Abstract
2-Aminomuconate, an intermediate in the metabolism of tryptophan in mammals, is also an intermediate in the biodegradation of nitrobenzene by Pseudomonas pseudoalcaligenes JS45. Strain JS45 hydrolyzes 2-aminomuconate to 4-oxalocrotonic acid, with the release of ammonia, which serves as the nitrogen source for growth of the microorganism. As an initial step in studying the novel deamination mechanism, we report here the purification and some properties of 2-aminomuconate deaminase. The purified enzyme migrates as a single band with a molecular mass of 16.6 kDa in 15% polyacrylamide gel electrophoresis under denaturing conditions. The estimated molecular mass of the native enzyme was 100 kDa by gel filtration and 4 to 20% gradient nondenaturing polyacrylamide gel electrophoresis, suggesting that the enzyme consists of six identical subunits. The enzyme was stable at room temperature and exhibited optimal activity at pH 6.6. The Km for 2-aminomuconate was approximately 67 μM, and the Vmax was 125 μmol · min−1 · mg−1. The N-terminal amino acid sequence of the enzyme did not show any significant similarity to any sequence in the databases. The purified enzyme converted 2-aminomuconate directly to 4-oxalocrotonate, rather than 2-hydroxymuconate, which suggests that the deamination was carried out via an imine intermediate.
2-Aminomuconate (2-aminohexa-2,4-diene-1,6-dioate), an α-amino acid with conjugated double bonds, was first found to be an intermediate in tryptophan metabolism in mammalian liver and kidney (8, 13). However, the compound was not isolated since it is readily converted to 2-hydroxymuconic acid under acidic conditions (8). Nishizuka et al. (13) proposed that a 2-aminomuconate reductase reductively deaminated 2-aminomuconate to 2-ketoadipate in the presence of NADH or NADPH. In the process, the amino group was removed and a double bond was reduced. However, the details of the catalytic mechanism and the identification of the product were not reported. The enzyme name, 2-aminomuconate reductase, has not been listed in the official book of enzyme nomenclature (21). A recent textbook of biochemistry proposed that two enzymes, a hydratase and a dehydrogenase, are involved in the transformation of 2-aminomuconate to 2-ketoadipate during tryptophan degradation (20). Although not specified, the intermediate of hydrolysis would have to be 4-oxalocrotonate (2-oxohex-3-ene-1,6-dioate) in the enzyme pathway.
In our investigations of the biodegradation of nitrobenzene by Pseudomonas pseudoalcaligenes JS45, we found that 2-aminomuconate is one of the intermediates in the catabolic pathway (7). The compound, isolated by anion exchange chromatography, was hydrolytically deaminated to 4-oxalocrotonate by crude extracts of P. pseudoalcaligenes JS45 in a reaction similar to the first step in the hypothetical two-enzyme pathway of degradation of 2-aminomuconate in mammals. We designated the enzyme 2-aminomuconate deaminase. Our preliminary experiments also indicated that crude extracts of JS45 catalyzed the transformation of 2-hydroxymuconate (2-hydroxyhexa-2,4-diene-1,6-dioate) to 4-oxalocrotonate (tautomerization of the enol form to the keto form of 4-oxalocrotonate). The presence of a 4-oxalocrotonate tautomerase activity in JS45 raised two questions about the deaminase. (i) Does a single enzyme catalyze both reactions? (ii) If there are two distinguishable enzymes, does the deaminase transform 2-aminomuconate to 4-oxalocrotonate directly or to 2-hydroxymuconate, which is subsequently converted to 4-oxalocrotonate by the tautomerase? To answer these questions and to provide insight into the mechanism of the deamination reaction, we have purified and characterized the 2-aminomuconate deaminase from P. pseudoalcaligenes JS45.
MATERIALS AND METHODS
Growth of bacteria.
P. pseudoalcaligenes JS45 was maintained and grown with nitrobenzene (12). Cells were harvested by centrifugation and washed with 25 mM potassium phosphate (pH 7.0), and the cell pellets were stored at −70°C until use.
Protein purification.
All purification procedures were carried out at 4°C in 25 mM potassium phosphate buffer (pH 7.0). Cells (9.5 g [wet weight]) were suspended in 50 ml of buffer and were broken by two passages through a French pressure cell at 135,000 kPa. The resulting suspension was centrifuged at 100,000 × g for 60 min, and the pellet was discarded. The supernatant (crude extract) was stored at −70°C until use.
Half of the crude extract (25 ml) was thawed and diluted to 200 ml with phosphate buffer and loaded onto a DEAE-Sepharose column (Pharmacia; 2.6 by 10 cm). The column was washed with 200 ml of buffer, and proteins were eluted with a linear NaCl gradient (0 to 0.4 M in buffer; 400 ml at 2 ml/min). The fractions (6 ml each) containing either 2-aminophenol 1,6-dioxygenase or 2-aminomuconate semialdehyde dehydrogenase activities were used to prepare 2-aminomuconate (see below). The fractions containing 2-aminomuconate deaminase activity were pooled and loaded onto a Hitrap Cu(II)-chelating column (Pharmacia; 2 by 5 ml). The column was washed with 30 ml of 0.5 M NaCl in buffer, and proteins were eluted with a linear EDTA gradient (0 to 50 mM in buffer containing 0.5 M NaCl; 60 ml at 1.5 ml/min). The fractions (3 ml each) containing 2-aminomuconate deaminase activity were pooled and concentrated in a Centriprep-10 tube (Amicon, Beverly, Mass.) to a final volume of 2.4 ml. The concentrated preparation was applied to a Sephacryl S-300 gel filtration column (Pharmacia; 1.6 by 100 cm) and eluted with phosphate buffer (1 ml/min). The active fractions (2 ml each) were pooled and loaded onto a Hitrap-Q column (Pharmacia; 5 ml). The column was washed with 40 ml of buffer, and proteins were eluted with an NaCl gradient (0 to 0.25 M NaCl in buffer; 60 ml at 1 ml/min). The active fractions (2 ml each) were pooled, the molarity was adjusted to 2.5 M NaCl, and then the solution was loaded onto a phenyl-Sepharose CL-4B column (Pharmacia; 2.6 by 4 cm). The column was washed with 40 ml of 2.5 M NaCl in buffer, and the proteins were eluted with a descending NaCl gradient (2.5 to 0.5 M in buffer; 150 ml at 1 ml/min). The active fractions (3 ml each) were pooled and concentrated in a Centriprep-10 tube and used for characterization studies.
Enzyme assays.
2-Aminomuconate deaminase activity was determined spectrophotometrically by monitoring the decrease in absorbance at 326 nm concomitant with the disappearance of 2-aminomuconate (ɛ = 16,500 M−1) (7, 8, 13). The reaction was started by the addition of the enzyme preparation (5 to 10 μl) to 2-aminomuconate solution (0.03 mM, 750 μl) in potassium phosphate buffer (25 mM, pH 8.0) containing 0.12 M NaCl, unless stated otherwise. The initial rate of deamination (in less than 1 min) was recorded. In inhibition experiments, the enzyme was incubated with various additives for 10 min in potassium phosphate buffer (200 mM, pH 7.0) prior to the addition of 2-aminomuconate solution. Tris-HCl buffer (100 mM, pH 7.0) was used in testing the effects of metal ions on the enzyme activity. Specific activities are expressed as micromoles of substrate transformed per minute per milligram of protein. For experiments to determine substrate specificity, the activity was measured by determining the release of ammonia with test kit 171-C from Sigma (St. Louis, Mo.) in order to measure the oxidation of NADPH in the presence of 2-ketoglutarate and glutamate dehydrogenase. The appearance of an absorbance peak at 375 nm (2-hydroxymuconic semialdehyde) was used to determine the deamination of 2-aminomuconic semialdehyde.
Protein determination.
Protein concentrations were determined by the Bradford method (1), with Coomassie Plus protein assay reagent (Pierce, Rockford, Ill.). Bovine serum albumin was used as a standard.
Estimation of molecular mass.
The purity and the molecular mass of 2-aminomuconate deaminase were examined by native-gradient (4 to 20%) polyacrylamide gel electrophoresis (PAGE) and sodium dodecyl sulfate (SDS)-PAGE (4). The molecular mass of the enzyme was determined by comparison with protein molecular mass standards. The molecular standards used in SDS-PAGE were bovine serum albumin (66 kDa), chicken egg ovalbumin (45 kDa), rabbit muscle glyceraldehyde-3-phosphate dehydrogenase (36 kDa), bovine erythrocyte carbonic anhydrase (29 kDa), soybean trypsin inhibitor (20 kDa), and bovine milk α-lactalbumin (14.2 kDa). The molecular mass standards used in native-gradient PAGE were jack bean urease (hexamer, 545 kDa; trimer, 272 kDa), and bovine serum albumin (dimer, 132 kDa; monomer, 66 kDa). The native molecular mass was also measured by gel filtration on a Sephacryl S-300 column (Pharmacia; 1.6 by 100 cm) with a flow rate of 1 ml of 25 mM potassium phosphate–0.1 M NaCl (pH 7.0) per min. The molecular mass standards used for gel filtration chromatography were horse spleen apoferritin (443 kDa), sweet potato β-amylase (200 kDa), yeast alcohol dehydrogenase (150 kDa), and bovine serum albumin (66 kDa).
N-terminal sequence.
Subunits of 2-aminomuconate deaminase were obtained on an SDS-PAGE gel and transferred to a polyvinylidene fluoride membrane (Trans-Blot; Bio-Rad). The blotted membrane was stained with Coomassie blue R-250. The N-terminal amino acid sequence was determined by the Protein Core Facility of the University of Florida, Gainesville.
Preparation of 2-aminomuconate.
2-Aminomuconate was prepared as described previously (7), but pooled fractions containing partially purified 2-aminophenol 1,6-dioxygenase and 2-aminomuconate semialdehyde dehydrogenase from the DEAE-Sepharose chromatography were used instead of crude extracts. The isolation column used was Hitrap-Q (2 by 5 ml). Generally, 2-aminomuconate was prepared daily. If the preparation of 2-aminomuconate was frozen at −70°C, the absorbance at 326 nm decreased about 50% after thawing. The compound was more stable under alkaline conditions (pH 13).
Chemicals.
2-Hydroxymuconic acid was prepared by the method of Lapworth (9) from the potassium salt of diethyl 2,4-hexadiene-5-hydroxy-1,6-dioate, which was obtained from condensation of diethyl oxalate and ethyl crotonate in the presence of potassium metal in toluene, as described by Wiley and Hart (23). All other chemicals were from Sigma or Aldrich (Milwaukee, Wis.), unless stated otherwise.
RESULTS
Purification of 2-aminomuconate deaminase.
A typical purification (Table 1) yielded a 222-fold purification with a recovery of 36% of the 2-aminomuconate deaminase activity. The preparation of the enzyme is colorless and does not have an absorbance peak above 300 nm.
TABLE 1.
Purification of 2-aminomuconate deaminase
| Step | Total protein (mg) | Total activity (μmol · min−1) | Sp act (μmol · min−1 · mg−1) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Cell extract | 420 | 72.4 | 0.17 | 1 | 100 |
| DEAE-Sepharose | 42.8 | 84.6 | 1.98 | 11.6 | 117 |
| Hitrap Cu(II)-chelating | 25.2 | 69.5 | 2.76 | 16.2 | 96 |
| Sephacryl S-300 | 12.4 | 59.2 | 4.77 | 28.1 | 82 |
| Hitrap-Q | 3.7 | 42.6 | 11.5 | 67.6 | 59 |
| Phenyl-Sepharose CL-4B | 0.69 | 26.1 | 37.8 | 222 | 36 |
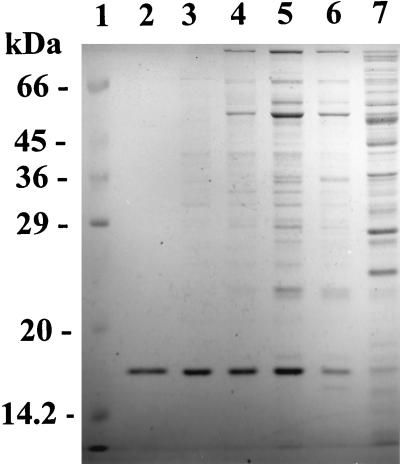
An analysis of the purified protein by SDS–15% PAGE revealed a single band corresponding to a molecular mass of 16.6 kDa (Fig. 1), which is bigger than that of 4-oxalocrotonate tautomerase (3.6 kDa) (2). Nondenaturing 4 to 20% gradient PAGE showed a single band at 100 kDa. The 2-aminomuconate deaminase activity was found in the band on the gel when the appropriate area of the gel was cut out and transferred to potassium phosphate buffer (100 mM, pH 7.5) prior to staining. Gel filtration chromatography also revealed a native molecular mass of 100 kDa. Therefore, the enzyme is apparently composed of six identical subunits, each with a molecular mass of 16.6 kDa.
FIG. 1.
SDS–15% PAGE of 2-aminomuconate deaminase. Purified 2-aminomuconate deaminase (lane 2; 2 μg) was compared with the crude extract (lane 7; 40 μg), the DEAE fraction (lane 6; 20 μg), the Cu(II)-chelating fraction (lane 5; 20 μg), the gel filtration fraction (lane 4; 20 μg), the Hitrap-Q fraction (lane 3; 7 μg), and protein molecular mass standards (lane 1).
Catalytic properties of the enzyme.
The 2-aminomuconate deaminase was stable when stored at room temperature for 3 days. However, 77% of the activity was lost when the enzyme was heat treated for 5 min at 60°C. 2-Aminomuconate deaminase exhibited optimal activity at pH 6.6 and was stable for at least 3 h between pH 5.7 and 8.8 at room temperature without significant loss of activity. For 2-aminomuconate, the Km was approximately 67 μM, the Vmax was 125 μmol · min−1 · mg−1, and the kcat was 208 s−1 at pH 6.6 (80 mM potassium phosphate–0.1 M NaCl) and 25°C.
No absorbance at 326 nm was observed when ammonia (0.8 mM) and 4-oxalocrotonate (0.14 mM) were incubated with 2-aminomuconate deaminase (0.01 mg/ml) in 50 mM potassium phosphate (pH 7.0) for 1 h. Therefore, the deamination reaction appears to be irreversible under the assay conditions.
Substrate specificity and inhibition.
2-Aminomuconate deaminase did not act on 2-aminomuconic semialdehyde, the precursor of 2-aminomuconate. It did not deaminate saturated α-amino acids, including glycine, alanine, aspartic acid, glutaric acid, and 2-aminoadipic acid (a saturated analog of 2-aminomuconate). On the other hand, the deamination of 2-aminomuconate was not inhibited by the presence of these amino acids (2 mM). The enzyme activity was not affected by changes in potassium phosphate buffer concentration from 12 to 260 mM. EDTA (25 mM) did not inhibit the enzyme activity, which indicated that divalent cations are not required for the enzyme activity. MgSO4 (2 mM) did not affect the enzyme activity. Both CuSO4 and MnCl2 inhibited the activity by 20%; ZnCl2 inhibited it by 70%. Phenylhydrazine (10 mM) decreased the activity to 20% of the activity with no inhibitors. Diethyl pyrocarbonate (2 mM) completely destroyed the activity, which indicated that a histidine residue may be involved in the catalytic mechanism.
Although the crude extracts of JS45 catalyzed the tautomerization of 2-hydroxymuconate to 4-oxalocrotonate (the half-life of 2-hydroxymuconate decreased from 7 min in spontaneous tautomerization to 2 min in tautomerization catalyzed by crude extracts of JS45 [0.1 mg of protein/ml]), the purified deaminase did not change the rate of spontaneous tautomerization. This result indicated that 2-aminomuconate deaminase is distinct from 4-oxalocrotonate tautomerase.
True products of deamination.
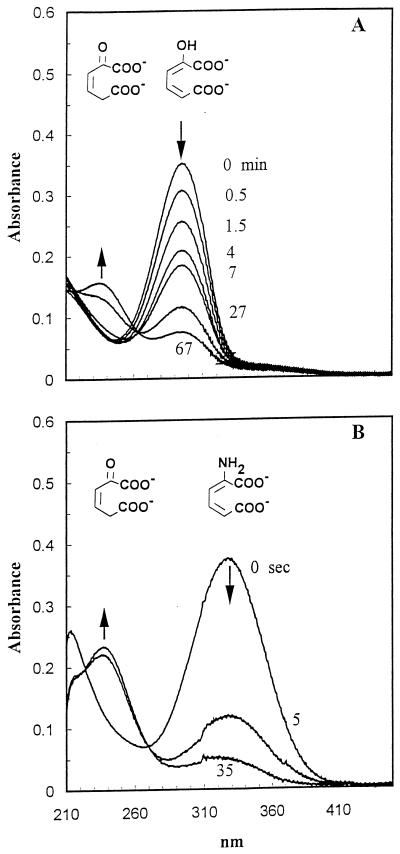
2-Hydroxymuconate and 4-oxalocrotonate are spectrally distinguishable. 2-Hydroxymuconate exhibits maximum absorbance at 296 nm, and 4-oxalocrotonate exhibits maximum absorbance at 237 nm (6, 15, 22). 2-Hydroxymuconate spontaneously converts to 4-oxalocrotonate in an aqueous solution, but the process is slow (about a 7-min half-life under the experimental conditions) (Fig. 2A). When 2-aminomuconate was deaminated by excess purified 2-aminomuconate deaminase, the absorbance at 326 nm decreased from 0.35 to about 0.1 in 5 s, concomitant with an increase in absorbance at 237 nm and with no increase in absorbance at 296 nm (Fig. 2B). These results clearly indicate that the true product of enzymatic deamination of 2-aminomuconate is 4-oxalocrotonate rather than 2-hydroxymuconate.
FIG. 2.
The spectral changes during the tautomerization of 2-hydroxymuconate and the deamination of 2-aminomuconate. (A) Tautomerization of 2-hydroxymuconate (296 nm) to 4-oxalocrotonate (236 nm) in 67 min, initiated by the addition of an ethanolic solution of 2-hydroxymuconate to potassium phosphate buffer (25 mM, pH 8.0, 0.15 M NaCl). (B) Deamination of 2-aminomuconate (326 nm) in the presence of the deaminase (0.0056 mg of protein/ml) in 35 s.
N-terminal sequence.
The N-terminal amino acid sequence for the first 25 residues of the subunit was STLSS NDAKV VDGKA TPLGS FPHVK. A search of the sequence databases (nonredundant GenBank CDS translations, PDB, SwissProt, Spupdate, and PIR) through the National Center for Biotechnology Information with BLAST software revealed no significant similarity between the sequence of 2-aminomuconate deaminase and any other known amino acid sequence.
DISCUSSION
The novel aminohydrolytic enzyme 2-aminomuconate deaminase from P. pseudoalcaligenes JS45 is the first purified deaminase found which acts on an unsaturated linear amino acid. A similar enzymatic deamination of trans-4-amino-6-carboxy-2-oxo-hexa-3,5-dienoate was reported for bacterial metabolism of 5-aminosalicylic acid (17), but the purification and the properties of the enzyme were not reported.
2-Aminomuconate deaminase acted specifically on the unsaturated α-amino acid 2-aminomuconate. The fact that the enzyme did not act on saturated α-amino acids and 2-aminomuconic 6-semialdehyde indicated that the double bonds and the distal carboxylate group are essential for enzyme activity. The absence of absorbance above 300 nm suggests that the enzyme does not contain a pyridoxal 5′-phosphate group, which would be characteristic of a classical aminotransferase.
On the basis of the general catalytic mechanism, the hydrolytic deamination of 2-aminomuconate could be base or acid catalyzed (Fig. 3). The fact that 2-aminomuconate hydrolyzes spontaneously to 4-oxalocrotonate at low pH suggests that the nonenzymatic reaction occurs by the acid-catalyzed mechanism. The two different direct products of hydrolysis of 2-aminomuconate are spectrally distinguishable. Our result (Fig. 2) clearly indicated that the direct product of enzymatic deamination is 4-oxalocrotonate, which provides strong evidence for the acid-catalyzed mechanism. In this mechanism, an active imine intermediate is formed and hydrolysis of the imine produces 4-oxalocrotonate. Imine bond formation (Schiff base) is a common mechanism of deamination or transamination (20). The imine can be formed by oxidation, as in the deamination of glutamate by glutamate dehydrogenase in the presence of NAD or NADP (16) and as in the deamination of various d-amino acids by d-amino acid oxidase in the presence of flavin adenine dinucleotide (19). The imine bond can also be formed with the help of pyridoxal 5′-phosphate, as for 1-aminocyclopropane 1-carboxylate deaminase (10) and as in the transamination catalyzed by aminotransferases (20). Aminoacrylate, an unsaturated α-amino acid, tautomerizes nonenzymatically to its imine form, which hydrolyzes spontaneously to pyruvate and ammonia (20). 2-Aminomuconate contains the conjugated double bonds which enable spontaneous tautomerization to the imine form, and it hydrolyzes to release ammonia as aminoacrylate does; however, the nonenzymatic process is slow. In the enzyme-catalyzed reaction, a proton donor could initiate and facilitate the tautomerization. The mechanism of tautomerization would be analogous to the conversion of 2-hydroxymuconate to 4-oxalocrotonate, which is catalyzed by 4-oxalocrotonate tautomerase (11, 14, 18, 22). The evidence for the involvement of a histidine in the active site of 2-aminomuconate deaminase would be consistent with a mechanism involving donation of a proton by the enzyme. The tautomerization of 2-aminomuconate to form an imine intermediate would provide an explanation for why a cofactor is not required for the 2-aminomuconate deaminase activity.
FIG. 3.
Alternative hypothetical mechanisms of deamination of 2-aminomuconate. (A) A hydroxyl group attacks the α-carbon of 2-aminomuconate to form 2-hydroxymuconate, which slowly tautomerizes to 4-oxalocrotonate (not observed). (B) A proton from the aqueous solution (spontaneous deamination) or from a proton donor (enzymatic catalysis) initiates tautomerization to form an imine intermediate which hydrolyzes spontaneously to 4-oxalocrotonate.
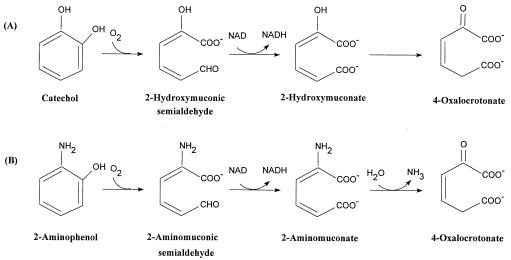
The direct conversion of 2-aminomuconate to 4-oxalocrotonate by 2-aminomuconate deaminase without the formation of the enol intermediate of 2-hydroxymuconate indicates that 4-oxalocrotonate tautomerase does not play a significant part in the degradation of nitrobenzene by P. pseudoalcaligenes JS45 even though the tautomerase activity is detectable in the crude extracts of JS45. The pathway of degradation of 2-aminophenol by JS45 (the downstream pathway of degradation of nitrobenzene) seems analogous to the meta cleavage pathway of catechol (Fig. 4) (3, 5–7, 15). The first two reactions are clearly analogous, and our results suggest that the third reaction is also similar. Additional work will be required to reveal how the two pathways are related at the enzymatic and genetic levels. Although 2-aminomuconate deaminase from JS45 does not catalyze the tautomerization of 4-oxalocrotonate and differs from 4-oxalocrotonate tautomerase in the molecular masses of its subunits and in N-terminal amino acid sequence (2), the tautomerase and deaminase may have similar catalytic mechanisms because the two reactions are similar and so are their substrate structures. The fact that both enzymes are hexamers suggests that the two enzymes may share some similarity in their tertiary and/or quaternary structures. Cloning and sequencing of the whole structural gene of the deaminase will allow a more complete understanding of this novel and interesting enzyme, which might also have a function in the metabolism of tryptophan in eukaryotes.
FIG. 4.
Comparison of the meta cleavage pathway of catechol (A) with the pathway for degradation of 2-aminophenol by JS45 (B).
ACKNOWLEDGMENTS
The work was supported in part by the U.S. Air Force Office of Scientific Research and the Strategic Environmental Defense Research Program. Z.H. is a recipient of the National Research Council Postdoctoral Research Associateship award in 1996–1998.
We thank S. F. Nishino and L. Nadeau for reviewing the manuscript.
REFERENCES
- 1.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 2.Chen L H, Kenyon G L, Curtin F, Harayama S, Bembenek M E, Hajipour G, Whitman C P. 4-Oxalocrotonate tautomerase, an enzyme composed of 62 amino acid residues per monomer. J Biol Chem. 1992;267:17716–17721. [PubMed] [Google Scholar]
- 3.Dagley S, Gibson D T. The bacterial degradation of catechol. Biochem J. 1965;95:466–474. doi: 10.1042/bj0950466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garfin E D. One-dimensional gel electrophoresis. Methods Enzymol. 1990;182:425–441. doi: 10.1016/0076-6879(90)82035-z. [DOI] [PubMed] [Google Scholar]
- 5.Harayama S, Rekik M. The meta cleavage operon of TOL degradative plasmid pWW0 comprises 13 genes. Mol Gen Genet. 1990;221:113–120. doi: 10.1007/BF00280375. [DOI] [PubMed] [Google Scholar]
- 6.Harayama S, Rekik M, Ngai K-L, Ornston L N. Physically associated enzymes produce and metabolize 2-hydroxy-2,4-dienoate, a chemically unstable intermediate formed in catechol metabolism via meta cleavage in Pseudomonas putida. J Bacteriol. 1989;171:6251–6258. doi: 10.1128/jb.171.11.6251-6258.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He Z, Spain J C. Studies of the catabolic pathway of degradation of nitrobenzene by Pseudomonas pseudoalcaligenes JS45: removal of the amino group from 2-aminomuconic semialdehyde. Appl Environ Microbiol. 1997;63:4839–4843. doi: 10.1128/aem.63.12.4839-4843.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ichiyama A, Nakamura S, Kawai H, Honjo T, Nishizuka Y, Hayashi O, Senoh S. Studies on the metabolism of the benzene ring of tryptophan in mammalian tissues. II. Enzymic formation of α-aminomuconic acid from 3-hydroxyanthranilic acid. J Biol Chem. 1965;240:740–749. [PubMed] [Google Scholar]
- 9.Lapworth A. The form of change in organic compounds, and the function of the alpha-meta-orientating groups. J Chem Soc. 1901;79:1265–1284. [Google Scholar]
- 10.Li K, Du W, Que N L S, Liu H. Mechanistic studies of 1-aminocyclopropane-1-carboxylate deaminase: unique covalent catalysis by coenzyme B6. J Am Chem Soc. 1996;118:8763–8764. [Google Scholar]
- 11.Lian H, Whitman C P. Ketonization of 2-hydroxy-2,4-pentadienoate by 4-oxalocrotonate tautomerase: implications for the stereochemical course and the mechanism. J Am Chem Soc. 1993;115:7978–7984. [Google Scholar]
- 12.Nishino S F, Spain J C. Degradation of nitrobenzene by a Pseudomonas pseudoalcaligenes. Appl Environ Microbiol. 1993;59:2520–2525. doi: 10.1128/aem.59.8.2520-2525.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nishizuka Y, Ichiyama A, Hayaishi O. Metabolism of the benzene ring of tryptophan (mammals) Methods Enzymol. 1970;17A:463–491. [Google Scholar]
- 14.Roper D I, Subramanya H S, Shingler V, Wigley D B. Preliminary crystallographic analysis of 4-oxalocrotonate tautomerase reveals the oligomeric structure of the enzyme. J Mol Biol. 1994;243:799–801. doi: 10.1016/0022-2836(94)90050-7. [DOI] [PubMed] [Google Scholar]
- 15.Sala-Trepat J M, Evans W C. The meta cleavage of catechol by Azotobacter species. Eur J Biochem. 1971;20:400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- 16.Smith E L, Austen B M, Blumenthal K M, Nyc J F. Glutamate dehydrogenases. In: Boyer P D, editor. The enzymes. 3rd ed. Vol. 11. New York, N.Y: Academic Press; 1975. pp. 293–367. [Google Scholar]
- 17.Stolz A, Knackmuss H-J. Bacterial metabolism of 5-aminosalicylic acid: enzymic conversion to l-malate, pyruvate and ammonia. J Gen Microbiol. 1993;139:1019–1025. doi: 10.1099/00221287-139-5-1019. [DOI] [PubMed] [Google Scholar]
- 18.Subramanya H S, Roper D I, Dauter Z, Dodson E J, Davies G J, Wilson K S, Wigley D B. Enzymatic ketonization of 2-hydroxymuconate: specificity and mechanism investigated by the crystal structures of two isomerases. Biochemistry. 1996;35:792–802. doi: 10.1021/bi951732k. [DOI] [PubMed] [Google Scholar]
- 19.Todone F, Vanoni M A, Mozzarelli A, Bolognesi M, Coda A, Curti B, Mattevi A. Active site plasticity in d-amino acid oxidase: a crystallographic analysis. Biochemistry. 1997;36:5853–5860. doi: 10.1021/bi9630570. [DOI] [PubMed] [Google Scholar]
- 20.Voet D, Voet J G. Biochemistry. New York, N.Y: Wiley; 1990. pp. 678–729. [Google Scholar]
- 21.Webb E C, editor. Enzyme nomenclature. New York, N.Y: Academic Press; 1992. [Google Scholar]
- 22.Whitman C P, Aird B A, Gillespie W R, Stolowich N J. Chemical and enzymatic ketonization of 2-hydroxymuconate, a conjugated enol. J Am Chem Soc. 1991;113:3154–3162. [Google Scholar]
- 23.Wiley R H, Hart A J. 2-Pyrones. IX. 2-Pyrone-6-carboxylic acid and its derivatives. J Am Chem Soc. 1954;76:1942–1944. [Google Scholar]