Abstract
Background
Circular RNAs (circRNAs) play a significant role in the tumorigenesis and progression of diverse human cancers, including lung adenocarcinoma. A previous study suggested that circ_0004140 expression was increased in lung adenocarcinoma cells. However, the molecular mechanism of circRNA circ_0004140 involved in lung adenocarcinoma is poorly defined.
Methods
Circ_0004140, microRNA‐330‐5p (miR‐330‐5p), and NOVA alternative splicing regulator 2 (NOVA2) expression were determined by real‐time quantitative polymerase chain reaction (RT‐qPCR). Cell proliferation, apoptosis, migration, invasion, and angiogenesis ability were assessed using 5‐ethynyl‐2’‐deoxyuridine (EdU), flow cytometry, wound healing, transwell, and capillary‐like network formation assays. Proliferating cell nuclear antigen (PCNA), cyclin D1, and NOVA2 protein levels were detected using Western blot assay. The interaction between miR‐330‐5p and circ_0004140 or NOVA2 was verified by dual‐luciferase reporter assay. Xenograft tumor model was utilized to assess the role of circ_0004140 in tumor growth in vivo.
Results
Circ_0004140 was upregulated in lung adenocarcinoma tissues and cell lines. Circ_0004140 silencing suppressed cell proliferation, migration, invasion and tube formation ability, and triggered the apoptosis of lung adenocarcinoma cells. Circ_0004140 acted as a molecular sponge for miR‐330‐5p, and miR‐330‐5p silencing largely reversed circ_0004140 knockdown‐induced effects in lung adenocarcinoma cells. NOVA2 was a target of miR‐330‐5p, and NOVA2 overexpression might largely overturn miR‐330‐5p overexpression‐induced influences in lung adenocarcinoma cells. Circ_0004140 upregulated NOVA2 expression via sponging miR‐330‐5p in lung adenocarcinoma cells. Circ_0004140 silencing restrained xenograft tumor growth in vivo.
Conclusion
Circ_0004140 knockdown might suppress the malignant biological behaviors of lung adenocarcinoma cells via miR‐330‐5p‐dependent regulation of NOVA2.
Keywords: lung adenocarcinoma, circ_0004140, miR‐330‐5p, NOVA2
Circ_0004140 targets miR‐330‐5p to upregulate NOVA2 expression, thereby promoting lung adenocarcinoma progression.
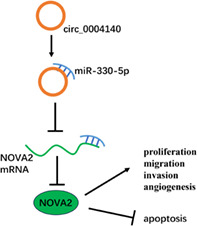
INTRODUCTION
Lung adenocarcinoma belongs to non‐small cell lung cancer (NSCLC). 1 Despite achievement in the diagnosis and treatment of lung adenocarcinoma, a high rate of recurrence and metastasis result in a dismal prognosis. 1 , 2 , 3 Hence, much hope is placed on identifying novel therapeutic targets to improve the outcome.
Circular RNAs (circRNAs) are stable circular molecules, and act as sponges of microRNAs (miRNAs) to regulate cell behaviors. 4 , 5 , 6 Increasingly studies have demonstrated the important regulatory functions of circRNAs in lung adenocarcinoma. For example, circ_0001588 aggravated lung adenocarcinoma development by positively regulating NACC1 expression via sponging miR‐524‐3p. 7 Circ‐ZNF609 accelerated growth in lung adenocarcinoma via modulating miR‐1224‐3p/ETV1 axis. 8 Notably, a recent report validated that circ_0004140 abundance was enhanced in lung adenocarcinoma, and acted as a carcinogenic factor by boosting cell proliferation and migration in vitro. 9 Yet, the pathogenesis of circ_0004140 involved in lung adenocarcinoma is still largely unknown.
MiRNAs modulate gene expression via direct binding to them, 10 and dysregulated miRNAs are relevant to lung adenocarcinoma progression. 11 MiR‐330‐5p has been shown to exhibit anticancer function in multiple malignancies. For instance, circ‐PTN accelerated glioma development via suppressing miR‐330‐5p and miR‐145‐5p. 12 Wang et al. demonstrated that miR‐330‐5p hampered proliferation capacity and invasion ability in osteosarcoma. 13 MiR‐330‐5p acted as a downstream target of EWSAT1 to suppress cervical cancer progression. 14 MiR‐330‐5p also blocked NSCLC progression by previous articles. 15 , 16 , 17 Based on bioinformatic prediction, miR‐330‐5p was a potential downstream molecule of circ_0004140. In this study, the target relation between miR‐330‐5p and circ_0004140 and their functional relevance in lung adenocarcinoma progression were explored.
NOVA alternative splicing regulator 2 (NOVA2) belongs to the NOVA family, and regulates the development of motor neurons. 18 NOVA2 has been previously reported to exhibit a protumor role in NSCLC. 19 , 20 , 21 , 22 Xiao et al. demonstrated that miR‐7‐5p restrained the motility of NSCLC cells by inhibiting NOVA2. 19 NOVA2 is a possible target of miR‐330‐5p via bioinformatic prediction. The relevance between miR‐330‐5p and NOVA2 in lung adenocarcinoma development has been investigated.
Here, the functions of circ_0004140 in lung adenocarcinoma were analyzed and the working mechanism of circ_0004140 was explored via bioinformatic analysis and rescue experiments.
METHODS
Human tissue specimens
Lung adenocarcinoma specimens (n = 33) and adjacent normal specimens (n = 33) were harvested from lung adenocarcinoma cases at West China Hospital, Sichuan University. Tissue specimens were preserved at −80°C. The detailed clinical characteristics of patients are described in Table 1. All patients signed their written informed consent. Human materials were collected and used with the permission of the ethics committee of West China Hospital, Sichuan University.
TABLE 1.
Clinicopathological features of patients with lung adenocarcinoma.
| Characteristic | Cases |
|---|---|
| Gender | |
| Male | 18 |
| Female | 15 |
| Age (years) | |
| <60 | 16 |
| ≥60 | 17 |
| Smoking status | |
| Nonsmoker | 14 |
| Smoker | 19 |
| Tumour differentiation | |
| Well/moderate | 20 |
| Poor | 13 |
| T stage | |
| T1/2 | 18 |
| T3 | 15 |
| TNM Stages | |
| I/II | 19 |
| III | 14 |
Cell lines
Normal cells (BEAS‐2B) and two lung adenocarcinoma cell lines (H522 and A549) were acquired in the BeNa culture collection company (Beijing, China). Cells were cultivated in Roswell Park Memorial Institute‐1640 (RPMI‐1640; Hyclone, Logan, UT, USA) with a supplement of 10% fetal bovine serum (FBS: Gibco) and 1% antibiotic mixture (Transgen) in a 37°C and 5% CO2 humidified atmosphere.
Real‐time quantitative polymerase chain reaction (RT‐qPCR)
Commercial TRizol solution (Invitrogen) was adopted to prepare RNA samples. RNA samples were utilized for DNA generation using the First Strand complementary DNA (cDNA) synthesis kit (Toyobo) and MiRNA reverse transcription reagents (Applied Biosystems). After that, cDNA was utilized as template DNA for RT‐qPCR reaction with SYBR mix (Toyobo) and specific primers (Table 2). With U6 or glyceraldehyde‐3‐phosphate dehydrogenase (GAPDH) as the reference gene, RNA expression was analyzed using the 2−∆∆Ct equation.
TABLE 2.
Primer sequences in RT‐qPCR assay.
| Gene name | Species | Direction | Sequence (5'‐3') |
|---|---|---|---|
| Circ_0004140 | Human | Forward | GGCAGGCAATGCGGAATATC |
| Reverse | TCATGGCTTGTTCCCATCCA | ||
| YAP1 | Human | Forward | TGACCCTCGTTTTGCCATGA |
| Reverse | GTTGCTGCTGGTTGGAGTTG | ||
| miR‐330‐5p | Human | Forward | GCCGAGTCTCTGGGCCTGTG |
| Reverse | CTCAACTGGTGTCGTGGA | ||
| NOVA2 | Human | Forward | GCAACACGGGAGTTTCCTCT |
| Reverse | TCTCGTCCGTTCCTCACAAC | ||
| U6 | Human | Forward | AATTGGAACGATACAGAGAAGATTAGC |
| Reverse | TATGGAACGCTTCACGAATTTG | ||
| GAPDH | Human | Forward | GAATGGGCAGCCGTTAGGAA |
| Reverse | AAAAGCATCACCCGGAGGAG |
Stability analysis of circ_0004140
Total RNA samples isolated from lung adenocarcinoma cells were digested with 100 μg/mL RNase R (Applied Biological Materials) for 25 min at 37°C. RNA expression of circ_0004140 and YAP1 messenger RNA (mRNA) was assessed by RT‐qPCR.
Cell transfection
Small interfering (si)RNA targeting circ_0004140 (si‐circ_0004140), negative control of siRNA (si‐NC), short hairpin (sh) RNA targeting circ_0004140 (sh‐circ_0004140), sh‐NC, overexpression plasmid of circ_0004140 (oe‐circ_0004140), pLO‐ciR vector (vector), miR‐330‐5p mimic: 5'‐UCUCUGGGCCUGUGUCUUAGGC‐3', miR‐330‐5p inhibitor: 5'‐ GCCTAAGACACAGGCCCAGAGA‐3', and their controls (miR‐NC mimic: 5'‐UUCUCCGAACGUGUCACGUTT‐3' and miR‐NC inhibitor: 5'‐CAGUACUUUUGUGUAGUACAA‐3'), si‐NC: 5'‐AAUUCUCCGAACGUGUCACGU‐3', si‐NOVA2: 5'‐UGUUGUAGCCGUAACUUGCCA‐3', NOVA2 overexpression plasmid in pcDNA vector (pcDNA‐NOVA2) and pcDNA‐NC were acquired from Geneseed (Guangzhou) and GenePharma (Shanghai). Oligonucleotides and plasmids were introduced to lung adenocarcinoma cells with commercial Lipofectamine 3000 reagent (Invitrogen).
5‐ethynyl‐2'‐deoxyuridine (EdU) assay
In summary, 4 × 103 transfected H522 and A549 cells in 96‐well plates were treated with 100 μL medium containing EdU (20 μM, RiboBio) for 2 h. After being fixed in 4% formaldehyde solution for 30 min, cells were permeabilized with 0.5% Triton X‐100. Ten minutes later, cells were reacted with Apollo reaction cocktail for 30 min. Then, 4′,6‐diamidino‐2‐phenylindole (DAPI) was used to stain the nuclei, which were visualized under a fluorescence microscope.
Western blot assay
Protein samples were loaded onto sodium dodecyl sulfate‐polyacrylamide gel electrophoresis (SDS‐PAGE), and separated protein bands were transferred to polyvinylidene difluoride (PVDF) membrane (Millipore). Sealing was applied using 5% skimmed milk, and the membrane was mixed with antibodies below to mark the target proteins. The primary antibodies contained antiproliferating cell nuclear antigen (PCNA; ab29; Abcam), anti‐cyclin D1 (ab16663; Abcam), anti‐NOVA2 (AV40400; Sigma) and anti‐β‐actin (ab6276; Abcam). The membrane was then mixed with the corresponding secondary antibody (Abcam). β‐Actin functioned as the reference. After exposure to enhanced chemiluminescence (ECL) reagent (Invitrogen), the protein bands were visualized.
Cell apoptosis analysis
Annexin V‐fluorescein isothiocyanate (FITC) reagent and PI reagent in a commercial apoptosis kit (BestBio) were incubated with lung adenocarcinoma cells. Apoptotic cells (cells with FITC‐positive and PI‐positive or negative) were identified using the FACS CantoII flow cytometer (BD Biosciences).
Wound healing assay
A wound scratch was created when cell confluence reached about 90% in six‐well plates. After creating the wound for 0 h and 24 h, the images of the wound were captured, and the relative migration distance was analyzed.
Transwell invasion assay
Matrigel (BD Biosciences) was added to the upper transwell compartments in order to analyze cell invasion ability. Transfected lung adenocarcinoma cells in medium (without serum) were added to the above compartments, with the lower compartments added with 20% FBS‐contained culture medium. Cells transferred to the lower surface were dyed via 0.5% crystal violet dye liquor (Sigma). Cell number was manually counted using the light microscope (Olympus) at 100×.
Capillary‐like network formation assay
Cell angiogenesis ability was assessed in this assay. Lung adenocarcinoma cells were added to Matrigel (BD Biosciences)‐coated 96‐well plates with 2 × 104 cells in each well. After 48‐h cultivation, the average number of branches of each node was analyzed.
Establishment of circ_0004140/miR‐330‐5p/NOVA2 axis
Bioinformatic software Circinteractome (https://circinteractome.irp.nia.nih.gov) was utilized to predict the target miRNAs of circ_0004140, whereas the possible target mRNAs of miR‐330‐5p were predicted based on the starBase database (http://starbase.sysu.edu.cn).
Dual‐luciferase reporter assay
The partial fragment of circ_0004140 or NOVA2, including the wild‐type or mutant predicted sequence with miR‐330‐5p, was cloned to reporter vector psiCHECK2 (Promega), termed as WT‐circ_0004140, MUT‐circ_0004140, WT‐NOVA2 3’ untranslated region (3' UTR) and MUT‐NOVA2 3' UTR. A total of 10 pmol miR‐330‐5p mimic or miR‐NC mimic was cointroduced with the reporter plasmids into H522 or A549 cells. The luciferase intensity was evaluated via commercial luciferase report analysis reagents (Promega).
RNA pulldown assay
In order to further verify the interaction between circ_0004140 and many potential target miRNAs, RNA pulldown assay was carried out in A549 cells. In brief, control probe or circ_0004140 probe was synthesized by Genepharma, and incubated with M‐280 streptavidin magnetic beads (Invitrogen) for 10 min to make probe‐coated beads. Then, A549 cells were incubated with the probe‐coated beads. Finally, RT‐qPCR assay was executed for the amount of bound RNAs measurement.
Xenografts in nude mice
The animal assay was approved via the Ethics Committee of West China Hospital, Sichuan University. Twelve BALB/c strain nude mice were obtained from Vital River Laboratory Animal Technology (Beijing, China). A549 cells (3 × 106 cells in 200 μL phosphate buffer saline [PBS; Sangon Biotech]) stably transfected with sh‐NC or sh‐circ_0004140 were subcutaneously inoculated into the right flank of the mice (n = 6). Tumor dimension was assessed every week as volume = length × width2 × 0.5. On the 28th day, mice were sacrificed and xenografts were resected. Tumor weight was recorded. RT‐qPCR and Western blot assay were adopted to analyze the abundance of circ_0004140, miR‐330‐5p, and NOVA2 protein. For immunohistochemical (IHC) staining assay, after being fixed, dehydrated, and embedded into paraffin, tumor tissue from the nude mice was cut into 5 μm slices, with an antibody specific for Ki67. Finally, a microscope was used to capture these images.
Statistical analysis
Data processing was applied via GraphPad software (GraphPad) and student's t‐test or one‐way analysis of variance (ANOVA), and analytical results are presented as a form of mean ± standard deviation (SD). p < 0.05 denoted a significant difference between the experimental and the control group.
RESULTS
Circ_0004140 expression is upregulated in lung adenocarcinoma tissues and cell lines
Circ_0004140 was generated from exons 4, 5, and 6 of the YAP1 gene, and the end of exons 4 and 6 was back‐spliced to form the circular structure (Figure 1a). The expression profile of circ_0004140 in lung adenocarcinoma was analyzed. Compared with adjacent normal tissues (n = 33), circ_0004140 was upregulated in lung adenocarcinoma tissues (n = 33) (Figure 1b). We also analyzed the level of circ_0004140 in BEAS‐2B and two lung adenocarcinoma cell lines. Compared with the BEAS‐2B cell line, a significant upregulation in circ_0004140 expression was observed in two lung adenocarcinoma cell lines (Figure 1c). Circ_0004140 was resistant to the digestion of RNase R, and its linear counterpart YAP1 mRNA was degraded in the RNase R treatment group (Figure 1d,e), suggesting that circ_0004140 stably distributed in lung adenocarcinoma cells. The abnormal expression of circ_0004140 might imply its important function in lung adenocarcinoma.
FIGURE 1.
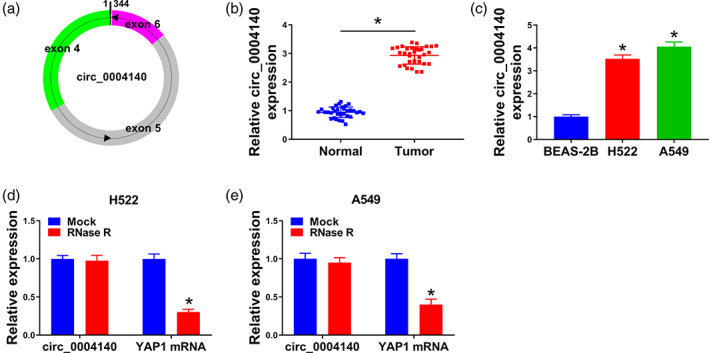
Circ_0004140 expression is upregulated in lung adenocarcinoma tissues and cell lines. (a) Circ_0004140 derived from back‐splicing of exons 4, 5, and 6 of the YAP1 gene. (b) Real‐time quantitative polymerase chain reaction (RT‐qPCR) was applied to measure the level of circ_0004140 in lung adenocarcinoma tissues (n = 33) and adjacent normal tissues (n = 33). (c) The expression of circ_0004140 in the BEAS‐2B cell line and two lung adenocarcinoma cell lines was detected by RT‐qPCR. (d and e) The stability of circ_0004140 was assessed via RNase R digestion. The linear counterpart (YAP1 mRNA) served as the control. *p < 0.05.
Circ_0004140 knockdown blocks cell proliferation, migration, invasion, the tube formation and promotes the apoptosis of lung adenocarcinoma cells
Si‐circ_0004140 was utilized to specifically knockdown circ_0004140 for loss‐of‐function experiments. Data exhibited that circ_0004140 expression was apparently reduced, rather than YAP1 mRNA expression, in si‐circ_0004140‐transfected H522 and A549 cells, suggesting that the interference efficiency of si‐circ_0004140 was high in two lung adenocarcinoma cell lines (Figure 2a and S1). With the silencing of circ_0004140, EdU positive rates were dramatically reduced (Figure 2b,c). Two cell proliferation‐associated markers were measured via Western blot assay. As shown in Figure 2d,e, with the interference of circ_0004140, the levels of PCNA and cyclin D1 were both reduced. Circ_0004140 silencing markedly increased the percentage of apoptotic cells (Figure 2f). The rate of wound closure was significantly reduced in the circ_0004140‐silenced group relative to the si‐NC‐transfected group (Figure 2g). The number of invaded lung adenocarcinoma cells was also reduced in the circ_0004140‐silenced group compared with the si‐NC‐transfected group (Figure 2h), suggesting that circ_0004140 knockdown restrained the invasion of lung adenocarcinoma cells. The tube formation ability of lung adenocarcinoma cells was tested by observing the capillary‐like structure via a capillary‐like network formation assay. As shown in Figure 2i, tube formation ability was markedly suppressed in circ_0004140‐silenced cells. Taken together, circ_0004140 silencing inhibited the malignant properties and induced the apoptosis of lung adenocarcinoma cells.
FIGURE 2.
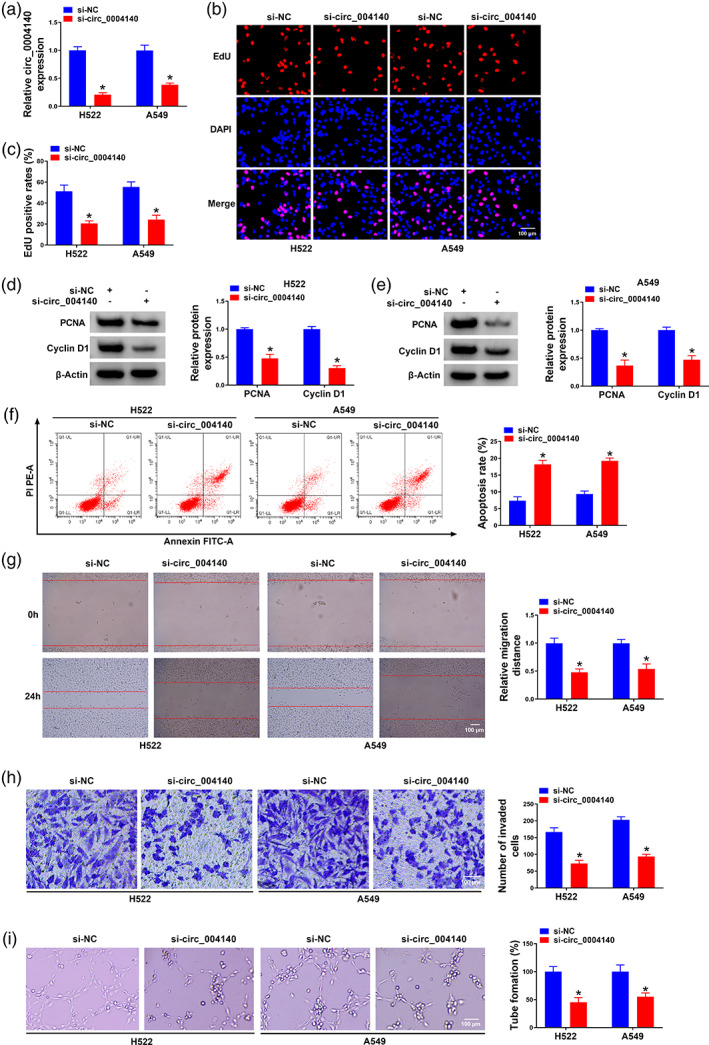
Circ_0004140 knockdown blocks cell proliferation, migration, invasion, tube formation and promotes the apoptosis of lung adenocarcinoma cells. (a–i) H522 and A549 cells were introduced with si‐NC or si‐circ_0004140. (a) The level of circ_0004140 was detected in transfected lung adenocarcinoma cells via RT‐qPCR. (b and c) Cell proliferation was assessed by 5‐ethynyl‐2'‐deoxyuridine (EdU) assay in transfected lung adenocarcinoma cells. (d and e) The expression of two cell proliferation‐associated markers (proliferating cell nuclear antigen [PCNA] and cyclin D1) was analyzed by Western blot assay. (f) Cell apoptosis was analyzed by flow cytometry. (g) The migration ability was assessed by wound healing assay. (h) Cell invasion ability was analyzed by transwell assay. (i) The capillary‐like structure in different groups via capillary‐like network formation assay was shown. *p < 0.05.
Circ_0004140 interacts with miR‐330‐5p in lung adenocarcinoma cells
Based on circinteractome database analysis, eight potential target miRNAs (miR‐1200, miR‐136, miR‐330‐5p, miR‐513‐3p, miR‐515‐5p, miR‐584, miR‐1270, and miR‐620) of circ_0004140 were predicted. For further selection, the miRNAs pulled down by biotinylated probes were purified and analyzed by RT‐qPCR. Among the eight candidate miRNAs, only miR‐330‐5p could be abundantly pulled down by circ_0004140 probe in A549 cells (Figure S1). Then, the binding sequence between circ_0004140 and miR‐330‐5p is shown in Figure 3a. Transfection with miR‐330‐5p mimic significantly upregulated miR‐330‐5p expression in H522 and A549 cells relative to miR‐NC mimic‐transfected group (Figure 3b). The luciferase intensity of reporter plasmid WT‐circ_0004140 was dramatically decreased with the overexpression of miR‐330‐5p, whereas the overexpression of miR‐330‐5p had almost no effect on the luciferase intensity of mutant reporter plasmid (Figure 3c), suggesting that miR‐330‐5p was a target of circ_0004140 in lung adenocarcinoma cells. RT‐qPCR verified the high transfection efficiency of oe‐circ_0004140 in H522 and A549 cells (Figure 3d). The overexpression of circ_0004140 decreased miR‐330‐5p level, whereas circ_0004140 silencing increased miR‐330‐5p level in H522 and A549 cells (Figure 3e), demonstrating the negative regulatory relationship between circ_0004140 and miR‐330‐5p in lung adenocarcinoma cells. MiR‐330‐5p expression was downregulated in lung adenocarcinoma tissues and cell lines (Figure 3f,g). Overall, circ_0004140 acted as a molecular sponge for miR‐330‐5p in lung adenocarcinoma cells.
FIGURE 3.
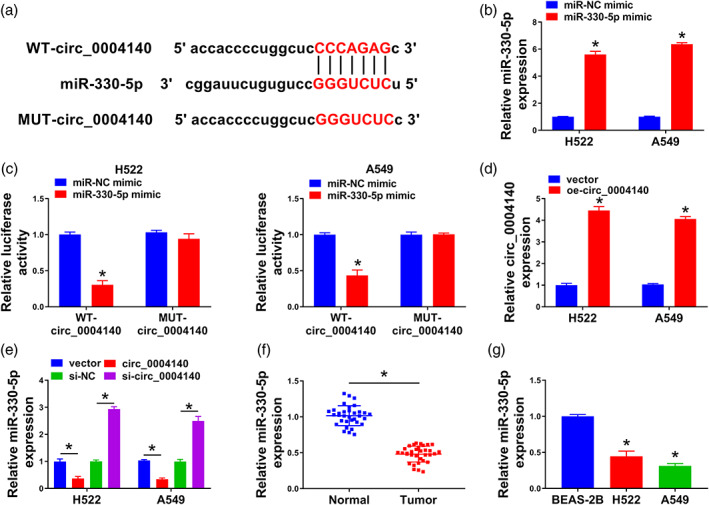
Circ_0004140 interacts with miR‐330‐5p in lung adenocarcinoma cells. (a) Bioinformatic software circinteractome predicted the potential binding sequence between circ_0004140 and miR‐330‐5p. (b) Real‐time quantitative polymerase chain reaction (RT‐qPCR) was applied to verify the transfection efficiency of miR‐330‐5p mimic in H522 and A549 cells. (c) Dual‐luciferase reporter assay was adopted to confirm if miR‐330‐5p was a target of circ_0004140 in lung adenocarcinoma cells. (d) The level of circ_0004140 was measured in H522 and A549 cells transfected with vector or oe‐circ_0004140 by RT‐qPCR. (e) The effect of circ_0004140 overexpression or knockdown on the level of miR‐330‐5p was tested by RT‐qPCR assay. (f) The level of miR‐330‐5p was analyzed in lung adenocarcinoma tissues and adjacent normal tissues by RT‐qPCR. (g) The expression of miR‐330‐5p was detected in the BEAS‐2B cell line and two lung adenocarcinoma cell lines by RT‐qPCR. *p < 0.05.
Circ_0004140 interference restrains the malignant properties of lung adenocarcinoma cells partly through elevating the miR‐330‐5p level
Circ_0004140 silencing increased miR‐330‐5p, and we cointroduced lung adenocarcinoma cells with si‐circ_0004140 and miR‐330‐5p inhibitor to perform compensation experiments (Figure 4a). The silencing of miR‐330‐5p largely rescued the cell proliferation ability in circ_0004140‐silenced lung adenocarcinoma cells (Figure 4b). Circ_0004140 knockdown‐induced downregulation in the levels of PCNA and cyclin D1 was largely overturned by the silencing of miR‐330‐5p (Figure 4c), suggesting that circ_0004140 knockdown suppressed cell proliferation partly through upregulating miR‐330‐5p. Circ_0004140 silencing‐induced apoptosis was largely counteracted by the knockdown of miR‐330‐5p in H522 and A549 cells (Figure 4e). Through performing wound healing and transwell assays, we found that circ_0004140 silencing‐induced suppressive effects on the migration and invasion of lung adenocarcinoma cells were largely attenuated by the addition of miR‐330‐5p inhibitor (Figure 4f,g). The introduction of miR‐330‐5p inhibitor largely rescued the tube formation ability of circ_0004140‐silenced lung adenocarcinoma cells (Figure 4h). Overall, circ_0004140 silencing restrained lung adenocarcinoma progression partly through upregulating miR‐330‐5p.
FIGURE 4.
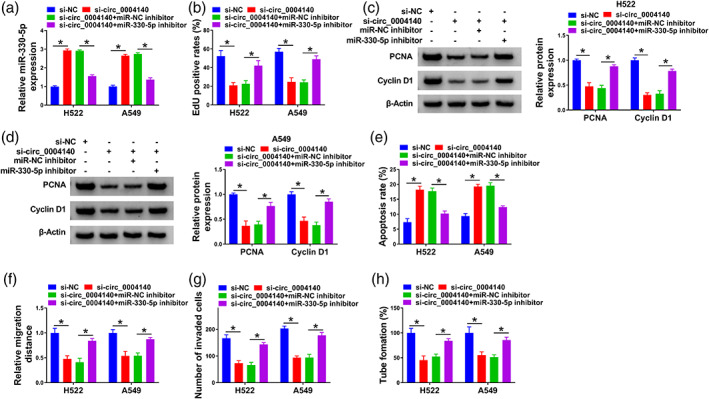
Circ_0004140 interference restrains the malignant properties of lung adenocarcinoma cells partly through elevating the miR‐330‐5p level. (a–i) H522 and A549 cells were transfected with si‐NC, si‐circ_0004140, si‐circ_0004140 + miR‐NC inhibitor or si‐circ_0004140 + miR‐330‐5p inhibitor. (a) The level of miR‐330‐5p was measured in lung adenocarcinoma cells by real‐time quantitative polymerase chain reaction (RT‐qPCR). (b) Cell proliferation capacity was tested by 5‐ethynyl‐2'‐deoxyuridine (EdU) assay. (c and d) The levels of cell proliferation‐associated proliferating cell nuclear antigen (PCNA) and cyclin D1 were detected in transfected lung adenocarcinoma cells by Western blot assay. (e) Flow cytometry was applied to analyze the ratio of apoptosis of transfected lung adenocarcinoma cells. (f) Wound healing assay was applied to assess cell migration ability. (g) Cell invasion ability was analyzed through performing transwell invasion assay. (h) Capillary‐like network formation assay was applied to analyze the tube formation ability of lung adenocarcinoma cells. *p < 0.05.
MiR‐330‐5p interacts with NOVA2 in lung adenocarcinoma cells
NOVA2 was a candidate target of miR‐330‐5p predicted by the starBase database (Figure 5a). The overexpression of miR‐330‐5p significantly reduced the luciferase intensity of wild‐type luciferase reporter plasmid (WT‐NOVA2 3' UTR) rather than mutant plasmid (MUT‐NOVA2 3' UTR) (Figure 5b), suggesting that NOVA2 was a target of miR‐330‐5p in lung adenocarcinoma cells. RT‐qPCR assay confirmed the high silencing efficiency of miR‐330‐5p inhibitor in lung adenocarcinoma cells (Figure 5c). NOVA2 was negatively regulated by miR‐330‐5p in H522 and A549 cells (Figure 5d). NOVA2 was upregulated in lung adenocarcinoma tissues in mRNA and protein levels (Figure 5e,f). Compared with the BEAS‐2B cell line, circ_0004140 was highly expressed in H522 and A549 cell lines (Figure 5g). Additionally, Western blot assay displayed that circ_0004140 silencing reduced NOVA2 protein expression, and its level was largely rescued in si‐circ_0004140 and miR‐330‐5p inhibitor cotransfected group in lung adenocarcinoma cells (Figure 5h–j), suggesting that circ_0004140 positively regulated NOVA2 expression through sponging miR‐330‐5p in lung adenocarcinoma cells. Overall, NOVA2 was a target of miR‐330‐5p in lung adenocarcinoma cells.
FIGURE 5.
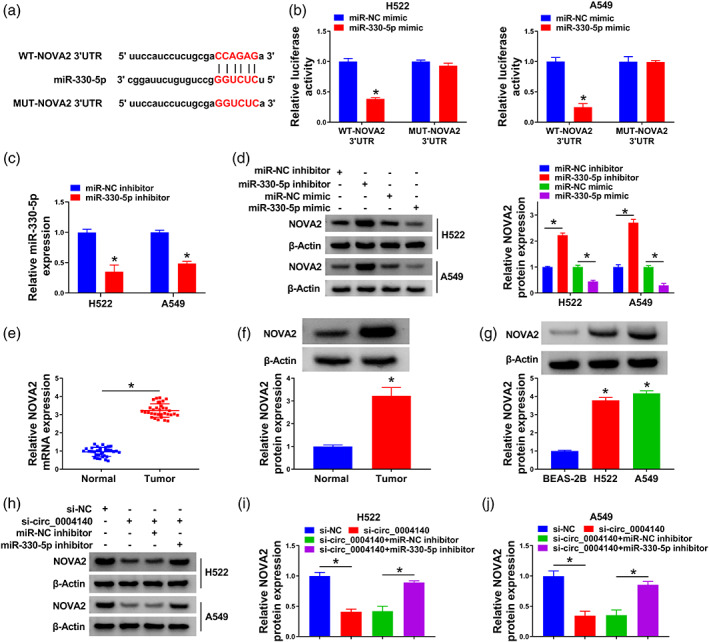
MiR‐330‐5p interacts with NOVA2 in lung adenocarcinoma cells. (a) The potential binding sites of miR‐330‐5p in the 3' UTR of NOVA2 were predicted by the starBase database. (b) The interaction between miR‐330‐5p and NOVA2 was tested by dual‐luciferase reporter assay. (c) The level of miR‐330‐5p was measured in H522 and A549 cells transfected with miR‐NC inhibitor or miR‐330‐5p inhibitor by real‐time quantitative polymerase chain reaction (RT‐qPCR). (d) The regulatory relation between miR‐330‐5p and NOVA2 was analyzed in lung adenocarcinoma cells. The protein level of NOVA2 was measured in H522 and A549 cells transfected with miR‐NC inhibitor, miR‐330‐5p inhibitor, miR‐NC mimic or miR‐330‐5p mimic by Western blot assay. (e and f) The RNA level and protein level of NOVA2 in lung adenocarcinoma tissues and adjacent normal tissues were analyzed by RT‐qPCR and Western blot assay, respectively. (g) The level of NOVA2 protein was detected in BEAS‐2B cells and two lung adenocarcinoma cell lines by Western blot assay. (a–c) The protein expression of NOVA2 was detected in H522 and A549 cells transfected with si‐NC, si‐circ_0004140, si‐circ_0004140 + miR‐NC inhibitor or si‐circ_0004140 + miR‐330‐5p inhibitor by Western blot assay. *p < 0.05.
NOVA2 overexpression largely overturns miR‐330‐5p overexpression‐mediated effects in lung adenocarcinoma cells
At first, our data exhibited that NOVA2 knockdown might repress lung adenocarcinoma cell proliferation and angiogenesis ability, and induce cell apoptosis, on the contrary, NOVA2 overexpression might promote lung adenocarcinoma cell proliferation and angiogenesis ability, and hinder cell apoptosis (Figure S2). Furthermore, miR‐330‐5p overexpression decreased NOVA2 protein expression in lung adenocarcinoma cells, and we rescued the level of NOVA2 through cotransfecting cells with miR‐330‐5p mimic and pcDNA‐NOVA2 (Figure 6a). MiR‐330‐5p overexpression hampered lung adenocarcinoma cell proliferation ability, and the introduction of NOVA2 plasmid largely rescued the malignant properties (Figure 6b). MiR‐330‐5p overexpression reduced the levels of PCNA and cyclin D1 in lung adenocarcinoma cells, and the addition of NOVA2 plasmid largely upregulated the levels of PCNA and cyclin D1 (Figure 6c,d). Cell apoptosis was triggered by miR‐330‐5p overexpression, and the overexpression of NOVA2 restrained miR‐330‐5p‐induced apoptosis in lung adenocarcinoma cells (Figure 6e). Through conducting wound healing assay and transwell assay, we found that miR‐330‐5p overexpression‐induced suppressive effect on the migration and invasion ability of lung adenocarcinoma cells was largely overturned by the overexpression of NOVA2 (Figure 6f,g). Beyond that, the angiogenesis ability of lung adenocarcinoma cells was restrained by miR‐330‐5p overexpression, and cell tube formation ability was largely rescued with the introduction of pcDNA‐NOVA2 plasmid (Figure 6h). In addition, our data exhibited that the upregulation of NOVA2 might partially abolish si‐circ_0004140‐ mediated cell proliferation and angiogenesis ability inhibition, and cell apoptosis promotion in lung adenocarcinoma cells (Figure S3). Taken together, miR‐330‐5p overexpression restrained lung adenocarcinoma progression partly through down‐regulating NOVA2.
FIGURE 6.
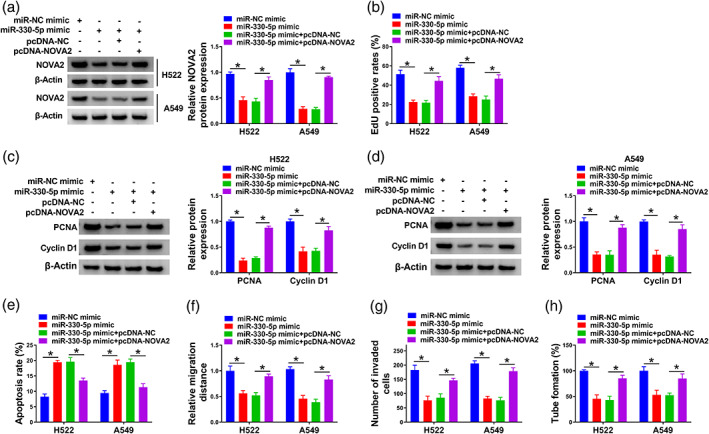
NOVA2 overexpression largely overturns miR‐330‐5p overexpression‐mediated effects in lung adenocarcinoma cells. (a–h) H522 and A549 cells were transfected with miR‐NC mimic, miR‐330‐5p mimic, miR‐330‐5p mimic + pcDNA‐NC or miR‐330‐5p mimic + pcDNA‐NOVA2. (a) Western blot assay was adopted to measure the protein level of NOVA2 in transfected lung adenocarcinoma cells. (b) The cell proliferation ability was analyzed by 5‐ethynyl‐2'‐deoxyuridine (EdU) assay. (c and d) The protein expression of proliferating cell nuclear antigen (PCNA) and cyclin D1 were analyzed via Western blot assay. (e) Flow cytometry was carried out to measure the percentage of apoptotic lung adenocarcinoma cells. (f) Cell migration ability of transfected lung adenocarcinoma cells was assessed via wound healing assay. (g) Cell invasion ability was assessed by transwell assay. (h) The tube formation ability of transfected lung adenocarcinoma cells was analyzed via capillary‐like network formation assay. *p < 0.05.
Circ_0004140 knockdown restrains tumor progression in vivo
The xenograft tumor model was utilized to analyze the in vivo role of circ_0004140. Circ_0004140 silencing markedly restrained tumor growth (Figure 7a). Tumor weight was significantly reduced in the circ_0004140‐silenced group relative to the sh‐NC group (Figure 7b). Circ_0004140 expression and NOVA2 protein level were significantly downregulated in the sh‐circ_0004140 group compared with the sh‐NC group (Figure 7c,d). The expression of miR‐330‐5p was markedly upregulated in the sh‐circ_0004140 group than that in the sh‐NC group (Figure 7c). In addition, IHC analysis verified that Ki67 (a proliferation marker) expression in the sh‐circ_0004140 group was significantly reduced compared with the sh‐NC group (Figure 7e). Besides, the Kaplan‐Meier survival curves demonstrated that fraction survival of mice with sh‐circ_0004140 had a lower survival rate than those in the sh‐NC group (Figure 7f). These findings manifested that circ_0004140 silencing suppressed tumor growth in vivo.
FIGURE 7.
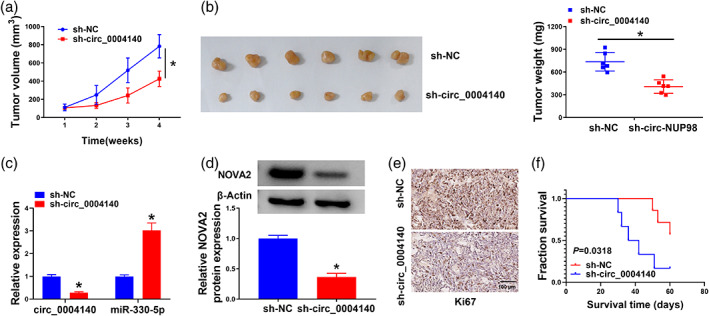
Circ_0004140 knockdown restrains tumor progression in vivo. (a) Tumor volume was monitored every week for four weeks. (b) Tumor weight was recorded at 4 week after inoculation. (c) The levels of circ_0004140 and miR‐330‐5p were analyzed in tumor tissues by real‐time quantitative polymerase chain reaction (RT‐qPCR). (d) Western blot assay was applied to measure the protein level of NOVA2 in tumor tissues. (e) Immunohistochemical staining was implemented to evaluate Ki67 content in removed tumor samples. (f) Fraction survival of mice with sh‐NC or sh‐circ_0004140 was determined by Kaplan‐Meier analysis.*p < 0.05.
DISCUSSION
Lung adenocarcinoma is a primary type of NSCLC, and circRNAs are involved in the regulation of lung adenocarcinoma progression. 1 We demonstrated that circ_0004140 aggravated lung adenocarcinoma via miR‐330‐5p/NOVA2 cascade. This study is the first to show the target relation between miR‐330‐5p and circ_0004140 or NOVA2.
Dysregulated circRNAs are associated with the nosogenesis of lung adenocarcinoma. For instance, circ‐TSPAN4 abundance was enhanced in lung adenocarcinoma, and its overexpression might aggravate progression in lung adenocarcinoma by modulating miR‐665/ZEB1 signaling. 23 Circ_0000326 expression was markedly upregulated in lung adenocarcinoma, and it contributed to lung adenocarcinoma development via sponging miR‐338‐3p. 24 Circ_0004140 level was elevated in lung adenocarcinoma, and circ_0004140 interference suppressed lung adenocarcinoma progression via the miR‐1184/CCL22. 9 Consistent with this former study, circ_0004140 abundance was found to be enhanced in lung adenocarcinoma. More importantly, circ_0004140 silencing repressed cell proliferation, migration, invasion, and angiogenesis and induced the apoptosis of lung adenocarcinoma cells. These results manifested that circ_0004140 exhibited an oncogenic role in lung adenocarcinoma.
CircRNAs act as miRNA sponges to regulate cell phenotypes in human diseases. 5 We predicted the interactions of circ_0004140‐miRNAs and identified that miR‐330‐5p was targeted by circ_0004140 in lung adenocarcinoma cells. MiR‐330‐5p was reversely modulated by circ_0004140, and it was downregulated in lung adenocarcinoma. Cui et al. demonstrated that PCAT6 contributed to malignant properties in NSCLC cells via suppressing miR‐330‐5p. 17 Bai et al. demonstrated that circ‐PRKCA aggravated NSCLC development through sponging miR‐330‐5p. 25 These studies manifested the antitumor role of miR‐330‐5p in NSCLC. Similarly, we found that miR‐330‐5p overexpression restrained the malignant behaviors of lung adenocarcinoma cells. Additionally, circ_0004140 knockdown‐induced effects were largely overturned by the silencing of miR‐330‐5p, indicating that circ_0004140 silencing suppressed lung adenocarcinoma progression partly through upregulating miR‐330‐5p.
MiR‐330‐5p‐mRNAs interactions were predicted by bioinformatic analysis, and we verified NOVA2 as a target of miR‐330‐5p in lung adenocarcinoma cells. NOVA2 played a protumor role in NSCLC. For instance, Xiao et al. found that miR‐7‐5p blocked the metastasis of NSCLC through downregulating NOVA2. 19 Tan et al. found that circ_0072088 accelerated the malignant properties of NSCLC cells through upregulating NOVA2 via sponging miR‐377‐5p. 20 Circ_0000376 contributed to the development of NSCLC by modulating miR‐1182/NOVA2 signaling. 21 Li et al. demonstrated that circ_0046263 aggravated NSCLC progression and metastasis through absorbing miR‐940 to upregulate NOVA2. 22 Here, we found that NOVA2 was reversely modulated by miR‐330‐5p in lung adenocarcinoma cells. NOVA2 was highly expressed in lung adenocarcinoma tissues and cell lines. NOVA2 overexpression largely counteracted miR‐330‐5p overexpression‐mediated effects in lung adenocarcinoma cells, demonstrating that miR‐330‐5p hampered the malignant behaviors of lung adenocarcinoma cells partly through reducing NOVA2 expression. Next, circ_0004140 was demonstrated to function as a miR‐330‐5p sponge to upregulate NOVA2 expression in lung adenocarcinoma cells.
Based on the oncogenic role of circ_0004140 in regulating cell phenotypes in vitro, we further confirmed the role of circ_0004140 in tumor growth in vivo. Circ_0004140 knockdown hampered xenografts growth in vivo, suggesting that circ_0004140 played a protumor role in vivo. In the future, more experiments need to be performed to analyze if the miR‐330‐5p/NOVA2 axis is essential for the oncogenic role of circ_0004140 in vivo.
In conclusion, circ_0004140 was highly expressed in lung adenocarcinoma tissues and cell lines. Circ_0004140 contributed to cell proliferation, migration, invasion, and angiogenesis, and restrained the apoptosis of lung adenocarcinoma cells via the miR‐330‐5p/NOVA2 axis. Our study suggests novel potential targets for lung adenocarcinoma therapy.
AUTHOR CONTRIBUTIONS
Deyun Cheng conceived, designed and revised the current study. Fan Xia analyzed the data and wrote the manuscript. Mei Xie and Jinqi He analyzed the data. All authors read and approved the final manuscript.
CONFLICT OF INTEREST STATEMENT
The authors declare that they have no conflicts of interest.
Supporting information
Figure S1. Many potential targets of circ0004140 were identified. (a) Relative levels of eight miRNAs in A549 lysates pulled down by circ0004140 probe or control probe. (b) YAP1 mRNA expression was assessed in H522 and A549 cells transfected with si‐NC or si‐circ0004140 using RT‐qPCR. *p < 0.05.
Figure S2. Effects of NOVA2 knockdown or overexpression on the proliferation, apoptosis, and angiogenesis ability in lung adenocarcinoma cells. (a–h) H522 and A549 cells were transfected with si‐NC, si‐NOVA2, pcDNA‐NC, or pcDNA‐NOVA2. (a and b) Western blot analysis of NOVA2 protein level in transfected lung adenocarcinoma cells. (c and d) The cell proliferation ability was analyzed by EdU assay. (e and f) Flow cytometry was performed to measure the percentage of apoptotic lung adenocarcinoma cells. (g and h) The tube formation ability of transfected lung adenocarcinoma cells was detected using capillary‐like network formation assay. *p<0.05.
Figure S3. Effects of circ0004140 and NOVA2 on the proliferation, apoptosis, and angiogenesis ability in lung adenocarcinoma cells. (A–D) H522 and A549 cells were transfected with si‐NC, si‐circ_0004140, si‐circ_0004140 + pcDNA‐NC, or pcDNA‐NOVA2. (a) Western blot analysis of NOVA2 protein level in transfected lung adenocarcinoma cells. (b) Cell proliferation ability was detected using EdU assay. (c) Flow cytometry was implemented to examine the percentage of apoptotic lung adenocarcinoma cells. (d) The tube formation ability of transfected lung adenocarcinoma cells was assessed using capillary‐like network formation assay. *p < 0.05.
Xia F, Xie M, He J, Cheng D. Circ_0004140 promotes lung adenocarcinoma progression by upregulating NOVA2 via sponging miR‐330‐5p. Thorac Cancer. 2023;14(35):3483–3494. 10.1111/1759-7714.15141
REFERENCES
- 1. Denisenko TV, Budkevich IN, Zhivotovsky B. Cell death‐based treatment of lung adenocarcinoma. Cell Death Dis. 2018;9:117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kuhn E, Morbini P, Cancellieri A, et al. Adenocarcinoma classification: patterns and prognosis. Pathologica. 2018;110:5–11. [PubMed] [Google Scholar]
- 3. Myers DJ, Wallen JM. Lung adenocarcinoma. In: StatPearls [Internet], Treasure Island (FL): StatPearls Publishing; 2023. [Google Scholar]
- 4. Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20:675–691. [DOI] [PubMed] [Google Scholar]
- 5. Kulcheski FR, Christoff AP, Margis R. Circular RNAs are miRNA sponges and can be used as a new class of biomarker. J Biotechnol. 2016;238:42–51. [DOI] [PubMed] [Google Scholar]
- 6. Panda AC. Circular RNAs Act as miRNA Sponges. Adv Exp Med Biol. 2018;1087:67–79. [DOI] [PubMed] [Google Scholar]
- 7. Sun Z. Circular RNA hsa_circ_0001588 promotes the malignant progression of lung adenocarcinoma by modulating miR‐524‐3p/NACC1 signaling. Life Sci. 2020;259:118157. [DOI] [PubMed] [Google Scholar]
- 8. Zuo Y, Shen W, Wang C, et al. Circular RNA Circ‐ZNF609 Promotes Lung Adenocarcinoma Proliferation by Modulating miR‐1224‐3p/ETV1 Signaling. Cancer Manag Res. 2020;12:2471–2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Liu Y, Zhang H, Zhang W, et al. circ_0004140 promotes LUAD tumor progression and immune resistance through circ_0004140/miR‐1184/CCL22 axis. Cell Death Discov. 2022;8:181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. [DOI] [PubMed] [Google Scholar]
- 11. Inamura K. Diagnostic and Therapeutic Potential of MicroRNAs in Lung Cancer. Cancers (Basel). 2017;9:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen J, Chen T, Zhu Y, et al. circPTN sponges miR‐145‐5p/miR‐330‐5p to promote proliferation and stemness in glioma. J Exp Clin Cancer Res. 2019;38:398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wang H, Liu L, Fang S. MicroRNA‐330‐5p inhibits osteosarcoma cell growth and invasion by targeting the proto‐oncogene survivin. Mol Med Rep. 2019;20:2236–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Zhou Q, Xie Y, Wang L, et al. LncRNA EWSAT1 upregulates CPEB4 via miR‐330‐5p to promote cervical cancer development. Mol Cell Biochem. 2020;471:177–188. [DOI] [PubMed] [Google Scholar]
- 15. Zhou ZF, Wei Z, Yao JC, et al. CircRNA_102179 promotes the proliferation, migration and invasion in non‐small cell lung cancer cells by regulating miR‐330‐5p/HMGB3 axis. Pathol Res Pract. 2020;216:153144. [DOI] [PubMed] [Google Scholar]
- 16. Wang Y, Xu R, Zhang D, et al. Circ‐ZKSCAN1 regulates FAM83A expression and inactivates MAPK signaling by targeting miR‐330‐5p to promote non‐small cell lung cancer progression. Transl Lung Cancer Res. 2019;8:862–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cui LH, Xu HR, Yang W, et al. lncRNA PCAT6 promotes non‐small cell lung cancer cell proliferation, migration and invasion through regulating miR‐330‐5p. Onco Targets Ther. 2018;11:7715–7724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Yano M, Hayakawa‐Yano Y, Mele A, et al. Nova2 regulates neuronal migration through an RNA switch in disabled‐1 signaling. Neuron. 2010;66:848–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Xiao H. MiR‐7‐5p suppresses tumor metastasis of non‐small cell lung cancer by targeting NOVA2. Cell Mol Biol Lett. 2019;24:60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Tan Z, Cao F, Jia B, et al. Circ_0072088 promotes the development of non‐small cell lung cancer via the miR‐377‐5p/NOVA2 axis. Thorac Cancer. 2020;11:2224–2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Li C, Liu H, Niu Q, et al. Circ_0000376, a Novel circRNA, Promotes the Progression of Non‐Small Cell Lung Cancer Through Regulating the miR‐1182/NOVA2 Network. Cancer Manag Res. 2020;12:7635–7647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Li G, Zhao C, Zhang H, et al. Hsa_circ_0046263 Drives the Carcinogenesis and Metastasis of Non‐Small Cell Lung Cancer Through the Promotion of NOVA2 by Absorbing Mir‐940 as a Molecular Sponge. Cancer Manag Res. 2020;12:12779–12790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ying X, Zhu J, Zhang Y. Circular RNA circ‐TSPAN4 promotes lung adenocarcinoma metastasis by upregulating ZEB1 via sponging miR‐665. Mol Genet Genomic Med. 2019;7:e991. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 24. Xu Y, Yu J, Huang Z, et al. Circular RNA hsa_circ_0000326 acts as a miR‐338‐3p sponge to facilitate lung adenocarcinoma progression. J Exp Clin Cancer Res. 2020;39:57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bai L, Peng X, Sun R. Knockdown of circPRKCA Restrained Cell Growth, Migration, and Invasion of NSCLC Cells Both in vitro and in vivo via Regulating miR‐330‐5p/PDK1/AKT Pathway. Cancer Manag Res. 2020;12:9125–9137. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Many potential targets of circ0004140 were identified. (a) Relative levels of eight miRNAs in A549 lysates pulled down by circ0004140 probe or control probe. (b) YAP1 mRNA expression was assessed in H522 and A549 cells transfected with si‐NC or si‐circ0004140 using RT‐qPCR. *p < 0.05.
Figure S2. Effects of NOVA2 knockdown or overexpression on the proliferation, apoptosis, and angiogenesis ability in lung adenocarcinoma cells. (a–h) H522 and A549 cells were transfected with si‐NC, si‐NOVA2, pcDNA‐NC, or pcDNA‐NOVA2. (a and b) Western blot analysis of NOVA2 protein level in transfected lung adenocarcinoma cells. (c and d) The cell proliferation ability was analyzed by EdU assay. (e and f) Flow cytometry was performed to measure the percentage of apoptotic lung adenocarcinoma cells. (g and h) The tube formation ability of transfected lung adenocarcinoma cells was detected using capillary‐like network formation assay. *p<0.05.
Figure S3. Effects of circ0004140 and NOVA2 on the proliferation, apoptosis, and angiogenesis ability in lung adenocarcinoma cells. (A–D) H522 and A549 cells were transfected with si‐NC, si‐circ_0004140, si‐circ_0004140 + pcDNA‐NC, or pcDNA‐NOVA2. (a) Western blot analysis of NOVA2 protein level in transfected lung adenocarcinoma cells. (b) Cell proliferation ability was detected using EdU assay. (c) Flow cytometry was implemented to examine the percentage of apoptotic lung adenocarcinoma cells. (d) The tube formation ability of transfected lung adenocarcinoma cells was assessed using capillary‐like network formation assay. *p < 0.05.


