Abstract
whiE is a complex locus that specifies the polyketide spore pigment in Streptomyces coelicolor A3(2). Two divergently oriented promoters, whiEP1 and whiEP2, were identified in the whiE gene cluster, and their activities were analyzed during colony development in wild-type and sporulation-deficient strains. Both promoters were developmentally regulated; whiEP1 and whiEP2 transcripts were detected transiently at approximately the time when sporulation septa were observed in the aerial hyphae, and transcription from both promoters depended on each of the six known “early” whi genes required for sporulation septum formation (whiA, -B, -G, -H, -I, and -J). Mutation of the late sporulation-specific sigma factor gene, sigF, had no effect on the activity of whiEP1 but blocked transcription from whiEP2. However, ςF-containing holoenzyme was not sufficient to direct transcription of whiEP2 in vitro. The whiEP2 promoter controls expression of whiE ORFVIII, encoding a putative flavin adenine dinucleotide-dependent hydroxylase that catalyzes a late tailoring step in the spore pigment biosynthetic pathway. Disruption of whiE ORFVIII causes a change in spore color, from grey to greenish (T.-W. Yu and D. A. Hopwood, Microbiology 141:2779–2791, 1995). Consistent with these observations, construction of a sigF null mutant of S. coelicolor M145 caused the same change in spore color, showing that disruption of sigF in S. coelicolor changes the nature of the spore pigment rather than preventing its synthesis altogether.
Spore pigmentation in Streptomyces coelicolor A3(2) arises from the synthesis of a grey compound. Attempts to purify the spore pigment have failed, possibly indicating its covalent attachment to a macromolecular component of the spore (7). Instead, the polyketide nature of the spore pigment was initially predicted from the sequence of the complex locus (whiE) that specifies it (14). The whiE genes encode proteins that closely resemble the components of type II polyketide synthases, which are involved in the synthesis of a variety of aromatic antibiotics, including tetracenomycin from Streptomyces glaucescens (3), granaticin from Streptomyces violaceoruber (39), oxytetracycline from Streptomyces rimosus (26), and actinorhodin from S. coelicolor itself (16). This prediction was confirmed and extended through the engineered expression of some of the whiE genes in S. coelicolor during vegetative growth, which led to the production of extracellular aromatic polyketides with carbon chain lengths of 22 and 24 (43).
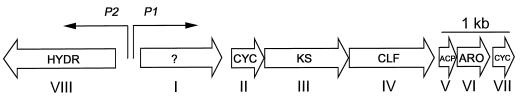
The extent of the whiE cluster known from available sequence is shown in Fig. 1. It consists of a likely operon of seven genes (ORFI to -VII) and at least one divergently transcribed gene, ORFVIII. The “minimal” polyketide synthase (PKS), responsible for the synthesis of the primary polyketide chain, is encoded by ORFIII to -V, while ORFII and ORFVII encode cyclases, ORFVI encodes an aromatase, and ORFVIII encodes a putative hydroxylase. The product of ORFI does not resemble proteins of known function but may be involved in retaining or targeting the spore pigment within the spore; artificially induced expression of ORFII to -VIII in the absence of ORFI led to copious production of presumed precursors in the medium, whereas the pigment remained within the mycelium when ORFI was present (44). There may well be other whiE genes lying outside of the sequenced region, encoding enzymes that catalyze other late tailoring steps in the biosynthetic pathway (44).
FIG. 1.
Organization of the whiE cluster (6, 14, 44). KS, ketosynthase; CLF, chain length factor; ACP, acyl carrier protein; ARO, aromatase; CYC, cyclase; HYDR, hydroxylase; ?, unknown.
Many other Streptomyces species have pigmented spores, and Southern blot analysis using probes from three different parts of the whiE cluster has suggested that homologs of whiE are widely distributed in streptomycetes (5). In Streptomyces halstedii, a locus (sch) that resembles whiE in both arrangement and sequence was identified and was shown to be required for synthesis of the spore pigment, which is green in this species (4–6). A partially sequenced and highly similar set of genes from Streptomyces curacoi is also likely to be involved in spore pigment production, but this has not been proven (1).
Spore pigmentation has been important in the genetic analysis of morphological differentiation in S. coelicolor because all of the original sporulation-deficient (whi) mutants were identified by virtue of their inability to synthesize wild-type levels of spore pigment, resulting in colonies that remained white on prolonged incubation instead of developing the normal grey color (10, 11, 19). These observations suggest that expression of whiE is under developmental control and hence that an understanding of the regulation of expression of these genes may be informative about the regulation of sporulation. Therefore, we have characterized two promoters within the whiE cluster and examined their activities during development in wild-type and sporulation-deficient strains of S. coelicolor.
MATERIALS AND METHODS
Strains, media, and culture conditions.
Bacterial strains are shown in Table 1. Unless stated otherwise, S. coelicolor strains were grown at 30°C on agar minimal medium (MM) (20) containing mannitol (0.5% [wt/vol]) instead of glucose as the carbon source, supplemented as necessary with histidine (125 μg/ml) and uracil (20 μg/ml). When mycelium was to be harvested from surface cultures for the preparation of RNA, cultures were inoculated onto sterile cellophane discs on the agar surface (41).
TABLE 1.
Derivatives of S. coelicolor A3(2) used in this work
| Strain | Genotype | Reference or source |
|---|---|---|
| Wild type | Pgl+ SCP1+ SCP2+ | 20 |
| C72 | whiA72 Pgl+ SCP1+ SCP2+ | 10 |
| C70 | whiB70 Pgl+ SCP1+ SCP2+ | 10 |
| C71 | whiG71 Pgl+ SCP1+ SCP2+ | 10 |
| C119 | whiH119 Pgl+ SCP1+ SCP2+ | 10 |
| C17 | whiI17 Pgl+ SCP1+ SCP2+ | 10 |
| C77 | whiJ77 Pgl+ SCP1+ SCP2+ | 12, 35 |
| J1508 | hisA1 strA1 uraA1 Pgl− SCP1NFa SCP2− | 22 |
| J1979 | hisA1 strA1 uraA1 sigF::tsr Pgl− SCP1NF SCP2− | 33 |
| M145 | Pgl+ SCP1− SCP2− | 20 |
| J1984 | sigF::tsr Pgl+ SCP1− SCP2− | This work |
| J1501 | hisA1 strA1 uraA1 Pgl− SCP1− SCP2− | 13 |
NF, normal fertility. The SCP1 plasmid is integrated into the “9 o’clock” region of the chromosome.
Construction of J1984, a sigF null mutant of S. coelicolor M145.
S. coelicolor J1979 total genomic DNA (2 μg) was alkaline denatured and used to transform S. coelicolor M145 to thiostrepton resistance according to the method of Oh and Chater (32). A representative transformant was designated J1984.
RNA isolation from surface-grown cultures of S. coelicolor.
Mycelium (0.5 to 1.0 g) grown on cellophane discs was harvested with a spatula and dispersed in 6 ml of P buffer (20) through vigorous vortexing with 4.5- to 5.5-mm-diameter glass beads. After lysozyme was added to a final concentration of 2 mg/ml, the samples were incubated at 30°C for 10 min before the addition of stock solutions of 0.5 M EDTA (pH 8.0) and 10% (wt/vol) sodium dodecyl sulfate to final concentrations of 0.15 M and 1% (wt/vol), respectively. In order to complete the lysis of all cells, samples were transferred to 65°C, 1.5 ml of phenol (saturated with 3% [wt/vol] sodium chloride) and 1.5 ml of chloroform were added, and, after brief vortexing, the samples were incubated at 65°C for a further 5 min. After vigorous vortexing for 3 to 4 min, samples were centrifuged (8,000 rpm in an SS34 rotor 10 min, 4°C) and the upper phases were removed and extracted repeatedly with phenol-chloroform. Nucleic acids were precipitated in the presence of sodium acetate (0.3 M, pH 7.0) with an equal volume of isopropanol. The remaining steps of RNA isolation were as described by Hopwood et al. (20), with DNase I used to remove DNA.
Molecular methods.
Methods for plasmid isolation, DNA manipulations, and radiolabelling of DNA fragments or oligonucleotide primers were as described by Sambrook et al. (37) or Hopwood et al. (20).
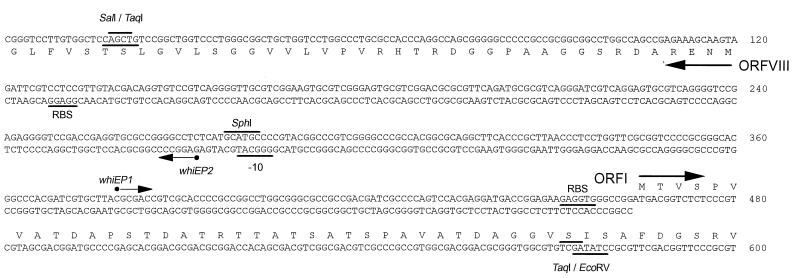
S1 nuclease mapping was based on the procedure of Bibb et al. (2). Forty micrograms of RNA (estimated spectrophotometrically) was used in each S1 nuclease protection experiment, with hybridization mixtures incubated at 45°C in Na-trichloroacetic acid buffer (30) after denaturation at 75°C for 10 min. All probes were labelled on the 5′ ends with [γ-32P]ATP (3,000 Ci/mmol) and T4 polynucleotide kinase. The probes used were probe A, a 295-bp EcoRV-SphI fragment (see Fig. 2) uniquely labelled at the EcoRV site; probe B, a 260-bp SalI-SphI fragment (see Fig. 2) uniquely labelled at the SalI site; probe C, a 555-bp TaqI fragment (see Fig. 2) labelled on both 5′ ends; the sigF probe, a 600-bp BssHII-PstI (polylinker site) fragment uniquely labelled at the BssHII site (25); and the hrdB probe, a 520-bp LspI-BglII (polylinker site) fragment uniquely labelled at the LspI site (9, 25). The chemical sequence ladder used for comparison with the whiEP1 S1 nuclease-protected fragment was generated from probe A, as described by Maxam and Gilbert (29).
FIG. 2.
Nucleotide sequence of the whiE ORFI to ORFVIII intergenic region and the 5′ ends of the genes. The sequences of both strands are shown in the intergenic region, but only the upper strand is shown for the DNA encoding ORFI and only the lower strand is shown for the DNA encoding ORFVIII (which runs from right to left). Filled circles indicate the most likely transcription start points for the whiEP1 and whiEP2 promoters, as determined by S1 nuclease mapping and/or reverse transcriptase-mediated primer extention. The potential −10 region for the whiEP2 promoter and potential ribosome binding sites (RBS) are underlined. Restriction sites used in the creation of S1 nuclease mapping probes are marked. Note: the sequence of the ORFI to ORFVIII intergenic region as originally published (GenBank accession no. X55942 [14] and X74213 [6]) contained five errors that have been corrected here.
Primer extension reactions were carried out according to Kelemen et al. (24) with oligonucleotides labelled at their 5′ ends with [γ-32P]ATP (3,000 Ci/mmol) and T4 polynucleotide kinase. The primers used were 5′-GGCCCACCTCTTCTCCGGTCATC-3′ (nucleotides 439 to 461, bottom strand [see Fig. 2]) for whiEP1 and 5′-GCGCGTCAGGGATCGTCAGG-3′ (nucleotides 204 to 223, top strand [see Fig. 2]) for whiEP2. Enzymatic sequence ladders were generated by dideoxy chain termination (38) with the same radiolabelled oligonucleotides used as primers.
Construction of a ςF overexpression plasmid.
The DNA upstream of a unique ScaI site 7 bp downstream of the natural sigF GTG start codon was replaced with two complementary oligonucleotides (5′-TATGCCGGCTTCT-3′ and 5′-AGAAGCCGGCA-3′) that introduced an NdeI half-site overlapping an ATG start codon and also replaced the third and fourth codons with synonymous codons commonly associated with highly expressed genes in Escherichia coli. The modified SigF gene was cloned into pET11c (40), which had been cut with NdeI and BamHI to generate pIJ5894, in which sigF is under the control of a T7 RNA polymerase promoter and an appropriately positioned Shine-Dalgarno sequence.
Overexpression and solubilization of ςF.
pIJ5894 was introduced into E. coli BL21(DE3), also carrying plasmid pLysS (40). A fresh transformant was used to inoculate a 50-ml overnight culture of L broth containing 100 μg of carbenicillin per ml and 10 μg of chloramphenicol per ml. This culture was innoculated late at night and grown at 30°C to prevent the cells from reaching stationary phase. The following morning, this culture was used to inoculate 1 liter of the same medium, which was grown at 37°C until it reached an optical density at 600 nm of 0.5. The culture was induced by the addition of IPTG (isopropyl-β-d-thiogalactopyranoside) to a final concentration of 1 mM and grown for a further 4 h before harvest. This procedure yielded approximately 1.5 g (wet weight) of cells.
ςF was recovered from inclusion bodies by a minor modification of the method of Nguyen et al. (31). The cell pellet was resuspended in 20 ml of lysis buffer (50 mM Tris-HCl [pH 8.0], 10 mM EDTA, 1 mM dithiothreitol [DTT], 50 mM NaCl, 1 mM phenylmethylsulfonyl fluoride, 0.2% [wt/vol] sodium deoxycholate [Na-DOC], 5% [vol/vol] glycerol, 200 μg of lysozyme per ml) and incubated on ice for 30 min before lysis was completed by three 30-s cycles of sonication. The cell lysate was then centrifuged for 20 min at 15,000 rpm in an SS34 rotor, and the supernatant was discarded. The inclusion bodies were purified by resuspension in 20 ml of wash buffer, which is TGED (50 mM Tris-HCl [pH 8.0], 5% [vol/vol] glycerol, 0.1 mM EDTA, 0.1 mM DTT] containing 50 mM NaCl and 2% (wt/vol) Na-DOC, followed by stirring at 4°C for 1 h and repeated sonication as before. The inclusion bodies were recovered by centrifugation, and the washing procedure was repeated. The purified inclusion bodies were solubilized by resuspension in 20 ml of solubilization buffer [TGED containing 50 mM NaCl and 0.25 (wt/vol) sarkosyl(N-lauroylsarcosine)] and stirred for 1 h at 4°C.
MonoQ anion-exchange column chromatography.
The solubilized material was dialyzed for at least 24 h against 2 liters of TGED containing 50 mM NaCl, with several changes of buffer to attempt the complete removal of sarkosyl, followed by centrifugation and filtration through a 0.2-μm-pore-size cellulose acetate filter (Sartorius GmbH). Protein was applied via a 50-ml Superloop (Pharmacia plc) to a MonoQ HR 5/5 FPLC anion-exchange column (Pharmacia plc) that had been equilibrated with the same buffer. The column was washed with 10 ml of this buffer and then eluted with a 50-ml linear gradient of 0.05 to 1.0 M NaCl at a flow rate of 0.5 ml/min. Fractions (1.0 ml) were collected from the start of sample application, and those containing ςF were dialyzed into storage buffer (50 mM Tris-HCl [pH 8.0], 0.1 mM EDTA, 0.1 mM DTT, 50 mM NaCl, 50% [vol/vol] glycerol) and stored at −20°C.
In vitro transcription.
Runoff transcription assays were performed with [α-32P]CTP (600 Ci/mmol) (New England Nuclear), as described by Buttner et al. (8). Transcription from the whiEP2 promoter region was assayed by using a 555-bp SalI-EcoRV fragment (see Fig. 2), and transcription from the Bacillus subtilis ctc promoter was assayed by using a 340-bp EcoRI-BamHI fragment from pMI340 (21). Transcripts were analyzed on 6% polyacrylamide–7 M urea gels with a heat-denatured, 32P-labelled HinFI digest of φX174 used as a size standard. E. coli core RNA polymerase was purchased from Epicentre Technologies (Madison, Wis.).
RESULTS
Identification of two promoters in the whiE cluster.
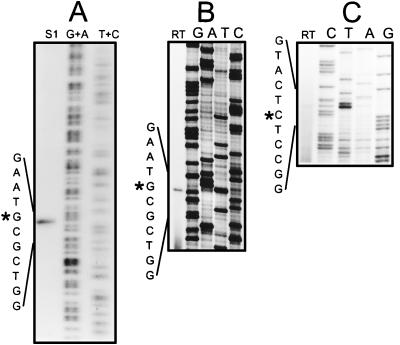
The promoter activity of the 343-bp intergenic region between ORFI and the divergent gene, ORFVIII (Fig. 2), was investigated. High-resolution S1 nuclease mapping of the rightward promoter (whiEP1) was performed by using probe A (see Materials and Methods) and RNA isolated from S. coelicolor J1501 containing pIJ556 (the multicopy vector pIJ486 carrying whiE ORFI to -VII on a 5.7-kb SphI fragment [14]) grown for 60 h on agar. A single RNA-protected DNA fragment, corresponding to a transcription start point at C379 (Fig. 3A), 85 bp upstream of the ORFI ATG start codon, was observed (Fig. 2). The observed transcript was not an artifact arising from multiple plasmid-borne copies of the operon, since reverse transcriptase-mediated primer extension of the whiEP1 transcript at its natural abundance, in RNA isolated from wild-type S. coelicolor grown for 72 h on agar, identified a transcription start point at the same nucleotide (Fig. 3B).
FIG. 3.
Mapping of the transcription start points for whiEP1 and whiEP2. (A) High-resolution S1 nuclease mapping of the 5′ end of the whiEP1 transcript by using probe A. G+A and T+C, chemical sequence ladders; S1, RNA-protected product of the S1 nuclease protection assay. RNA was isolated from J1501 carrying pIJ556 (the multicopy vector pIJ486 carrying whiE ORFI to -VII on a 5.7-kb SphI fragment [14]) grown for 60 h on agar. (B) Primer extension analysis of the 5′ end of the whiEP1 transcript. RNA isolated from wild-type S. coelicolor grown for 72 h on agar was used as a template for avian myeloblastosis virus (AMV) reverse transcriptase (RT) with the oligonucleotide primer 5′-GGCCCACCTCTTCTCCGGTCATC-3′ (nucleotides 439 to 461, bottom strand [Fig. 2]). (C) Primer extension analysis of the 5′ end of the whiEP2 transcript. RNA isolated from wild-type S. coelicolor grown for 72 h on agar was used as a template for AMV RT with the oligonucleotide primer 5′-GCGCGTCAGGGATCGTCAGG-3′ (nucleotides 204 to 223, top strand [Fig. 2]).
S1 nuclease mapping of the leftward promoter, whiEP2, was performed by using probe B (see Materials and Methods) and RNA isolated from wild-type S. coelicolor grown for 72 h on agar, and the approximate size of the protected DNA fragment was determined by comparison with a heat-denatured, radiolabelled HpaII digest of pBR322. A single RNA-protected DNA fragment of approximately 250 nucleotides was detected. For more precise mapping of the 5′ end of the ORFVIII transcript, reverse transcriptase-mediated primer extension of the same RNA was used, identifying a transcription start point at G272 (Fig. 2), 152 nucleotides upstream of the ORFVIII ATG start codon (Fig. 3C).
The whiEP1 and whiEP2 promoters are developmentally regulated.
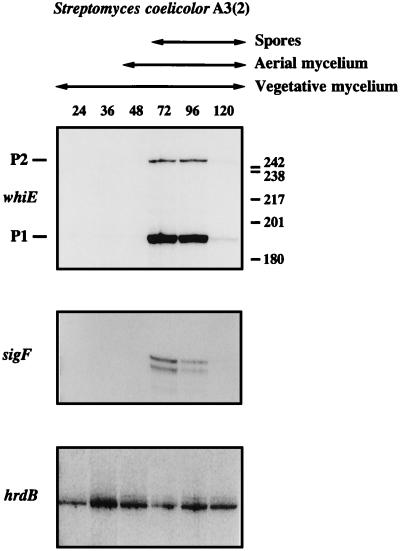
The pattern of transcription from whiEP1 and whiEP2 during development of wild-type S. coelicolor was monitored by S1 nuclease protection. Following previous work (25), we used a time course of RNA samples that had already been used to assess the developmental pattern of transcription of the late sporulation-specific sigma factor gene, sigF. The data for sigF are reproduced here for comparison, along with the data for hrdB, encoding the principal (essential) sigma factor of S. coelicolor, designed to act as a positive internal control. No attempt was made to fractionate the harvested cell material used for RNA preparation; thus, for example, the late samples contained vegetative and aerial mycelium as well as spores (25).
To simplify the assessment of the relative levels of the whiEP1 and whiEP2 transcripts, a single TaqI fragment, radiolabelled on both 5′ ends (probe C [see Materials and Methods]), was used. The whiEP1 and whiEP2 promoters were found to be developmentally regulated: both transcripts first appeared at 72 h, the time at which sporulation was first detected in the culture, and were present at similar levels 24 h later (Fig. 4). The whiE transcripts were seen neither during vegetative growth nor during aerial mycelium formation and were almost undetectable at 120 h in mature colonies. It is possible that the decrease in signals seen at 120 h results from a difficulty in recovering RNA quantitatively from spore compartments that have undergone wall thickening, although the RNA isolation protocol used (25) was developed especially to try to ensure a representative yield from all cellular compartments. At the level of resolution of this experiment, the developmental profiles of the two whiE transcripts were very similar to that of the sigF transcript (Fig. 4).
FIG. 4.
S1 nuclease protection analysis of transcription of whiE, sigF, and hrdB during development in surface-grown MM cultures of wild-type S. coelicolor A3(2). The time points (in hours) at which mycelium was harvested for RNA isolation and the presence of vegetative mycelium, aerial mycelium, and spores, as judged by microscopic examination, are shown. The positions of the size markers (a 32P-labelled HpaII digest of pBR322) are indicated on the right of the whiE panel. The sigF and hrdB panels from this figure were published previously (25) and are shown here for comparison with the whiE data.
The whiEP1 and whiEP2 promoters depend on each of the early whi loci required for sporulation septum formation.
To reveal any dependence of the whiEP1 and whiEP2 promoters on other whi genes, we analyzed their activities in representative mutants of the six early whi genes required for sporulation septum formation, whiA, whiB, whiG, whiH, whiI, and whiJ, again by using S1 nuclease mapping. All of the whi mutants were of the same (wild-type) genetic background (Table 1).
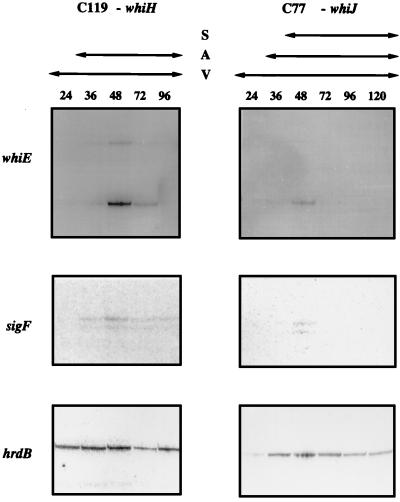
Mutations in whiA, whiB, whiG, and whiI completely blocked transcription from both whiEP1 and whiEP2 (data not shown). In contrast, transcription from both whiEP1 and whiEP2 was still detectable in the whiH and whiJ mutants (Fig. 5), but the level of these transcripts was more than 10-fold lower than in the wild-type (it should be noted that the whiE and sigF panels in Fig. 5 were deliberately overexposed and are therefore not directly comparable with the whiE and sigF panels in Fig. 4 corresponding to the wild type). As expected, transcription of the control gene, hrdB, was unaffected in all six whi mutants (Fig. 5) (25).
FIG. 5.
S1 nuclease protection analysis of transcription of whiE, sigF, and hrdB during development in surface-grown MM cultures of the two early whi mutants C119 (whiH) and C77 (whiJ). The time points (in hours) at which mycelium was harvested for RNA isolation and the presence of vegetative mycelium (V), aerial mycelium (A), and spores (S), as judged by microscopic examination, are shown. The whiE and sigF panels in this figure have been deliberately overexposed so that these extremely weak signals are visible; therefore, the band intensities are not directly comparable to those shown for the wild type (Fig. 4). The level of the whiE transcripts in the whiH and whiJ mutants was more than 10-fold lower than in the wild type. The sigF and hrdB panels from this figure were published previously (25) and are shown here for comparison with the whiE data.
A very similar dependence pattern was previously obtained in a study of sigF expression; sigF transcripts were undetectable in whiA, whiB, whiG, and whiI mutants and were severely reduced, but still detectable, in whiH and whiJ mutants (Fig. 5) (25). In principle, the faint signals corresponding to the whiEP1 and whiEP2 transcripts could have arisen because these promoters are not absolutely dependent on whiH and whiJ and/or because the mutant alleles used in these experiments (whiH119 and whiJ77) are not null mutations. Recently, the whiH gene was cloned and a null mutant was constructed (36). The null mutant is pale grey, not pure white, implying that some spore pigment synthesis occurs. It therefore seems likely that the whiEP1 and whiEP2 promoters are strongly, but not absolutely, dependent on whiH. Since the known whiJ mutants are also pale grey, it is also likely that the whiEP1 and whiEP2 promoters are not absolutely dependent on whiJ.
whiEP2, but not whiEP1, depends on the late sporulation-specific RNA polymerase sigma factor, ςF.
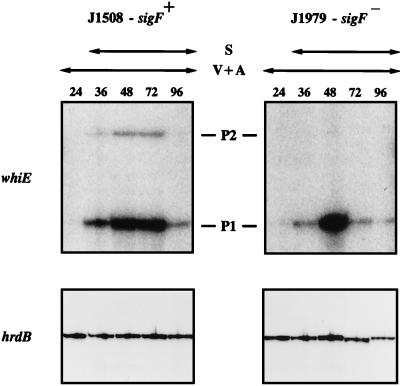
sigF encodes the late sporulation-specific RNA polymerase sigma factor, ςF (25, 33). The spores of a constructed sigF null mutant strain, J1979, are white (33), suggesting that expression of one or more of the whiE genes might depend, directly or indirectly, on sigF. To address this question, we examined transcription of whiE in J1979 and its congenic sigF+ parent, J1508, by the same methods as before (J1979 and J1508 have a different background genotype from the wild-type and whi mutant strains discussed above [Table 1]). In the sigF+ strain J1508, appearance of the whiEP1 and whiEP2 transcripts coincided with sporulation (Fig. 6), as it did in wild-type S. coelicolor (Fig. 4), although the particularly rapid development of J1508 in this experiment meant that aerial mycelium had already developed by the earliest time point (24 h). While disruption of sigF did not affect transcription from whiEP1, transcription from whiEP2 was undetectable in the sigF mutant (Fig. 6). The previously published control data (25) for hrdB in J1508 and J1979, derived from the same RNA time courses, are reproduced here for comparison.
FIG. 6.
S1 nuclease protection analysis of transcription of whiEP1, whiEP2, and hrdB during development in surface-grown MM cultures of the sigF null mutant J1979 and its congenic sigF+ parent strain, J1508. The time points (in hours) at which mycelium was harvested for RNA isolation and the presence of vegetative and aerial mycelium (V+A) and spores (S), as judged by microscopic examination, are shown. The exposure times for panels corresponding to a given probe were identical. The hrdB panels from this figure were published previously (25) and are shown here for comparison with the whiE data.
A sigF null mutant of S. coelicolor M145 makes a greenish spore pigment.
Disruption of the whiE ORFVIII causes a change in the color of the spore pigment from grey to greenish (44). Because we found that whiEP2, but not whiEP1, depends on sigF, and because whiE ORFVIII is transcribed from whiEP2, we would predict that sigF mutants should be greenish (with the caveat that the sequence of the whiE cluster is incomplete and therefore there may be other, unidentified whiE genes that also depend on sigF). However, J1979, the constructed sigF null mutant of J1508, is white (33). To see if this might be a consequence of the genetic background, we moved our constructed sigF null mutation (in which part of the coding sequence was replaced by the thiostrepton resistance gene, tsr) from J1979 into the prototrophic, plasmid-free strain M145 by using a recently developed method for transformation of S. coelicolor with denatured total chromosomal DNA (32). The spores of the resulting strain, J1984 (Table 1), were greenish, showing that disruption of sigF alters the nature of the spore pigment rather than blocking its synthesis altogether. Disruption of the sigF gene of Streptomyces aureofaciens also changes the color of the spore pigment, in that case from the wild-type grey-pink to green (34a).
Promoter specificity of ςF in vitro.
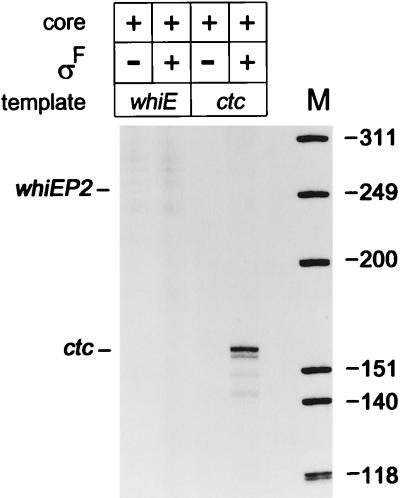
In an attempt to determine if whiEP2 is a direct biochemical target for ςF-containing holoenzyme (EςF), we overexpressed and purified ςF as described in Materials and Methods. Because ςF formed inclusion bodies, the protein was solubilized and refolded by the method of Nguyen et al. (31). Approximately 10 μg of purified ςF was subjected to sequential Edman degradation to determine the sequence of the first 10 N-terminal residues. The result—PASTAPQAPP—showed complete agreement with that predicted from the DNA sequence of sigF (33) and showed that the N-terminal N-formylmethionine had been removed. EςF was not sufficient to direct transcription of whiEP2 in vitro (Fig. 7).
FIG. 7.
Promoter specificity of ςF-containing holoenzyme in vitro. Transcription from the whiEP2 promoter region was assayed with a 555-bp SalI-EcoRV fragment and was expected to generate a transcript of 251 nucleotides (Fig. 2), and transcription from the B. subtilis ctc promoter was assayed with a 340-bp EcoRI-BamHI fragment from pMI340 (21) and was expected to generate a transcript of 155 nucleotides. Transcripts were analyzed on 6% polyacrylamide–7 M urea gels with a heat-denatured, 32P-labelled HinFI digest of φX174 as size standards.
In the absence of a characterized ςF-dependent promoter, we needed a promoter to use as a positive control to ensure that the refolded ςF preparation was active. Of the sigma factors in the databases, ςF is most similar to ςB of B. subtilis, particularly in regions 2.4 and 4.2, the two regions known to interact with promoters at the −10 and −35 regions, respectively (33). It was therefore likely that ςF-dependent promoters would closely resemble B. subtilis ςB-dependent promoters such as ctc (21). Accordingly, we attempted to use the B. subtilis ctc promoter as a positive control and found that it was indeed recognized by EςF in vitro (Fig. 7).
DISCUSSION
Addition of two promoters from the whiE cluster to the dependence pathway for sporulation genes.
We have identified two divergently oriented promoters, whiEP1 and whiEP2, within the whiE gene cluster. Both promoters are developmentally regulated, the transient appearance of both transcripts coinciding with the onset of sporulation septum formation. Transcription from both promoters depends on each of the six known early whi genes required for sporulation septum formation (whiA, -B, -G, -H, -I, and -J). In contrast, mutation of the late sporulation-specific sigma factor gene, sigF, has no effect on the activity of whiEP1 but abolishes transcription from whiEP2.
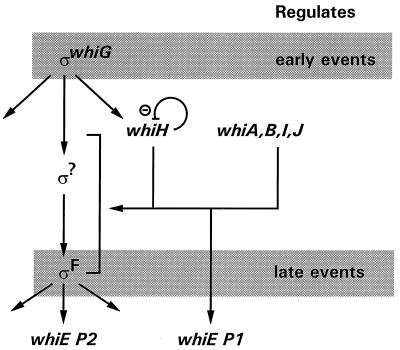
The whiEP1 and whiEP2 promoters can now be added to the genetic hierarchy controlling sporulation in the aerial hyphae of S. coelicolor (Fig. 8). The dependence of sigF expression on each of the six early whi genes (Fig. 8) (25) provides a likely explanation for the dependence of whiEP2 on these same six genes. In contrast, the dependence of whiEP1 on the six early whi genes (Fig. 8) cannot be explained in the same way, since whiEP1 is still active in a sigF null mutant. The whiEP1 promoter does not conform to the well-established consensus sequence for promoters transcribed by the early sporulation-specific sigma factor, ςWhiG (41, 42); consequently, it seems clear that whiEP1 is not transcribed by either of the known sporulation-specific sigma factors, ςWhiG and ςF.
FIG. 8.
Dependence hierarchy of genes controlling sporulation in the aerial hyphae of S. coelicolor, adapted from reference 25 to include the whiEP1 and whiEP2 promoters. The hierarchy also shows that whiH expression is negatively regulated, directly or indirectly, by whiH itself (23). The dependence of whiH on ςWhiG is known to be direct (42).
Spatial location of whiE expression.
Previous work has suggested that whiE expression is confined to a subset of cell types. In addition to the whiE pigment, S. coelicolor is known to make a second polyketide compound, the blue antibiotic actinorhodin specified by the act cluster (16). A mutation in any one of the three act minimal PKS subunit genes results in loss of actinorhodin production, showing that such mutations are not complemented in trans by the naturally expressed equivalent subunits from the whiE gene cluster. However, when the whiE PKS genes were expressed artificially under control of the thiostrepton-inducible promoter PtipA in mutant strains of S. coelicolor from which any one of the three act minimal PKS subunit genes had been individually deleted, a functional hybrid PKS was formed in which the spore pigment PKS subunits substituted for their homologs from the actinorhodin PKS and actinorhodin production was restored (27, 44). Similarly, mutations in any of the three whiE minimal PKS subunit genes are not, in the natural situation, complemented by the corresponding act genes, but this can again be corrected by placing the relevant genes under the control of the PtipA promoter (44).
Taken together, these results indicated that, under normal circumstances, the absence of “cross-talk” between the two PKS complexes arises because the two gene sets are expressed in separate cell types within the developing colony rather than by any biochemical incompatibility between the two. Although the work presented here deals with the timing of whiE expression, rather than its spatial location, the observations that in wild-type S. coelicolor whiEP1 and whiEP2 are active only when sporulation is detected in the aerial mycelium and that their activity depends on each of the six known whi genes required for sporulation septum formation are consistent with whiE expression being confined to the spore compartments.
Greenish mutants of S. coelicolor.
whiE ORFVIII almost certainly encodes a tailoring enzyme for a late step in the spore pigment pathway, in which a hydroxyl group is introduced into the cyclized polyketide. The predicted product of ORFVIII is homologous to flavin adenine dinucleotide-linked hydroxylases (6), and disruption of the gene caused a change in the color of the spore pigment from grey to greenish (44). Disruption of the ORFVIII homolog in the sch cluster of S. halstedii also changed the spore pigment color, in that case from the wild-type green to lilac (6). Further, the original whiE clone isolated by Davis and Chater (14) carried ORFI to -VII but only part of ORFVIII. Introduction of this plasmid into a whiE mutant caused production of a greenish spore pigment, presumably arising from an accumulation of the substrate of the hydroxylase encoded by ORFVIII, thereby mimicking the phenotype of ORFVIII disruption.
During early screens for whi mutants of S. coelicolor, six mutants with greenish spores were also isolated (18). Of these six mutants, five (C1, C22, C23, C24, and C42) mapped approximately halfway between strA and cysD, a chromosomal location compatible with the later-mapped whiE locus (18). It therefore seems likely that these five strains carried mutations in whiE ORFVIII. Interestingly, the remaining greenish mutant, C27, mapped in the pheA-to-strA interval but closer to pheA (18), a location compatible with that of sigF (34). Given the greenish phenotype of the constructed sigF null mutant J1984, C27 may well have carried a mutation in sigF.
Why does the original sigF null mutant, J1979, have a whi phenotype?
The greenish phenotype of the sigF null mutant J1984 can be explained by the observation that whiEP2, the promoter which drives transcription of whiE ORFVIII, depends on sigF. The reason why the spores of the original sigF null mutant, J1979, are white, not greenish, is unclear. The presence of a second site mutation in J1979 can be excluded because the white phenotype is fully complemented by a DNA fragment carrying sigF alone (see Fig. 4 in reference 33). We think that the most likely explanation for the white phenotype of J1979 is that although J1984 and J1979 make the same polyketide molecule, there is a considerably lower overall level of pigment synthesis in J1979. Given that the greenish spore color of J1984 is very pale, it is therefore likely to be undetectable in J1979. Indeed, this difference can be seen in the level of wild-type spore pigment produced by the sigF+ parents of these two strains: M145 is considerably darker than J1508. In addition, the levels of the whiE transcripts detected in J1508 were much lower than in wild-type S. coelicolor (the experiments shown in Fig. 4 and 6 were done at the same time with the same probe preparation, but the whiE panel in Fig. 4 is taken from an autoradiograph while the two whiE panels in Fig. 6 are taken from a phosphorimager).
ςF is active as a primary translation product.
Both S. coelicolor and Streptomyces aureofaciens ςF possess a poorly conserved, repetitive, proline-rich amino-terminal extension that is absent from B. subtilis ςB and other ς factors (33). Apart from this N-terminal region, the two ςF sequences are very similar throughout their length. These observations led to speculation that ςF might be synthesized as an inactive, pro-sigma factor (33) by analogy with the mother cell-specific sigma factors of B. subtilis, ςE and ςK, which are activated posttranslationally by proteolysis of the N-terminal 29 and 20 amino acids, respectively (15, 28). The activity of the recombinant, full-length ςF overexpressed in E. coli argues strongly against this possibility.
Is whiEP2 a direct biochemical target for the EςF holoenzyme?
whiEP2 depends on sigF in vivo, but EςF is not sufficient to direct transcription of whiEP2 in vitro. One possible explanation for these observations is that the dependence is indirect and whiEP2 is not a direct biochemical target for the EςF holoenzyme. On the other hand, of the sigma factors in the databases, ςF is most similar to ςB of Bacillus subtilis, particularly in regions 2.4 and 4.2, the two regions known to interact with promoters at the −10 and −35 regions, respectively (33), and EςF recognized the B. subtilis ςB-dependent promoter ctc in vitro. It is therefore likely that ςF-dependent promoters will be found to closely resemble ςB-dependent promoters. The −10 region of whiEP2 (GGGCAT) clearly resembles the −10 consensus for ςB (GGGTAT) (17), but there are no sequences in the −35 region of whiEP2 that resemble the −35 consensus for ςB (GTTTAA) (17). Therefore, another possibility is that whiEP2 is transcribed by the EςF holoenzyme in conjunction with a transcription activator.
ACKNOWLEDGMENTS
We thank Mark Paget, Mervyn Bibb, and David Hopwood for helpful comments on the manuscript.
This work was supported by BBSRC grant CAD 04355 (to M.J.B.), by a Lister Institute research fellowship (to M.J.B.), and by a grant-in-aid to the John Innes Centre from the BBSRC.
REFERENCES
- 1.Bergh S, Uhlén M. Analysis of a polyketide synthesis-encoding gene cluster of Streptomyces curacoi. Gene. 1992;117:131–136. doi: 10.1016/0378-1119(92)90501-f. [DOI] [PubMed] [Google Scholar]
- 2.Bibb M J, Janssen G R, Ward J M. Cloning and analysis of the promoter region of the erythromycin resistance gene (ermE) of Streptomyces erythraeus. Gene. 1986;41:E357–E368. doi: 10.1016/0378-1119(85)90220-3. [DOI] [PubMed] [Google Scholar]
- 3.Bibb M J, Biró S, Montamedi H, Collins J F, Hutchinson C R. Analysis of the nucleotide sequence of the Streptomyces glaucescens tcml genes provides key information about the enzymology of polyketide antibiotic biosynthesis. EMBO J. 1989;8:2727–2736. doi: 10.1002/j.1460-2075.1989.tb08414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanco G, Pereda A, Méndez C, Salas J A. Cloning and disruption of a DNA fragment of Streptomyces halstedii involved in the biosynthesis of a spore pigment. Gene. 1992;112:59–65. doi: 10.1016/0378-1119(92)90303-7. [DOI] [PubMed] [Google Scholar]
- 5.Blanco G, Brian P, Pereda A, Méndez C, Salas J A, Chater K F. Hybridisation and DNA sequence analysis suggest an early evolutionary divergence of related biosynthetic gene sets encoding polyketide antibiotics and spore pigments in Streptomyces spp. Gene. 1993;130:107–116. doi: 10.1016/0378-1119(93)90352-4. [DOI] [PubMed] [Google Scholar]
- 6.Blanco G, Pereda A, Brian P, Méndez C, Chater K F, Salas J A. A hydroxylase-like gene product contributes to synthesis of a polyketide spore pigment in Streptomyces halstedii. J Bacteriol. 1993;175:8043–8048. doi: 10.1128/jb.175.24.8043-8048.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brian P. A developmentally regulated spore pigment locus from Streptomyces coelicolor A3(2). Ph.D. thesis. Norwich, United Kingdom: University of East Anglia; 1992. [Google Scholar]
- 8.Buttner M J, Fearnley I M, Bibb M J. The agarase gene (dagA) of Streptomyces coelicolor A3(2): nucleotide sequence and transcriptional analysis. Mol Gen Genet. 1987;209:101–109. doi: 10.1007/BF00329843. [DOI] [PubMed] [Google Scholar]
- 9.Buttner M J, Chater K F, Bibb M J. Cloning, disruption, and transcriptional analysis of three RNA polymerase sigma factor genes of Streptomyces coelicolor A3(2) J Bacteriol. 1990;172:3367–3378. doi: 10.1128/jb.172.6.3367-3378.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chater K F. A morphological and genetic mapping study of white colony mutants of Streptomyces coelicolor. J Gen Microbiol. 1972;72:9–28. doi: 10.1099/00221287-72-1-9. [DOI] [PubMed] [Google Scholar]
- 11.Chater K F. Genetics of differentiation in Streptomyces. Annu Rev Microbiol. 1993;47:685–713. doi: 10.1146/annurev.mi.47.100193.003345. [DOI] [PubMed] [Google Scholar]
- 12.Chater K F, Merrick M J. Approaches to the study of differentiation in Streptomyces coelicolor A3(2) In: MacDonald K D, editor. Second international symposium on the genetics of industrial microorganisms. London, United Kingdom: Academic Press; 1976. pp. 583–593. [Google Scholar]
- 13.Chater K F, Bruton C J, King A A, Suarez J E. The expression of Streptomyces and Escherichia coli drug resistance determinants cloned into the Streptomyces phage φC31. Gene. 1982;19:21–32. doi: 10.1016/0378-1119(82)90185-8. [DOI] [PubMed] [Google Scholar]
- 14.Davis N K, Chater K F. Spore colour in Streptomyces coelicolor A3(2) involves the developmentally regulated synthesis of a compound biosynthetically related to polyketide antibiotics. Mol Microbiol. 1990;4:1679–1691. doi: 10.1111/j.1365-2958.1990.tb00545.x. [DOI] [PubMed] [Google Scholar]
- 15.Errington J. Bacillus subtilis sporulation: regulation of gene expression and control of morphogenesis. Microbiol Rev. 1993;57:1–33. doi: 10.1128/mr.57.1.1-33.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernández-Moreno M A, Martínez E, Boto L, Hopwood D A, Malpartida F. Nucleotide sequence and deduced functions of a set of co-transcribed genes of Streptomyces coelicolor A3(2) including the polyketide synthase for the antibiotic actinorhodin. J Biol Chem. 1992;267:19278–19290. [PubMed] [Google Scholar]
- 17.Hecker M, Schumann W, Völker U. Heat-shock and general stress response in Bacillus subtilis. Mol Microbiol. 1996;19:417–428. doi: 10.1046/j.1365-2958.1996.396932.x. [DOI] [PubMed] [Google Scholar]
- 18.Hopwood, D. A., and H. M. Kieser. Personal communication.
- 19.Hopwood D A, Wildermuth H, Palmer H M. Mutants of Streptomyces coelicolor defective in sporulation. J Gen Microbiol. 1970;61:397–408. doi: 10.1099/00221287-61-3-397. [DOI] [PubMed] [Google Scholar]
- 20.Hopwood D A, Bibb M J, Chater K F, Kieser T, Bruton C J, Kieser H M, Lydiate D J, Smith C P, Ward J M, Schrempf H. Genetic manipulation of Streptomyces: a laboratory manual. Norwich, United Kingdom: The John Innes Foundation; 1985. [Google Scholar]
- 21.Igo M M, Losick R. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J Mol Biol. 1986;191:615–624. doi: 10.1016/0022-2836(86)90449-3. [DOI] [PubMed] [Google Scholar]
- 22.Ikeda H, Seno E T, Bruton C J, Chater K F. Genetic mapping, cloning and physiological aspects of the glucose kinase gene of Streptomyces coelicolor. Mol Gen Genet. 1984;196:501–507. doi: 10.1007/BF00436199. [DOI] [PubMed] [Google Scholar]
- 23.Kelemen, G. H. Unpublished data.
- 24.Kelemen G H, Cundliffe E, Financsek I. Cloning and characterisation of gentamicin-resistance genes from Micromonospora purpurea and Micromonospora rosea. Gene. 1991;98:53–60. doi: 10.1016/0378-1119(91)90103-i. [DOI] [PubMed] [Google Scholar]
- 25.Kelemen G H, Brown G L, Kormanec J, Potúčková L, Chater K F, Buttner M J. The positions of the sigma factor genes, whiG and sigF, in the hierarchy controlling the development of spore chains in the aerial hyphae of Streptomyces coelicolor A3(2) Mol Microbiol. 1996;21:593–603. doi: 10.1111/j.1365-2958.1996.tb02567.x. [DOI] [PubMed] [Google Scholar]
- 26.Kim E-S, Bibb M J, Buter M J, Hopwood D A, Sherman D H. Nucleotide sequence of the oxytetracycline (otc) polyketide synthase genes from Streptomyces rimosus. Gene. 1994;14:141–142. doi: 10.1016/0378-1119(94)90144-9. [DOI] [PubMed] [Google Scholar]
- 27.Kim E-S, Hopwood D A, Sherman D H. Analysis of type II polyketide β-ketoacyl synthase specificity in Streptomyces coelicolor A3(2) by trans complementation of actinorhodin synthase mutants. J Bacteriol. 1994;176:1801–1804. doi: 10.1128/jb.176.6.1801-1804.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Losick R, Stragier P. Crisscross regulation of cell-type-specific gene expression during development in B. subtilis. Nature. 1992;355:601–604. doi: 10.1038/355601a0. [DOI] [PubMed] [Google Scholar]
- 29.Maxam A, Gilbert W. Sequencing end-labelled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 30.Murray M G. Use of sodium trichloroacetate and mung bean nuclease to increase sensitivity and precision during transcript mapping. Anal Biochem. 1986;158:165–170. doi: 10.1016/0003-2697(86)90605-6. [DOI] [PubMed] [Google Scholar]
- 31.Nguyen L H, Jensen D B, Burgess R R. Overexpression and purification of ς32, the Escherichia coli heat shock transcription factor. Protein Expr Purif. 1993;4:425–433. doi: 10.1006/prep.1993.1056. [DOI] [PubMed] [Google Scholar]
- 32.Oh S-H, Chater K F. Denaturation of circular or linear DNA facilitates targeted integrative transformation of Streptomyces coelicolor A3(2): possible relevance to other organisms. J Bacteriol. 1997;179:122–127. doi: 10.1128/jb.179.1.122-127.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Potúčková L, Kelemen G H, Findlay K C, Lonetto M A, Buttner M J, Kormanec J. A new RNA polymerase sigma factor, ςF, is required for the late stages of morphological differentiation in Streptomyces sp. Mol Microbiol. 1995;17:37–48. doi: 10.1111/j.1365-2958.1995.mmi_17010037.x. [DOI] [PubMed] [Google Scholar]
- 34.Redenbach M, Kieser H M, Denapaite D, Eichner A, Cullum J, Kinashi H, Hopwood D A. A set of ordered cosmids and a detailed genetic and physical map for the 8Mb Streptomyces coelicolor A3(2) chromosome. Mol Microbiol. 1996;21:77–96. doi: 10.1046/j.1365-2958.1996.6191336.x. [DOI] [PubMed] [Google Scholar]
- 34a.Řežuchova B, Barák I, Kormanec J. Disruption of a sigma factor gene, sigF, affects an intermediate stage of spore pigment production in Streptomyces aureofaciens. FEMS Microbiol Lett. 1997;153:371–377. doi: 10.1111/j.1574-6968.1997.tb12598.x. [DOI] [PubMed] [Google Scholar]
- 35.Ryding N J. Analysis of sporulation genes in Streptomyces coelicolor A3(2). Ph.D. thesis. Norwich, United Kingdom: University of East Anglia; 1995. [Google Scholar]
- 36.Ryding, N. J. Personal communication.
- 37.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 38.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sherman D H, Malpartida F, Bibb M J, Kieser H, Bibb M J, Hopwood D A. Structure and deduced function of the granaticin-producing polyketide synthase gene cluster of Streptomyces violaceoruber Tü22. EMBO J. 1989;8:2717–2725. doi: 10.1002/j.1460-2075.1989.tb08413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Studier W F, Rosenberg A H, Dunn J J, Dubendorff J W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 41.Tan H, Chater K F. Two developmentally controlled promoters of Streptomyces coelicolor A3(2) that resemble the major class of motility-related promoters in other bacteria. J Bacteriol. 1993;175:933–940. doi: 10.1128/jb.175.4.933-940.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whatling, C. A. Personal communication.
- 43.Yu T-W. Physical and functional studies of polyketide synthase genes of Streptomyces. Ph.D. thesis. Norwich, United Kingdom: University of East Anglia; 1995. [Google Scholar]
- 44.Yu T-W, Hopwood D A. Ectopic expression of the Streptomyces coelicolor whiE genes for polyketide spore pigment synthesis and their interaction with the act genes for actinorhodin biosynthesis. Microbiology. 1995;141:2779–2791. doi: 10.1099/13500872-141-11-2779. [DOI] [PubMed] [Google Scholar]