: 10.1128/jb.173.17.5385-5395.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neidle E L, Hartnett C, Ornston L N, Bairoch A, Rekik M, Harayama S. cis-Diol dehydrogenases encoded by the TOL pWWO plasmid xylL gene and the Acinetobacter calcoaceticus chromosomal benD gene are members of the short-chain alcohol dehydrogenase superfamily. Eur J Biochem. 1992;204:113–120. doi: 10.1111/j.1432-1033.1992.tb16612.x. [DOI] [PubMed] [Google Scholar]
- 23.Neidle E L, Ornston L N. Benzoate and muconate, structurally dissimilar metabolites, induce expression of catA in Acinetobacter calcoaceticus. J Bacteriol. 1987;169:414–415. doi: 10.1128/jb.169.1.414-415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neidle E L, Shapiro M K, Ornston L N. Cloning and expression in Escherichia coli of Acinetobacter calcoaceticus genes for benzoate degradation. J Bacteriol. 1987;169:5496–5503. doi: 10.1128/jb.169.12.5496-5503.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ngai K-L, Neidle E L, Ornston L N. Catechol and chlorocatechol 1,2-dioxygenases. Methods Enzymol. 1990;188:120–126. doi: 10.1016/0076-6879(90)88022-3. [DOI] [PubMed] [Google Scholar]
- 26.Ornston L N, Neidle E L. Evolution of genes for the β-ketoadipate pathway in Acinetobacter calcoaceticus. In: Towner K J, Bergogne-Berezin E, Fewson C A, editors. The biology of Acinetobacter. New York, N.Y: Plenum Press; 1991. pp. 201–237. [Google Scholar]
- 27.Perez-Martin J, Rojo F, de Lorenzo V. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol Rev. 1994;58:268–290. doi: 10.1128/mr.58.2.268-290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pickup R W, Williams P A. Spontaneous deletions in the TOL plasmid pWW20 which give rise to the B3 regulatory mutants of Pseudomonas putida MT20. J Gen Microbiol. 1982;128:1385–1390. doi: 10.1099/00221287-128-7-1385. [DOI] [PubMed] [Google Scholar]
- 29.Prentki P, Krisch H M. In vitro insertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 30.Romero-Arroyo C E, Schell M A, Gaines III G L, Neidle E L. catM encodes a LysR-type transcriptional activator regulating catechol degradation in Acinetobacter calcoaceticus. J Bacteriol. 1995;177:5891–5898. doi: 10.1128/jb.177.20.5891-5898.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothmel R K, Aldrich T L, Houghton J E, Coco W M, Ornston L N, Chakrabarty A M. Nucleotide sequencing and characterization of Pseudomonas putida catR: a positive regulator of the catBC operon is a member of the LysR family. J Bacteriol. 1990;172:922–931. doi: 10.1128/jb.172.2.922-931.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 33.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 34.Shanley M S, Neidle E L, Parales R E, Ornston L N. Cloning and expression of Acinetobacter calcoaceticus catBCDE genes in Pseudomonas putida and Escherichia coli. J Bacteriol. 1986;165:557–563. doi: 10.1128/jb.165.2.557-563.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Simon R, Priefer U, Puhler A. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology. 1983;1:37–45. [Google Scholar]
- 35a.Tyrrell R, Verschueren K H G, Dodson E J, Murshudov G N, Addy C, Wilkinson A J. The structure of the cofactor-binding fragment of the LysR family member, CysB: a familiar fold with a surprising subunit arrangement. Structure. 1997;5:1017–1032. doi: 10.1016/s0969-2126(97)00254-2. [DOI] [PubMed] [Google Scholar]
- 36.van der Meer J R, Frijters A C J, Leveau J H J, Eggen R I L, Zehnder A J B, de Vos W M. Characterization of the Pseudomonas sp. strain P51 gene tcbR, a LysR-type transcriptional activator of the tcbCDEF chlorocatechol oxidative operon, and analysis of the regulatory region. J Bacteriol. 1991;173:3700–3708. doi: 10.1128/jb.173.12.3700-3708.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]