Abstract
Strategies to target specific protein cysteines are critical to covalent probe and drug discovery. 3-Bromo-4,5-dihydroisoxazole (BDHI) is a natural product-inspired, synthetically accessible electrophilic moiety that has previously been shown to react with nucleophilic cysteines in the active site of purified enzymes. Here, we define the global cysteine reactivity and selectivity of a set of BDHI-functionalized chemical fragments using competitive chemoproteomic profiling methods. Our study demonstrates that BDHIs capably engage reactive cysteine residues in the human proteome and the selectivity landscape of cysteines liganded by BDHI is distinct from that of haloacetamide electrophiles. Given its tempered reactivity, BDHIs showed restricted, selective engagement with proteins driven by interactions between a tunable binding element and the complementary protein sites. We validate that BDHI forms covalent conjugates with glutathione S-transferase Pi (GSTP1) and peptidyl-prolyl cis−trans isomerase NIMA-interacting 1 (PIN1), emerging anticancer targets. BDHI electrophile was further exploited in Bruton’s tyrosine kinase (BTK) inhibitor design using a single-step late-stage installation of the warhead onto acrylamide-containing compounds. Together, this study expands the spectrum of optimizable chemical tools for covalent ligand discovery and highlights the utility of 3-bromo-4,5-dihydroisoxazole as a cysteine-reactive electrophile.
Graphical Abstract
INTRODUCTION
Covalent ligands that combine chemical reactivity and molecular recognition present a powerful strategy for chemical probes and drug development.1 Cysteine-directed covalent warheads are already a component of many FDA-approved drugs, and advanced chemoproteomic studies have uncovered thousands of additional cysteines that can be covalently modified by small molecules.2−5 However, these sites have been identified using a relatively limited number of electrophilic chemotypes, most notably haloacetamides, nitriles, and acrylamides.6 Thus, there remains an unmet need for novel electrophilic scaffolds whose reactivity and selectivity can berationally tuned to address protein targets of interest.
Electrophilic natural products have proven a remarkable source of chemical innovation and antipathogenic/tumorigenic activity.7,8 One unique naturally occurring electrophile is the 3-chloro-4,5-dihydroisoxazole heterocycle, a five-membered ring found in the natural product acivicin. Acivicin was originally isolated from the fermentation broth of Streptomyces sviceus and exhibits anti-inflammatory, antibacterial, antiparasitic, and anticancer activities.9,10 Mechanistic studies have found that the 3-halo-4,5-dihydroisoxazole scaffold of acivicin can react with cysteine residues in enzymes such as glutamine amidotransferase to form a covalent bond by displacement of the chlorine atom (Figure 1A).11−16 It has been hypothesized that protein binding-associated activation and stabilization precede the covalent modification of acivicin’s targets and confer specificity to this heterocyclic warhead.11,17 Inspired by acivicin’s intriguing chemical and biological properties, the 3-halo-4,5-dihydroisoxazole warhead has been explored to covalently target a few specific enzymes.16 However, besides a single effort to identify direct targets of acivicin itself,18 the potential of the 3-halo-4,5-dihydroisoxazole scaffold to globally address unique targets in the human proteome has not been assessed. Given its power to broadly and rapidly map reactive amino acid sites and corresponding ligands directly in complex biological systems, we envisioned that the chemoproteomic approach would provide a foundation for the utility of this unique electrophile in covalent inhibitor design.
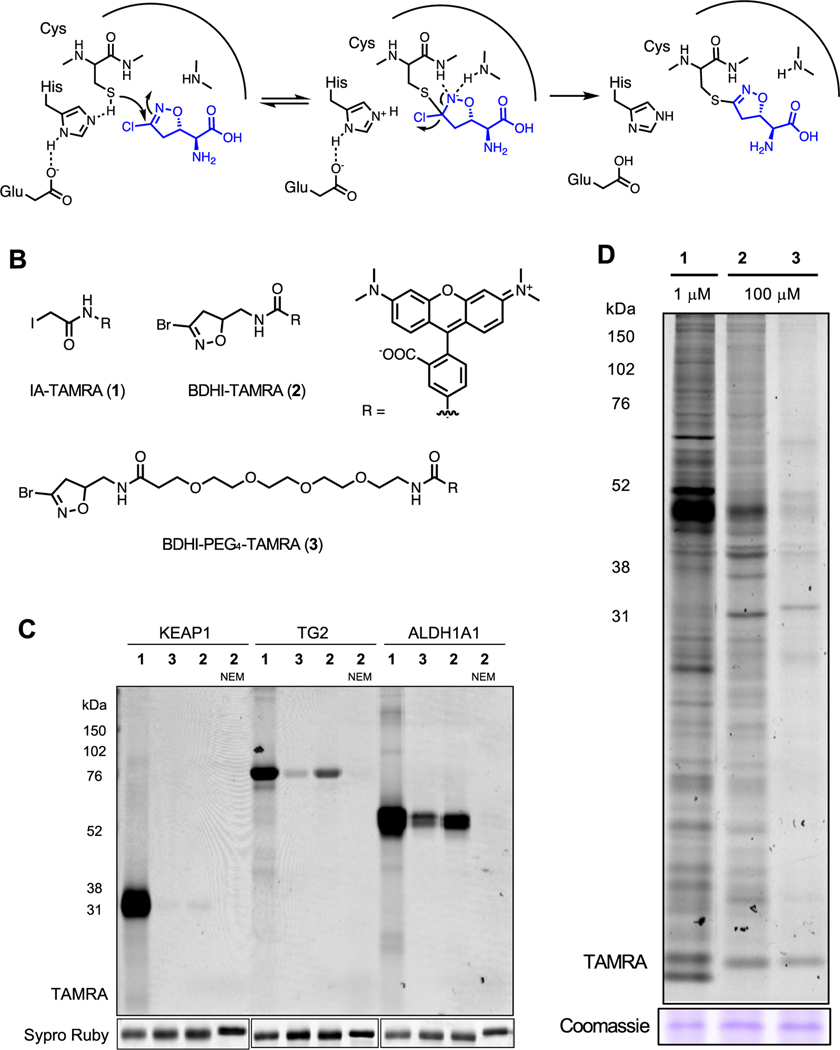
Figure 1.
BDHI electrophile reacts with proteins in the human proteome. (A) Covalent inactivation mechanism of acivicin for glutamine amidotransferase. (B) Structures of fluorescent-BDHI probes. (C) In-gel fluorescence scanning of recombinant KEAP1, TG2, and ALDH1A1 labeled with BDHI-TAMRA (2) and BDHI-PEG4-TAMRA (3) probes (1 μM, 2 h). (D) In-gel fluorescence image depicting protein bands labeled with fluorescent-BDHI probes in the human proteome. Soluble proteome from Jurkat cells was treated with the indicated probes for 2 h, followed by SDS-PAGE and in-gel fluorescence scanning.
Here, we set out to define and manipulate the reactivity and selectivity of this natural product-inspired electrophilic scaffold using a combined rational design and proteome-wide covalent ligand screening approach. First, we explore the attenuated reactivity of a 3-bromo-4,5-dihydroisoxazole (BDHI) scaffold toward human proteins. Analysis of this electrophile’s quantum chemical properties enables the design of chemoproteomic probes with orthogonal glutathione reactivity, solution stability, and proteomic reactivity. Applying this knowledge, we synthesize a small panel of BDHI analogues and analyze their properties using competitive chemoproteomic profiling approach. This reveals the ability of selected fragments to drive site-specific engagement of a distinct subset of cysteines in the human proteome, whose functional significance could be validated via biochemical assays and structural modeling. Overall, our studies demonstrate the potential for natural product-inspired electrophiles to empower inverse drug design approaches and offer a strategy for further applying the BDHI scaffold to advance ligand discovery efforts.
RESULTS AND DISCUSSION
Selective Cysteine Reactivity of the BDHI Electrophile.
We selected the 3-bromo-4,5-dihydroisoxazole (BDHI) moiety as a subject for our studies because it is slightly more reactive than the chloride-substituted counterpart but is not expected to alter target preferences.19 To benchmark the reactivity of the BDHI electrophile, we compared a general fluorescent cysteine labeling reagent (1) to two fluorescent-BDHI probes (2 and 3; Figure 1B). As recombinant protein targets, we chose Kelch-like ECH-associated protein 1 (KEAP1), a protein that forms adducts with many electrophilic small molecules and contains 24 cysteine residues, as well as transglutaminase 2 (TG2) and aldehyde dehydrogenase 1A1 (ALDH1A1), two enzymes that have been previously reported to interact with the BDHI electrophile, which contain 20 and 11 cysteine residues, respectively.13,19 Proteins were individually incubated with 1 μM 1−3 for 2 h at 37 °C and analyzed by SDS-polyacrylamide gel electrophoresis (SDS-PAGE). In-gel fluorescence scanning indicated that iodoacetamide (IA) probe 1 capably reacts with all three proteins, whereas BDHI probes 2 and 3 only demonstrate visible labeling of TG2 and ALDH1A1 (Figure 1C). The hydrophobic BDHI-tetramethylrhodamine (TAMRA) analogue 2 exhibited more intense protein labeling than BDHI-PEG4-TAMRA 3. Of note, N-ethylmaleimide (NEM) pretreatment completely abolished the protein labeling by probe 2, consistent with cysteine-directed labeling. When incubated with proteomic lysates of Jurkat cells (a human T cell lymphoma line), BDHI-TAMRA probes 2 and 3 formed conjugates with numerous proteins, with a few showing preferential engagement relative to IA-TAMRA 1 (Figure 1D). BDHI probes labeled proteins to a much lesser extent compared to promiscuous iodoacetamide probe and required a relatively higher concentration, likely due to low or slow reactivity of the BDHI electrophile toward target proteins with a nonoptimal binding element. It is also possible that multiple cysteines present in each protein are modified with a more reactive IA electrophile, whereas the BDHI electrophile only reacts more selectively with cysteine(s) in a defined binding pocket. These studies provide the first human proteome-wide characterization of the natural product-inspired BDHI scaffold and led us to further explore its tunable selectivity.
Computational Analysis Guides the Tuning of BDHI Reactivity.
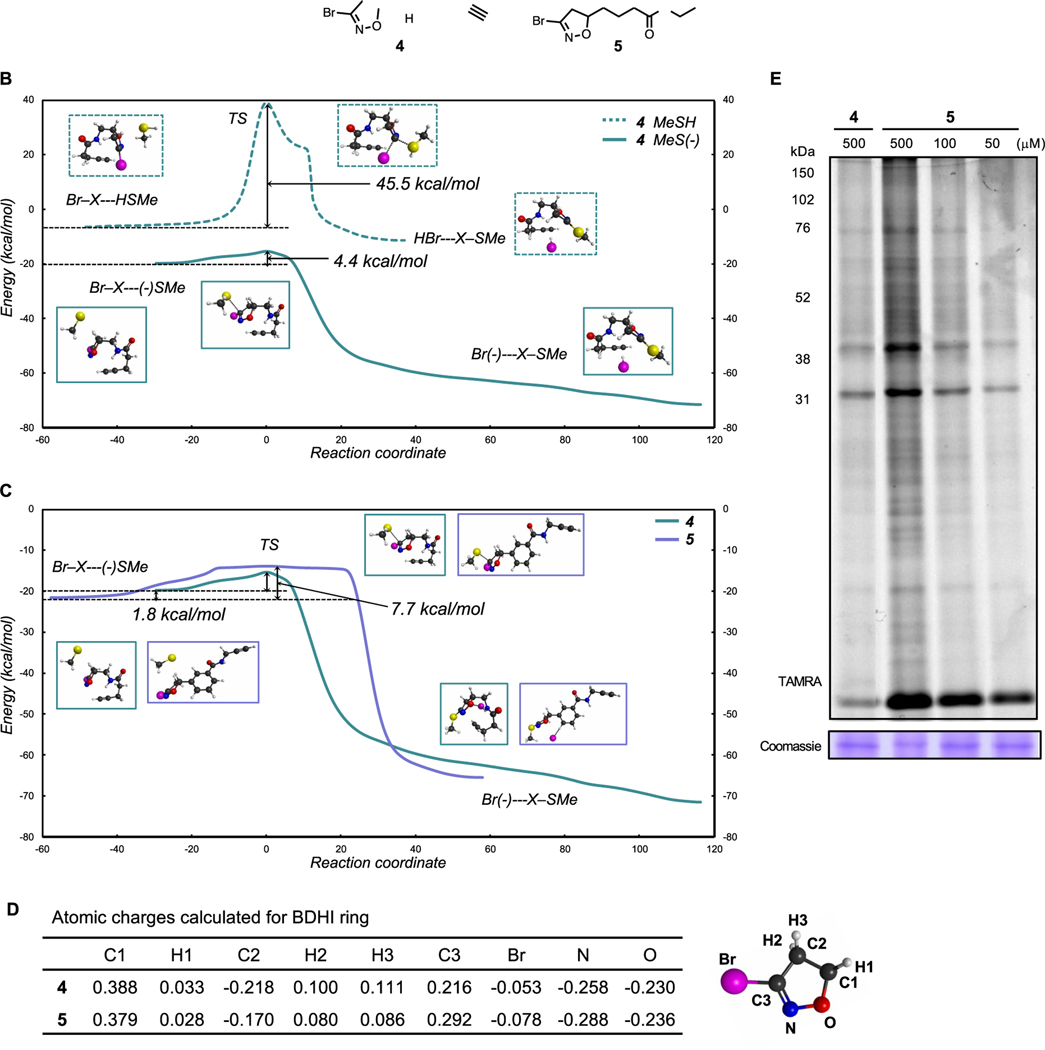
The five-membered heterocyclic ring of BDHI is unique relative to smaller electrophiles (e.g., haloacetamides) in that it is of similar size to a pharmacophore and has the potential to contribute to molecular recognition of targets itself. Acivicin inhibits glutamine amidotransferase by placing a 3-chloro-4,5-dihydroisoxazole directly adjacent to an amino acid backbone, which can influence both the electrophilicity and molecular recognition of protein surfaces by its covalent warhead. Considering this, we hypothesized that the computationally informed structure−function analysis may be able to similarly inform the interplay of these properties in the BDHI electrophile. For these studies, we designed two structurally distinct BDHI probes, 4 and 5 (Figure 2A). Compound 4 contains an α amido-BDHI, which is structurally identical to the warhead used in our fluorescent probes. Compound 5 replaces the amide with a substituted phenyl ring, which confers greater lipophilicity and electron-withdrawing properties. Each compound was also designed to incorporate a bioorthogonal alkyne handle to enable experimental analyses of protein labeling. Density functional theory (DFT) calculations were used to predict the susceptibility of these compounds toward nucleophilic attack by thiols and understand possible changes in reactivity resulting from differential substitution. Analyzing the reaction energy profiles (relative to separated reactants) of each compound with a simple methanethiol, we found that there are large transition-state barriers (45.5 kcal/mol for compound 4 and 44.3 kcal/mol for compound 5) for reaction with MeSH (Figure 2B). This is in good agreement with the observation that BDHIs do not readily react with glutathione (GSH) at pH 7.4 where the equilibrium lies on the thiol side given the pKa of GSH (9.17).20 On the other hand, nucleophilic addition of the thiolate anion (MeS−) is feasible and likely spontaneous since full reaction pathways (including transition states) lie below energies of separated reactants (Figure 2C). Notably, initial binding energies to MeS− are computed to be around 20 kcal/mol, thus very favorable, and energy barriers relative to the initial reactant complexes are low (ΔE‡ = 4.4 and 7.7 kcal/mol for 4 and 5, respectively). These results implicate thiol deprotonation as the likely rate-limiting step, and accordingly, we speculate that BDHIs may preferentially react with hyper-reactive cysteines with reduced pKa (heightened nucleophilicity).21 Overall energy profiles are very similar between 4 and 5; however, 5 has slightly stronger binding energy for the initial reactant complex with MeS− (by 1.8 kcal/mol).
Figure 2.
BDHI-alkyne probes. (A) Structures of BDHI-alkyne probes 4 and 5. (B, C) Computed reaction energy profiles of the BDHI electrophile with thiol nucleophiles using DFT calculations (relative to separated reactants). (B) Comparison of the reactions between MeSH and MeS− with 4 and (C) comparison of the reactions between 4 and 5 with MeS−. (D) Charge analysis of indicated atoms in the BDHI ring of compounds 4 and 5. (E) In-gel fluorescence image depicting protein bands labeled with BDHI-alkyne probes in the human proteome. Soluble proteome from Jurkat cells was treated with the indicated probes for 2 h and subsequently subjected to CuAAC with TAMRA-N3, followed by SDS-PAGE and in-gel fluorescence scanning.
Next, we calculated the atomic charges of the BDHI ring of the two probes by fitting to the electrostatic potentials. Importantly, the N−O bond of the dihydroisoxazole (DHI) ring is highly electronegative and contributes to the polarization of the C3−C2 bond, and the C3 imino carbon features a positive electrostatic profile (numbering shown in Figure 2D). The electronegative potential of the DHI ring may potentiate binding to oppositely charged protein pockets and ultimately allow the stabilization of a tetrahedral intermediate formed upon nucleophilic attack. Comparison of 4 and 5 observed an overall similar electrostatic potential, whereas an obvious difference was detected at the C3 carbon. Compound 5 exhibited a higher electropositive potential at the C3 compared to 4, suggesting slightly heightened susceptibility to nucleophilic attack. This is consistent with our analysis of the MeS−/BDHI reaction coordinate. To experimentally assess the proteome reactivity of these two probes, we synthesized each and treated Jurkat proteomes with BDHI-alkyne probes 4 and 5 followed by conjugation with a fluorescent tag using a copper-catalyzed azide−alkyne cycloaddition (CuAAC) chemistry. Each compound displayed efficient labeling of multiple proteins (Figure 2E). However, consistent with its predicted heightened reactivity and lipophilicity, probe 5 showed a broader labeling profile than 4. These studies demonstrate a computationally guided strategy for manipulating the reactivity of a natural product-inspired electrophile and provide access to a set of reporters for assessing the protein reactivity of two orthogonal BDHI chemotypes.
Proteome Reactivity of BDHI-Containing Fragments.
Based on our analysis of fluorescent and biorthogonal BDHI probes, we envisioned that BDHI warheads could be used to produce small molecules capable of selectively reacting with distinct subsets of the human proteome. To explore this notion, we installed the BDHI electrophile onto a set of chemical fragments (so-called “scout fragments”) previously shown to drive covalent protein engagement with different electronic and steric properties2 and assessed their proteomic reactivity. BDHI fragments were synthesized using 1,3-dipolar cycloaddition reactions with a stable precursor dibromoformaldoxime to afford a racemic mixture of 5-substituted 3-bromo-isoxazole derivatives (Scheme S1 and Figure 3A).22,23 This synthetic procedure also allows us to directly convert acrylamide-bearing covalent ligands to corresponding BDHI analogues (6−10). Analysis of the electrostatic potential surface of this small panel of BDHI analogues again identified slight differences in the electrostatic potential profile at the BDHI ring, with more substantial differences at the C1 carbon as well as at the substituted aromatic moieties (Figure S1 and Table S1). This suggests that 5-substitution may contribute not only to the steric effect (by altering how binding moieties present the BDHI electrophile) but also to the electronic effect for binding.
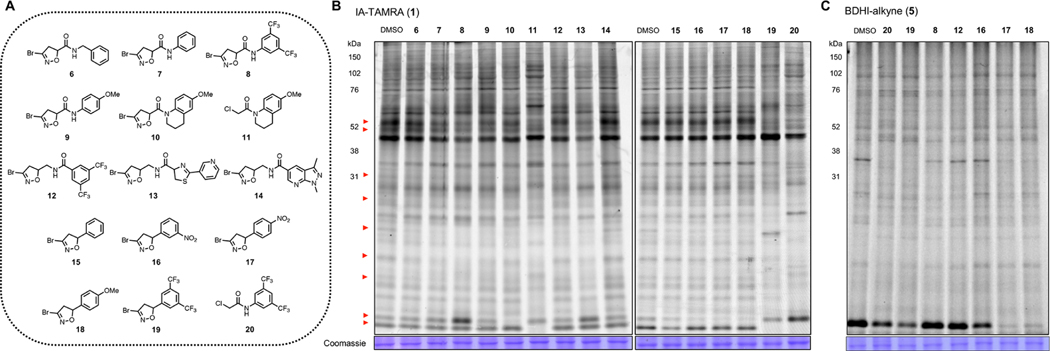
Figure 3.
BDHI electrophile-containing fragments competitively block the probe labeling of proteins. (A) Structures of BDHI-functionalized fragments. (B) Initial competitive analysis of the proteomic reactivity of fragments using an IA-TAMRA. Soluble proteome from Jurkat cells was treated with the indicated fragments (500 μM each) for 4 h, followed by labeling with IA-TAMRA (1 μM, 1 h) and analysis by SDS-PAGE and ingel fluorescence scanning. Red arrows highlight protein bands that showed diminished IA-TAMRA labeling. (C) BDHI ligands also competitively block the labeling of proteins by BDHI-alkyne 5 (100 μM, 2 h, subsequently conjugated with N3-TAMRA).
To quickly interrogate the cysteine reactivity of these compounds, we used a competitive gel-based screening, incubating Jurkat cell lysates with each compound followed by labeling of cysteine with IA-TAMRA. In this approach, compound pretreatment that competitively blocks probe labeling results in reduced fluorescent labeling of individual proteins. Given their low molecular weight and racemic nature, we screened them at a single high concentration (500 μM) similar to those used in fragment-based ligand discovery.24 Visual inspection of protein bands (Figure 3B, red arrows, left to right) revealed a discrete subset of proteins whose IA-TAMRA labeling was diminished in intensity upon pretreatment of BDHI analogues. However, compared to α-chloroacetamide (CA)-containing compounds 11 (KB02) or 20 (KB03),2 BDHI analogues exhibited more restricted and selective blockade of IA-TAMRA−protein interactions. Overall, IA-TAMRA labeling was altered most substantially by pretreatment with 8 and 13. In contrast, compound 14 did not show any visible evidence of competition, demonstrating the high dependence of BDHI reactivity on cognate fragment scaffold. When compared to fragments that contain a mildly reactive acrylamide electrophile (21−23; Figure S2), slightly enhanced proteomic reactivity was observed from the BDHI-containing fragments. Blockade of IA-TAMRA−protein interactions by 5-phenyl derivatives (15−19) was overall less pronounced. Considering the possibility that this reflects differential labeling kinetics of IA-TAMRA vs BDHI electrophiles, we also assessed competitive labeling of proteins by BDHI-TAMRA probe 2 (Figure S3 ) and alkyne probe 5 (Figure 3C). This experiment confirmed competitive labeling of many proteins by 5-phenyl analogues, with 18 showing the broadest level of proteome-wide reactivity. As we noticed more pronounced competition with compounds 7−9 (N-phenyl amide substitution), additional alkyne-functionalized analogues (24−26; Figure S4A) were synthesized and directly examined for their ability to label proteins. Compound 25 efficiently labeled proteins with enhanced reactivity compared to 5, both in cell lysates and in cells (Figure S4B,C). DFT calculations indicated that 25 has stronger binding energy for the initial reactant complex with MeS− compared to 5 (Figure S4D).
Supporting the attenuated reactivity of BDHI electrophile toward sulfhydryl groups in solution and a requirement for molecular recognition in BDHI labeling, incubating three BDHI analogues (8, 10, and 18) with GSH at pH 7.4 did not show any formation of a covalent adduct, whereas CA-containing compounds 11 and 20 produced more than 50% of corresponding adducts within 4 h of incubation (Figure S5A). To our surprise, BDHI 19 with a bis(trifluoromethyl)phenyl group was found to be unstable in phosphate buffer (Figure S5B). As such, we excluded compound 19 from further studies. Together, these data suggest that BDHI is a mildly reactive electrophile that does not readily react with GSH but can act as a covalent protein modifier upon interaction within protein binding pockets (useful as a latent electrophile) and its proteomic reactivity and selectivity can be largely modulated by fragments appended to the warhead.
Competitive Reactivity Profiling Defines a Subset of Proteomic Cysteines Targeted by BDHI Analogues.
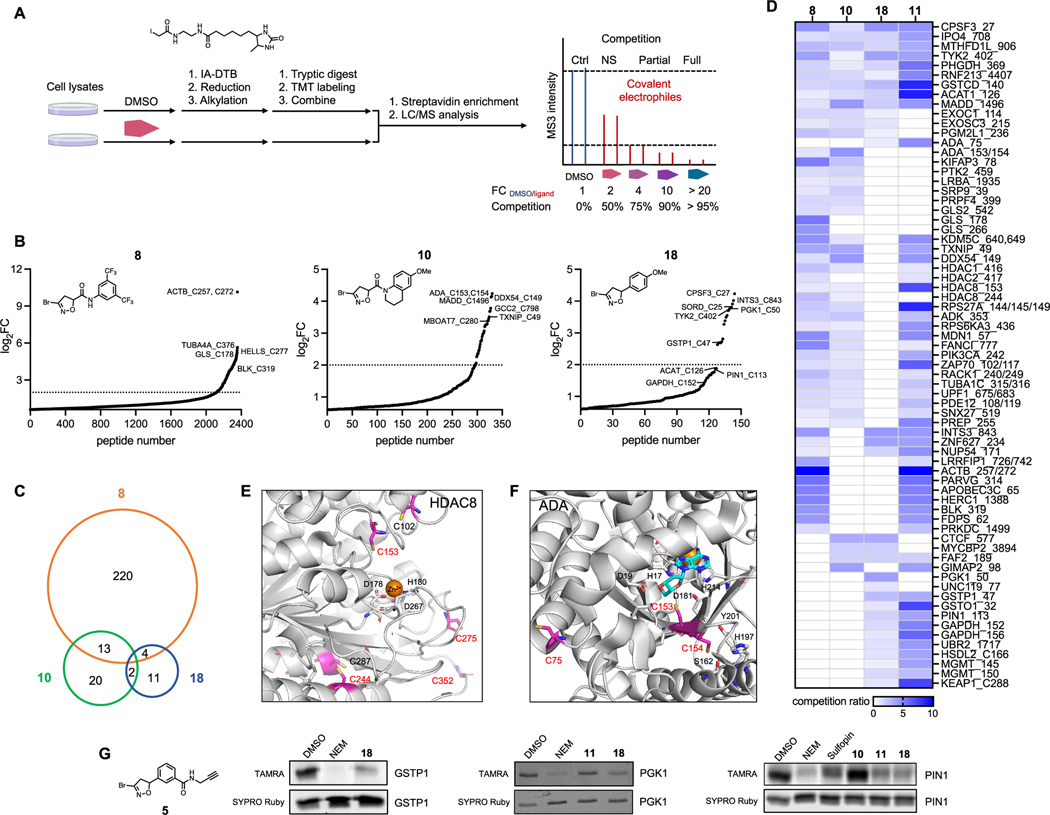
Based on our observation of competitive labeling using IA-TAMRA, we next performed mass spectrometry (MS)-based chemoproteomic profiling to globally map the protein targets of our BDHI probes. In brief, cell lysates were treated with either vehicle (DMSO) or a BDHI-containing ligand (Figure 4A). Samples were then labeled with a cysteine-reactive IA-desthiobiotin (DTB) probe,4 followed by digestion and tandem mass tag (TMT) labeling. After combining samples, labeled peptides were enriched by streptavidin beads, eluted, and subjected to liquid chromatography-mass spectrometry/MS (LC-MS/MS) analysis. Cysteines that show a reduction in TMT signals for BDHI-treated samples compared to DMSO control (calculated as a competition ratio) are considered candidate covalent targets. We selected IA−DTB as the chemoproteomic probe (as opposed to 4 or 5) due to its broad validation and to avoid potential technical issues resulting from poor ionization and fragmentation of the BDHI-adduct.4 Based on gel-based data, we selected compounds 8, 10, 13, 14, and 18 for analysis and compared their reactivity to promiscuous control compounds KB02 (11) and KB03 (20). Experiments were carried out by incubating compounds (500 μM, 4 h, in triplicate) in Jurkat whole cell lysates. Cysteine occupancy was determined by log2 fold change in the abundance of TMT reporter ion signals (Table S2). Using this approach, we identified 237, 35, 9, 0, and 17 liganded cysteine sites that showed >75% reduction in TMT signals (log2 FC > 2, p value <0.05) after incubation with BDHI 8, 10, 13, 14, and 18, respectively (Figures 4B and S6A). This number is significantly less than that observed for haloacetamide 11 (2209 sites) or 20 (3306 sites), again reflecting the more restricted blockade of IA probe−protein interactions by BDHI derivatives. Consistent with gel-based analyses, IA−DTB profiling revealed 8 and 14 to be the most and least reactive analogues, respectively. BDHIs were found to target functional proteins including both enzymes and nonenzymes. A large fraction of noncatalytic proteins identified were proteins that are involved in mediating protein−protein or protein−nucleic acid interactions. We found 21 BDHI-liganded sites among the top 150 most reactive cysteine residues identified from a previous study,25 including the active site cysteines of glutathione S-transferase omega-1 (GSTO1) and methylated DNA protein cysteine methyltransferase (MGMT) (Figure S6B). Moreover, compounds were observed to engage discrete sets of both overlapping and distinct cysteines (Figures 4C,D and S6C,D). A brief survey of these targets follows:
Figure 4.
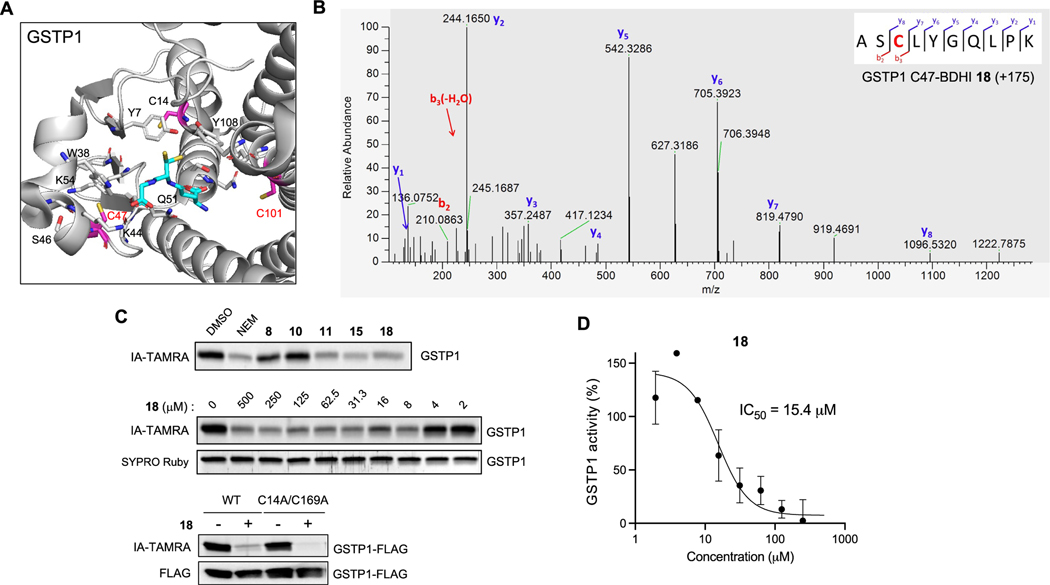
Competitive MS-based reactivity profiling defines cysteines liganded by BDHI analogues. (A) Workflow for competitive measurement of cysteine ligandability with fragment electrophiles using IA−DTB probe. Steps include the treatment of cell lysate with compounds (500 μM, 4 h), treatment with IA−DTB probe (500 μM, 2 h), trypsin digestion, TMT labeling, combination, avidin enrichment for biotinylated cysteine-containing peptides, and analysis of competition ratio. (B) Structures and competition ratios (log2 FC values) from Jurkat cell proteome treated with BDHI 8, 10, and 18. (C) Venn diagram representation of the number of cysteine sites significantly liganded (log2 FC > 2) by BDHIs. Results were obtained by comparing the site overlap at a given competition ratio threshold for each ligand. (D) Heatmap of competition ratios for representative cysteines and fragments. (E) Crystal structure of HDAC8 (PDB: 2V5W). The zinc binding domain is shown along with reactive cysteines (depicted in magenta). (F) Crystal structure of substrate-bound, human ADA (PDB: 3IAR) with possible cysteine sites for covalent modification. Adenosine is depicted in cyan. (G) Competition binding assay between BDHI 18 and BDHI-alkyne 5. Recombinantly expressed and purified PGK1, GSTP1, and PIN1 were treated with each compound (500 μM, 4 h), labeled with BDHI 5 (500 μM, 2 h), and conjugated with TAMRA-N3 for in-gel fluorescence scanning.
Cysteines that were liganded by both haloacetamide 11 and a subset of BDHI analogues include C27 of cleavage and polyadenylation specificity factor subunit 3 (CPSF3), C708 of importin-4 (IPO4), C906 of mitochondrial monofunctional C1-tetrahydrofolate synthase (MTHFD1L, also known as formyltetrahydrofolate synthetase), and C140 of glutathione S-transferase C-terminal domain-containing protein (GSTCD). However, we found that while the CA scout fragments 11 and 20 target multiple cysteines in each protein, BDHI derivatives appear to selectively engage only one cysteine. For example, BDHI 18 showed preferential engagement with the active site C126 among the four reactive cysteines (C119, C126, C196, and C413) found in the mitochondrial acetyl-CoA acetyltransferase (ACAT1, Figure S7A). On the other hand, for glyceraldehyde 3-phosphate dehydrogenase (GAPDH), BDHI 18 engaged with both C152 and C156 residues (Figure S7B).
Among the liganded sites that were uniquely observed with BDHIs were C178 and C266 of kidney-type glutaminase (GLS) and C244 of histone deacetylase 8 (HDAC8) with 8, C153/154 of adenosine deaminase (ADA) with 10, and C50 of phosphoglycerate kinase 1 (PGK1) with 18. It is noteworthy that acivicin as a glutamine mimetic has been reported to bind to the active site of GLS and other glutamine-binding enzymes.12,26,27
HDAC8, which belongs to the human histone deacetylase family consisting of 11 isozymes, is a validated target for T cell lymphoma.28 In addition, pharmacological inhibition of HDAC8 has been demonstrated to enhance antitumor immunity and efficacy of immunotherapy.29 The enzymatic activity of HDAC8 is regulated by a reversible thiol/disulfide redox switch involving C102 and C153 residues (Figure 4E).30 Three additional pairs of cysteines (C125/C131, C244/C287, and C275/C352) are in proximity that could also potentially form further disulfide bonds. C244, which is unique to HDAC8, is one of the most buried cysteines with the highest calculated pKa value.31 It is thus surprising that this cysteine with theoretically low reactivity showed ligandability with BDHI compound 8. Interestingly, a screening of maleimide analogues appended with the 3,5-bis(trifluoromethyl)phenyl group (similar to 8) has shown to label C244 and C275 of HDAC8 and inhibit the enzyme activity.32 Further validation and investigation will be required to understand the role of C244 in ligand binding and modulation of HDAC8 activity.
ADA plays an important role in purine metabolism by catalyzing the hydrolytic deamination of adenosine to inosine. It is highly expressed in T lymphocytes and known to regulate T cell coactivation through its interaction with CD26 (DPP4) at the cell surface.33,34 While covalent modification of C75 by electrophilic small molecules has been reported to allosterically inhibit the enzymatic activity of ADA and lead to antiproliferation of lymphocytic cells,35 there was no covalent modifier identified to target C153 or C154 that is located close to the active site of ADA (Figure 4F).
PGK1 is the first ATP-generating enzyme in the glycolytic pathway associated with one-carbon metabolism and cellular redox regulation. PGK1 is involved in shaping the inflamed tumor microenvironment and mediating interaction between tumor metabolism and immunoediting. Among the seven cysteines found in PGK1 (Figure S7C), C50 has been reported to undergo S-sulfinylation (−SO2H) during oxidative stress.36 Under hypoxic condition that triggers endogenous H2O2 production, PGK1 is translocated into the mitochondria.37 Whether the modification of this cysteine has any impact on mediating mitochondrial translocation and/or kinase activity of PGK1 remains to be investigated.
To provide orthogonal validation for target interactions, we employed BDHI-alkyne probe 5. When incubated with the recombinant protein followed by a click reaction with TAMRA−azide, compound 5 showed detectable labeling of PGK1, GSTP1, and PIN1, which in each case was blocked by pretreatment with the cognate BDHI fragment 18 (Figures 4G and S8). Overall, these data provide evidence that the BDHI electrophile, when coupled to a suitable binding element, has the potential to site-specifically target cysteines. Our analysis also highlights a small set of cysteines that show exclusive engagement with the BDHI electrophile.
Functional Engagement of Proteins by the BDHI Fragment.
Next, we sought to define the functional effects of covalent adduction by the BDHI electrophile. For these studies, we prioritized targets based on their therapeutic relevance, availability of structures in the Protein Data Bank, and overall abundance, which lends further confidence to chemoproteomic identifications. This led us to focus on glutathione S-transferase Pi (GSTP1) and peptidyl-prolyl cis−trans isomerase NIMA-interacting 1 (PIN1). GSTP1 is the most prevalent cytosolic glutathione transferases, which catalyzes the conjugation of glutathione to electrophilic compounds as a cellular detoxification process.38 GSTP1 is also involved in regulating cellular redox state and intracellular signal transduction.39 Owing to its overexpression in a variety of malignant cells and confirmed role in promoting tumorigenesis, GSTP1 inhibitors have emerged as promising cancer therapeutic agents.40−42 Among the four cysteine residues found in GSTP1, C47 and C101 have each been demonstrated to be solvent-accessible, highly reactive, and targeted by irreversible inhibitors (Figure 5A).43 Specifically, the chemical modification of C47, a residue located near the active site, has been shown to cause loss of enzyme activity.44,45 To confirm the site of modification by the BDHI compound, recombinant GSTP1 was incubated with 18 at 37 °C for 4 h and subjected to trypsin digestion and LC-MS/MS analysis. All data were searched for a differential modification mass of 175.06 (covalent adduct formation through the Br replacement of BDHI 18). This led to the identification of two cysteine residues, the active site residue C47 and C101, as modified by 18 in GSTP1 (Figures 5B and S9). The reason why C101 was not initially identified as one of the 18-liganded sites from the competition experiment is because IA−DTB labeled only C47 in GSTP1 when using 500 μM in Jurkat cell lysates. We confirmed that BDHI 18 and the related 5-phenyl-substituted analogue 15, but not 8 or 10, inhibit the labeling of GSTP1 by IA-TAMRA (Figure 5C), consistent with the competition observed in MS-based profiling experiments. When we mutated the other two cysteines to alanine (C14A/C169A; Figure S10), pretreatment with 18 led to a complete loss of IA-TAMRA labeling. Consequently, we found that BDHI 18 inhibits GSTP1 enzyme activity in vitro with an IC50 of 15 μM, determined by monitoring the transfer of GSH to 1-bromo-2,4-dinitrobenzene (BDNB) (Figure 5D). To further validate and understand this interaction, we obtained multiple crystal structures from GSTP1 crystals soaked with compound 18. While the position of 18 was not unambiguously determinable, structures showed a loss of electron density in the α2 helix (residues 35−50) and an opening of the interface between the two monomers upon treatment with 18 (Figure S11 and Table S3). The loss of electron density was increased with fragment concentration and soaking time. This observation is consistent with the fragment interacting at the C47 in the α2 helix and possibly with C101 at the dimer interface. Similar structural changes have been observed in GSTP1 with compounds that bind C47 and/or C101 in the absence of GSH.46−48 Mass spectrometric analysis of crystals soaked with 18 alone or back-soaked with GSH indicated that a majority of the protein in the crystals is bound to one or two molecules of 18 (Table S4), again consistent with these effects being driven by ligand binding.
Figure 5.
Validation of GSTP1 as the BDHI-reactive protein. (A) Crystal structure of GSTP1 (PDB: 6LLX). Reactive cysteine sites (in magenta) are shown along with the binding of glutathione depicted in cyan. (B) Annotated MS2 fragmentation spectra analysis of GSTP1 liganded with BDHI 18 at Cys47, highlighted in red. (C) Competitive and concentration-dependent inhibition of IA-TAMRA labeling of GSTP1 by 18. GSTP1 was preincubated with each compound (500 μM, 4 h), labeled with IA-TAMRA (1 μM, 1 h), and subsequently analyzed by SDS-PAGE and in-gel fluorescence. (D) Inhibition of GSTP1 activity determined by monitoring the transfer of GSH to 1-bromo-2,4-dinitrobenzene.
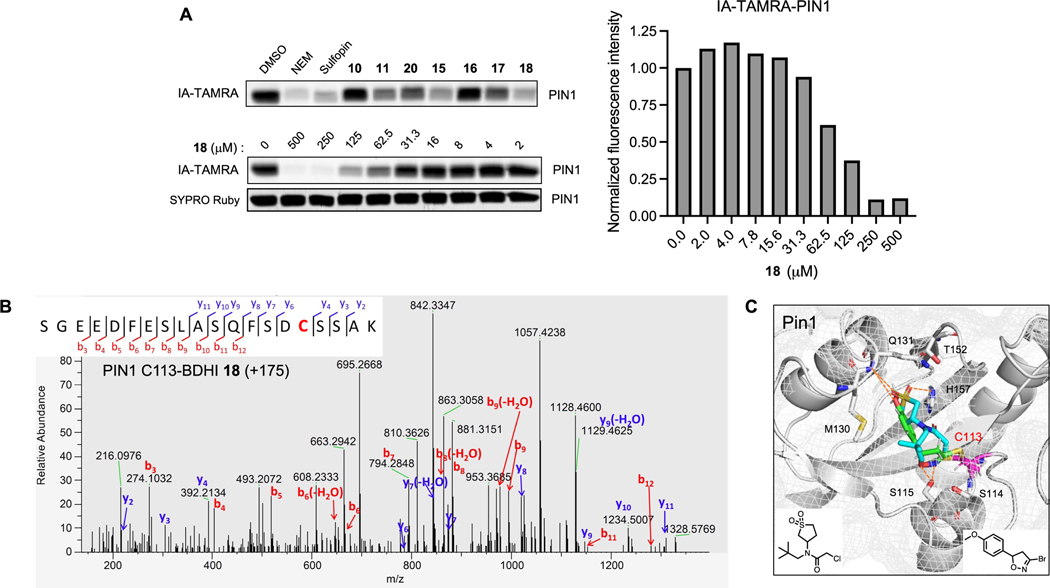
As a second target, we analyzed the binding of BDHI 18 to PIN1. PIN1 is a human peptidyl-prolyl cis−trans isomerase (PPIase) that is overactivated in numerous tumor types and whose aberrant activation has been associated with tumorigenesis.49,50 The PIN1 active site contains a nucleophilic cysteine residue (C113) that can be targeted by covalent inhibitors. These molecules have been shown to exhibit antiproliferative effects in various cancer cell lines including neuroblastoma and pancreatic ductal adenocarcinoma.51,52 Using a gel-based assay with recombinant protein, we found that BDHI 18 inhibits PIN1 labeling by IA-TAMRA in a dose-dependent manner (Figure 6A). Compound 18 also inhibited the catalytic activity of PIN1 at the tested concentrations (1, 10 μM) after 14 h of incubation at 4 °C, as measured using a chymotrypsin-coupled peptidyl-prolyl isomerization assay53 (PPIase assay; Figure S12). To further confirm that BDHI 18 site-specifically engages PIN1, we performed LC-MS/MS analysis and validated the catalytic residue Cys113 as the site of labeling (Figure 6B). In order to better understand the structural basis for PIN1 engagement, we employed in silico modeling to predict the potential binding mode of BDHI 18 to PIN1. Comparing the lowest energy binding pose of 18 to that of sulfopin,52 an inhibitor of PIN1 that contains haloacetamide warhead, we found that the BDHI warhead can covalently engage C113 with binding driven by hydrogen bond interactions of the DHI ring with neighboring S115 (Figure 6C). In this pose, the 4-methoxyphenyl group of 18 occupies the hydrophobic binding pocket surrounded by M130, Q131, T152, and H157 in a manner near identical to the sulfolane ring of sulfopin while also making a hydrogen bond interaction with the backbone amide of Q131. These data confirm that BDHI electrophile targets and covalently modifies cysteine residues in the allosteric and catalytic sites of GSTP1 and PIN1. Although further optimization is needed to improve the potency of these compounds, our study demonstrates that BDHI compounds have the potential for the development of novel covalent inhibitors of GSTP1 and PIN1, emerging anticancer targets.
Figure 6.
Validation of PIN1 as the BDHI-reactive protein. (A) Competitive and concentration-dependent inhibition of IA-TAMRA labeling of PIN1 by 18. Recombinant PIN1 was preincubated with each compound (500 μM, 4 h), labeled with IA-TAMRA (1 μM, 1 h), and subsequently analyzed by SDS-PAGE and in-gel fluorescence. (B) Annotated MS2 fragmentation spectra analysis of PIN1 liganded with BDHI 18 at the catalytic active site Cys113, highlighted in red. (C) Docking prediction of 18 binding to PIN1 (PDB: 7EKV). The predicted binding pose of 18 (green) is superimposed with the crystal structure of PIN1 with a covalently bound inhibitor sulfopin (cyan) (PDB: 6VAJ).
BDHI-Based BTK/BLK Inhibitors.
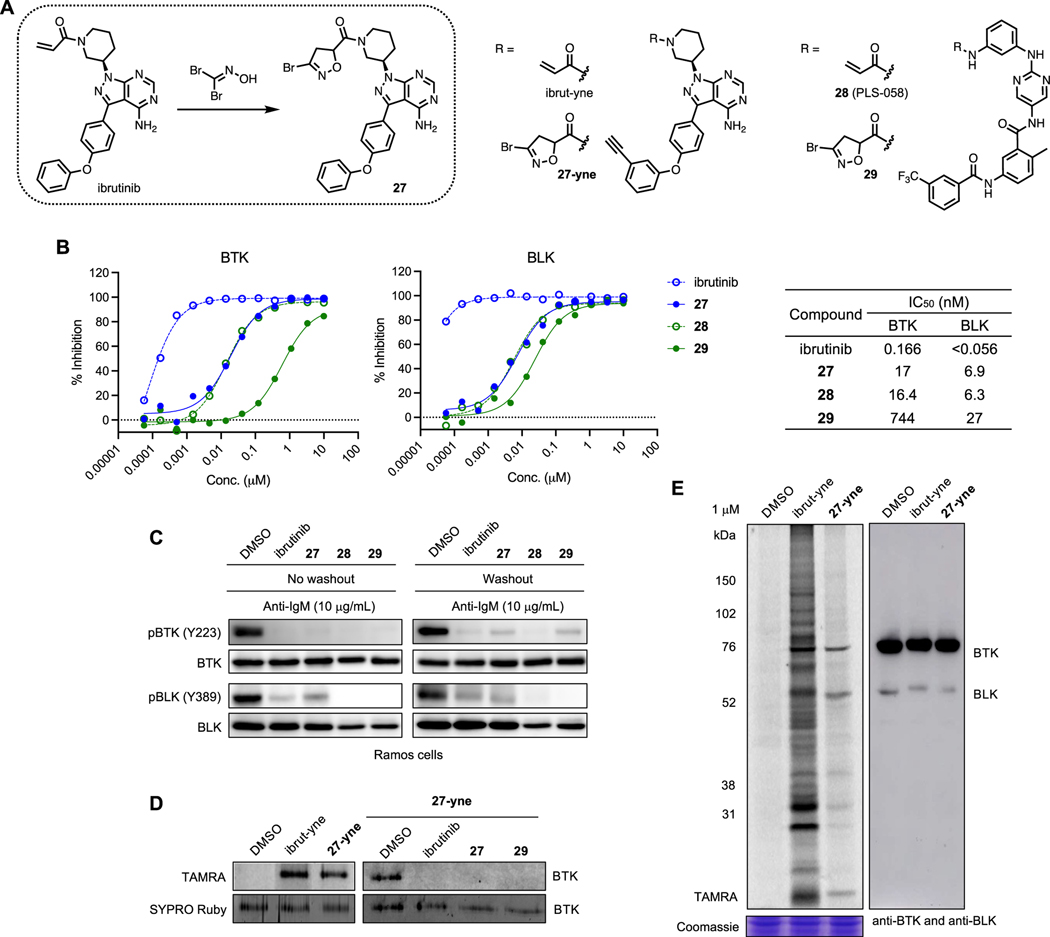
As Cys319 of B-lymphoid tyrosine kinase (BLK) is one of the BDHI-liganded sites identified by compound 8 (Figure 4B,D), we also sought to demonstrate the utility of BDHI warhead in targeted covalent inhibitor design for protein tyrosine kinases and investigate how the BDHI warhead performs on a known inhibitor scaffold. Primarily expressed in B-lineage cells, BLK plays a role in B-cell receptor signaling and cell development.54 BLK is one of 11 kinases in the human kinome including Bruton’s tyrosine kinase (BTK), IL-2-inducible T cell kinase (ITK), and EGFR family kinases that possesses a cysteine at the identical position (Cys319 in BLK and Cys481 in BTK), and many irreversible BTK inhibitors (e.g., ibrutinib) also target BLK.55,56 Since we can directly convert an acrylamide-bearing compound to a corresponding BDHI analogue using a single-step cycloaddition reaction, we selected two covalent kinase inhibitors ibrutinib57 and compound 28 (PLS-058)58 that are known to inhibit both BTK and BLK and prepared aliphatic and aromatic BDHI amides 27 and 29 (Figure 7A). To examine their inhibitory activity, we conducted in vitro kinase assay (Figure 7B). Although not as potent as acrylamides, both 27 and 29 strongly inhibited the kinase activity of BTK (IC50 = 17 and 744 nM, respectively) and BLK (IC50 = 6.9 and 27 nM, respectively). Given their racemic nature, this decrease in potency was not entirely surprising to us. In cells, all of these compounds potently inhibited autophosphorylation of BTK (Tyr223) and BLK (Tyr389) at 1 μM after 4 h of incubation (Figure 7C). Compounds 27 and 29 mostly maintained their inhibitory effects in cells even after 2 h of washout with fresh media, suggesting that it is likely through irreversible binding. To assess covalent engagement and selectivity, we further synthesized an alkyne analogue of BDHI 27 (27-yne) and carried out proteomic labeling experiments. 27-yne efficiently reacted with BTK at 1 μM, which was completely blocked by pretreatment with 10 μM of ibrutinib, 27, or 29 (Figure 7D). Additionally, in situ proteomic labeling in Ramos cells suggested that probe 27-yne remains relatively more selective than ibrut-yne for the protein at the appropriate molecular weight of BTK when both probes were used at 1 μM for 24 h although its labeling efficiency was slightly lower (Figure 7E). We performed docking study to model the potential binding mode of BDHI 27 to BTK (Figure S13). Although the lowest energy binding pose indicated that 27 can fit in the binding site quite similarly to ibrutinib59 with (R)-conformation being more compatible with covalent bond formation, we hypothesize that it is possible to orient the BDHI warhead better toward C481 by adopting more flexible liker in the place of pyrimidine ring and improve the covalent modification efficiency based on the molecular architecture of ibrutinib. In the future, it will be interesting to explore other chemotypes of BTK inhibitors or other tyrosine kinase inhibitors to gain more insight into how the proteome reactivity and selectivity could be altered by the installation of BDHI electrophile.
Figure 7.
BDHI-based BTK/BLK inhibitors. (A) Structures of acrylamide- and BDHI-based inhibitors. (B) In vitro kinase inhibitory activity against BTK and BLK. (C) Inhibition of BTK/BLK phosphorylation in Ramos cells. Cells were incubated with inhibitors at 1 μM for 4 h (for washout experiments, cells were washed and resuspended in fresh medium for 2 h), followed by activation with anti-IgM for 10 min. (D) Competitive BTK labeling. Recombinant BTK was incubated with DMSO or compound (10 μM, 4 h) and labeled with 27-yne (1 μM, 4 h). TAMRA-N3 was conjugated onto labeled protein via CuAAC prior to SDS-PAGE. (E) In-gel fluorescence scanning image for labeling of proteins in whole Ramos cells by ibrut-yne and BDHI 27-yne (1 μM, 24 h). Anti-BTK/anti-BLK Western blotting (right panel) was used to determine protein expression levels and relative position on a gel.
CONCLUSIONS
Covalent modification of proteins by small-molecule electrophiles has been widely validated in drug discovery, protein engineering, imaging, and chemical proteomic applications. However, there remains a need for novel reactive functionalities that possess selectivity suitable for ligand discovery and pharmacological optimization. In this study, we used a computationally guided approach to design and characterize a library of covalent fragments functionalized with a natural product-inspired 3-bromo-4,5-dihydroxazole (BDHI) electrophile. In vitro studies as well as proteome-wide competitive profiling experiments revealed that BDHIs engage a restricted subset of reactive cysteine residues in the human proteome. BDHI was found to possess tempered chemical reactivity relative to haloacetamide electrophiles as it does not readily react with GSH. Our data implicate the identity of the reversible binding element, the composition and geometry of the complementary protein binding sites, and stabilization of the covalent intermediate as critical factors driving covalent cysteine labeling by this chemotype. Our studies also demonstrated the ability of BDHI-containing molecules to functionally regulate protein activity, for example, via engagement of PIN1 and GSTP1. BDHI warhead was further exploited to develop BTK inhibitors using a late-stage installation onto the acrylamide-bearing covalent ligands. Finally, we highlight some limitations of our study and future work that is being performed to address them. First, the focus of our initial study was on the design and reactivity of this natural product-inspired electrophile rather than biological screening. However, in preliminary studies, we have found that BDHI 18 can inhibit surface expression of activation markers (CD25 and CD69) and reduce cytokine secretion (IFN-γ, IL-2, and IL-6) from primary human T-cells while maintaining cell viability (Figure S14). In the future, we anticipate that the chemoproteomic probes developed here will enable in situ analysis of target engagement by 18 as well as other biologically active BDHI molecules. Second, given their low molecular weight, the potencies of the molecules characterized in this study are relatively modest (high micromolar range) compared to many covalent small molecules, which can limit the applicability and increase concern about off-target binding. Thus, their medicinal chemistry optimization, especially on the recognition element, is the subject of ongoing research. Alternatively, substituting BDHI as warhead in known covalent drugs may provide a strategy to understand the pharmacological properties of this electrophile as well as the interplay between noncovalent and covalent interactions as we have shown with the example of BTK inhibitors. A unique property of the BDHI electrophile not explored in this study that differs from the natural product is the presence of a stereocenter, which will likely influence the reactivity for different protein binding sites. We anticipate the development of synthetic/purification routes to enantiomerically pure BDHIs may provide a facile strategy not only for optimizing ligands for potency and selectivity but also for differentiating new ligandable sites in proteins. Related to this, acivicin has been reported to form a covalent bond with an active site threonine residue in Escherichia coli γ-glutamyltranspeptidase (GGT).12 While our studies indicated that BDHI does not produce corresponding adducts when reacted with Boc-Ser-OMe or Boc-Lys-OMe in solution (at pH 7.4) and BDHI labeling of proteins validated here is cysteine-directed, in the future it will be interesting to assess whether tighter-binding BDHIs may modify other amino acid targets. Overall, our studies demonstrate how natural products, computation, and chemoproteomic profiling can augment electrophile design, providing a foundation for inhibitor discovery and optimization.
Supplementary Material
ACKNOWLEDGMENTS
The authors gratefully thank the staff members at the CCR-Frederick Biophysics Resource (BR) and Flow Cytometry Core Facility for their assistance, technical consultation, and instrument maintenance. The authors also thank Jordan Meier and Martin Schnermann for the critical reading of the manuscript. This work was supported by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, the Center for Cancer Research (ZIABC011961). This project was also funded in part under contract No. HHSN261200800001E. Part of this work was carried out on the MX1 and MX2 beamlines at the Australian Synchrotron, part of the Australian Nuclear Science and Technology Organization, and made use of the Australian Cancer Research Foundation detector on the MX2 beamline. The authors thank the beamline staff for their assistance. The authors acknowledge the use of the Mass Spectrometry and Proteomics Facility, Bio21 Institute, The University of Melbourne. Funding from the Victorian Government Operational Infrastructure Support Scheme to St Vincent’s Institute is acknowledged. MWP is a National Health and Medical Research Council (NHMRC) of Australia Investigator (APP1117183).
Footnotes
Complete contact information is available at: https://pubs.acs.org/10.1021/jacs.3c00598
The authors declare no competing financial interest.
Contributor Information
David P. Byun, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Frederick, Maryland 21702, United States
Jennifer Ritchie, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Frederick, Maryland 21702, United States.
Yejin Jung, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Frederick, Maryland 21702, United States.
Ronald Holewinski, Protein Characterization Laboratory, Frederick National Laboratory for Cancer Research, Leidos Biochemical Research, Frederick, Maryland 21702, United States.
Hong-Rae Kim, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Frederick, Maryland 21702, United States.
Ravichandra Tagirasa, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Frederick, Maryland 21702, United States.
Joseph Ivanic, Advanced Biomedical Computational Science, Frederick National Laboratory for Cancer Research, Leidos Biomedical Research, Frederick, Maryland 21702, United States.
Claire M. Weekley, Department of Biochemistry and Pharmacology, Bio21 Molecular Science and Biotechnology Institute, The University of Melbourne, Parkville, Victoria 3010, Australia
Michael W. Parker, Department of Biochemistry and Pharmacology, Bio21 Molecular Science and Biotechnology Institute, The University of Melbourne, Parkville, Victoria 3010, Australia; Australian Cancer Research Foundation Rational Drug Discovery Centre, St. Vincent’s Institute of Medical Research, Fitzroy, Victoria 3065, Australia
Thorkell Andresson, Protein Characterization Laboratory, Frederick National Laboratory for Cancer Research, Leidos Biochemical Research, Frederick, Maryland 21702, United States.
Euna Yoo, Chemical Biology Laboratory, Center for Cancer Research, National Cancer Institute, National Institutes of Health, Frederick, Maryland 21702, United States.
REFERENCES
- (1).Singh J; Petter RC; Baillie TA; Whitty A. The resurgence of covalent drugs. Nat. Rev. Drug Discovery 2011, 10, 307–317. [DOI] [PubMed] [Google Scholar]
- (2).Backus KM; Correia BE; Lum KM; Forli S; Horning BD; González-Páez GE; Chatterjee S; Lanning BR; Teijaro JR; Olson AJ; Wolan DW; Cravatt BF Proteome-wide covalent ligand discovery in native biological systems. Nature 2016, 534, 570–574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Vinogradova EV; Zhang X; Remillard D; Lazar DC; Suciu RM; Wang Y; Bianco G; Yamashita Y; Crowley VM; Schafroth MA; Yokoyama M; Konrad DB; Lum KM; Simon GM; Kemper EK; Lazear MR; Yin S; Blewett MM; Dix MM; Nguyen N; Shokhirev MN; Chin EN; Lairson LL; Melillo B; Schreiber SL; Forli S; Teijaro JR; Cravatt BF An activity-guided map of electrophile-cysteine interactions in primary human T cells. Cell 2020, 182, 1009–1026.e29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (4).Kuljanin M; Mitchell DC; Schweppe DK; Gikandi AS; Nusinow DP; Bulloch NJ; Vinogradova EV; Wilson DL; Kool ET; Mancias JD; Cravatt BF; Gygi SP Reimagining high-throughput profiling of reactive cysteines for cell-based screening of large electrophile libraries. Nat. Biotechnol. 2021, 39, 630–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Spradlin JN; Zhang E; Nomura DK Reimagining druggability using chemoproteomic platforms. Acc. Chem. Res. 2021, 54, 1801–1813. [DOI] [PubMed] [Google Scholar]
- (6).Du J; Yan X; Liu Z; Cui L; Ding P; Tan X; Li X; Zhou H; Gu Q; Xu J. cBinderDB: a covalent binding agent database. Bioinformatics 2017, 33, 1258–1260. [DOI] [PubMed] [Google Scholar]
- (7).Gersch M; Kreuzer J; Sieber SA Electrophilic natural products and their biological targets. Nat. Prod. Rep. 2012, 29, 659–682. [DOI] [PubMed] [Google Scholar]
- (8).Gehrtz P; London N. Electrophilic natural products as drug discovery tools. Trends Pharmacol. Sci. 2021, 42, 434–447. [DOI] [PubMed] [Google Scholar]
- (9).Hanka LJ; Martin DG; Neil GL A new antitumor antimetabolite, (alphaS,5S)-alpha-amino-3-chloro-4,5-dihydro-5-iso-xazoleacetic acid (NSC-163501): antimicrobial reversal studies and preliminary evaluation against L1210 mouse leukemia in vivo. Cancer Chemother. Rep. 1973, 57, 141–148. [PubMed] [Google Scholar]
- (10).Mendiola AS; Ryu JK; Bardehle S; Meyer-Franke A; Ang KK; Wilson C; Baeten KM; Hanspers K; Merlini M; Thomas S; Petersen MA; Williams A; Thomas R; Rafalski VA; Meza-Acevedo R; Tognatta R; Yan Z; Pfaff SJ; Machado MR; Bedard C; Rios Coronado PE; Jiang X; Wang J; Pleiss MA; Green AJ; Zamvil SS; Pico AR; Bruneau BG; Arkin MR; Akassoglou K. Transcriptional profiling and therapeutic targeting of oxidative stress in neuroinflammation. Nat. Immunol. 2020, 21, 513–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Chittur SV; Klem TJ; Shafer CM; Davisson VJ Mechanism for acivicin inactivation of triad glutamine amidotransferases. Biochemistry 2001, 40, 876–887. [DOI] [PubMed] [Google Scholar]
- (12).Wada K; Hiratake J; Irie M; Okada T; Yamada C; Kumagai H; Suzuki H; Fukuyama K. Crystal structures of Escherichia coli gamma-glutamyltranspeptidase in complex with azaserine and acivicin: novel mechanistic implication for inhibition by glutamine antagonists. J. Mol. Biol. 2008, 380, 361–372. [DOI] [PubMed] [Google Scholar]
- (13).Dafik L; Khosla C. Dihydroisoxazole analogs for labeling and visualization of catalytically active transglutaminase 2. Chem. Biol. 2011, 18, 58–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Klöck C; Herrera Z; Albertelli M; Khosla C. Discovery of potent and specific dihydroisoxazole inhibitors of human transglutaminase 2. J. Med. Chem. 2014, 57, 9042–9064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Ettari R; Tamborini L; Angelo IC; Grasso S; Schirmeister T; Lo Presti L; De Micheli C; Pinto A; Conti P. Development of rhodesain inhibitors with a 3-bromoisoxazoline warhead. ChemMedChem 2013, 8, 2070–2076. [DOI] [PubMed] [Google Scholar]
- (16).Pinto A; Tamborini L; Cullia G; Conti P; De Micheli C. Inspired by nature: The 3-halo-4,5-dihydroisoxazole moiety as a novel molecular warhead for the design of covalent inhibitors. ChemMedChem 2016, 11, 10–14. [DOI] [PubMed] [Google Scholar]
- (17).Miles BW; Thoden JB; Holden HM; Raushel FM Inactivation of the amidotransferase activity of carbamoyl phosphate synthetase by the antibiotic acivicin. J. Biol. Chem. 2002, 277, 4368–4373. [DOI] [PubMed] [Google Scholar]
- (18).Kreuzer J; Bach NC; Forler D; Sieber SA Target discovery of acivicin in cancer cells elucidates its mechanism of growth inhibition. Chem. Sci. 2015, 6, 237–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Orth R; Böttcher T; Sieber SA The biological targets of acivicin inspired 3-chloro- and 3-bromodihydroisoxazole scaffolds. Chem. Commun. 2010, 46, 8475–8477. [DOI] [PubMed] [Google Scholar]
- (20).Jöst C; Nitsche C; Scholz T; Roux L; Klein CD Promiscuity and selectivity in covalent enzyme inhibition: a systematic study of electrophilic fragments. J. Med. Chem. 2014, 57, 7590–7599. [DOI] [PubMed] [Google Scholar]
- (21).Weerapana E; Wang C; Simon GM; Richter F; Khare S; Dillon MB; Bachovchin DA; Mowen K; Baker D; Cravatt BF Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 2010, 468, 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Castelhano AL; Billedeau R; Pliura DH; Bonaventura BJ; Krantz A. Synthesis, chemistry, and absolute-configuration of novel transglutaminase inhibitors containing a 3-halo-4,5-dihydroisoxazole. Bioorg. Chem. 1988, 16, 335–340. [Google Scholar]
- (23).Deamici M; Magri P; Demicheli C; Cateni F; Bovara R; Carrea G; Riva S; Casalone G. Nitrile oxides in medicinal chemistry. 4. Chemoenzymatic synthesis of chiral heterocyclic-derivatives. J. Org. Chem. 1992, 57, 2825–2829. [Google Scholar]
- (24).Scott DE; Coyne AG; Hudson SA; Abell C. Fragment-based approaches in drug discovery and chemical biology. Biochemistry 2012, 51, 4990–5003. [DOI] [PubMed] [Google Scholar]
- (25).Weerapana E; Wang C; Simon GM; Richter F; Khare S; Dillon MBD; Bachovchin DA; Mowen K; Baker D; Cravatt BF Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature 2010, 468, 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Xu X; Meng Y; Li L; Xu P; Wang J; Li Z; Bian J. Overview of the development of glutaminase inhibitors: achievements and future directions. J. Med. Chem. 2019, 62, 1096–1115. [DOI] [PubMed] [Google Scholar]
- (27).Zhu M; Fang JZ; Zhang JJ; Zhang Z; Xie JH; Yu Y; Ruan JJ; Chen Z; Hou W; Yang GS; Su WK; Ruan BH Biomolecular interaction assays identified dual inhibitors of glutaminase and glutamate dehydrogenase that disrupt mitochondrial function and prevent growth of cancer cells. Anal. Chem. 2017, 89, 1689–1696. [DOI] [PubMed] [Google Scholar]
- (28).Balasubramanian S; Ramos J; Luo W; Sirisawad M; Verner E; Buggy JJ A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia 2008, 22, 1026–1034. [DOI] [PubMed] [Google Scholar]
- (29).Yang W; Feng Y; Zhou J; Cheung OK; Cao J; Wang J; Tang W; Tu Y; Xu L; Wu F; Tan Z; Sun H; Tian Y; Wong J; Lai PB-S; Chan SL; Chan AW-H; Tan PB-O; Chen Z; Sung JJ-Y; Yip KY-L; To K-F; Cheng AS-L A selective HDAC8 inhibitor potentiates antitumor immunity and efficacy of immune checkpoint blockade in hepatocellular carcinoma. Sci. Transl. Med. 2021, 13, No. eaaz6804. [DOI] [PubMed] [Google Scholar]
- (30).Jänsch N; Meyners C; Muth M; Kopranovic A; Witt O; Oehme I; Meyer-Almes F-J The enzyme activity of histone deacetylase 8 is modulated by a redox-switch. Redox Biol. 2019, 20, 60–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Jänsch N; Sugiarto WO; Muth M; Kopranovic A; Desczyk C; Ballweg M; Kirschhofer F; Brenner-Weiss G; Meyer-Almes F-J Switching the switch: Ligand induced disulfide formation in HDAC8. Chem. - Eur. J. 2020, 26, 13249–13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (32).Petri L; Abranyi-Balogh P; Timea I; Palfy G; Perczel A; Knez D; Hrast M; Gobec M; Sosic I; Nyiri K; Vertessy BG; Jansch N; Desczyk C; Meyer-Almes FJ; Ogris I; Grdadolnik SG; Iacovino LG; Binda C; Gobec S; Keserű, G. M. Assessment of tractable cysteines for covalent targeting by screening covalent fragments. ChemBioChem 2021, 22, 743–753. [DOI] [PubMed] [Google Scholar]
- (33).Pacheco R; Martinez-Navio JM; Lejeune M; Climent N; Oliva H; Gatell JM; Gallart T; Mallol J; Lluis C; Franco R. CD26, adenosine deaminase, and adenosine receptors mediate costimulatory signals in the immunological synapse. Proc. Natl. Acad. Sci. U.S.A. 2005, 102, 9583–9588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Weihofen WA; Liu J; Reutter W; Saenger W; Fan H. Crystal structure of CD26/dipeptidyl-peptidase IV in complex with adenosine deaminase reveals a highly amphiphilic interface. J. Biol. Chem. 2004, 279, 43330–43335. [DOI] [PubMed] [Google Scholar]
- (35).Zambaldo C; Vinogradova EV; Qi XT; Iaconelli J; Suciu RM; Koh M; Senkane K; Chadwick SR; Sanchez BB; Chen JS; Chatterjee AK; Liu P; Schultz PG; Cravatt BF; Bollong MJ 2-Sulfonylpyridines as tunable, cysteine-reactive electrophiles. J. Am. Chem. Soc. 2020, 142, 8972–8979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Akter S; Fu L; Jung Y; Conte ML; Lawson JR; Lowther WT; Sun R; Liu K; Yang J; Carroll KS Chemical proteomics reveals new targets of cysteine sulfinic acid reductase. Nat. Chem. Biol. 2018, 14, 995–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Li XJ; Jiang YH; Meisenhelder J; Yang WW; Hawke DH; Zheng YH; Xia Y; Aldape K; He J; Hunter T; Wang LW; Lu ZM Mitochondria-translocated PGK1 functions as a protein kinase to coordinate glycolysis and the TCA cycle in tumorigenesis. Mol. Cell 2016, 61, 705–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Nebert DW; Vasiliou V. Analysis of the glutathione S-transferase (GST) gene family. Hum. Genomics 2004, 1, No. 460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Laborde E. Glutathione transferases as mediators of signaling pathways involved in cell proliferation and cell death. Cell Death Differ. 2010, 17, 1373–1380. [DOI] [PubMed] [Google Scholar]
- (40).Ruzza P; Rosato A; Rossi CR; Floreani M; Quintieri L. Glutathione transferases as targets for cancer therapy. Anti-Cancer Agents Med. Chem. 2009, 9, 763–777. [DOI] [PubMed] [Google Scholar]
- (41).Crawford LA; Weerapana E. A tyrosine-reactive irreversible inhibitor for glutathione S-transferase Pi (GSTP1). Mol. Biosyst. 2016, 12, 1768–1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (42).Louie SM; Grossman EA; Crawford LA; Ding L; Camarda R; Huffman TR; Miyamoto DK; Goga A; Weerapana E; Nomura DK GSTP1 Is a driver of triple-negative breast cancer cell metabolism and pathogenicity. Cell Chem. Biol. 2016, 23, 567–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (43).Quesada-Soriano I; Primavera A; Casas-Solvas JM; Tellez-Sanz R; Baron C; Vargas-Berenguel A; Lo Bello M; Garcia-Fuentes L. Identifying and characterizing binding sites on the irreversible inhibition of human glutathione S-transferase P1–1 by S-thiocarbamoylation. ChemBioChem 2012, 13, 1594–1604. [DOI] [PubMed] [Google Scholar]
- (44).Grek CL; Zhang J; Manevich Y; Townsend DM; Tew KD Causes and consequences of cysteine S-glutathionylation. J. Biol. Chem. 2013, 288, 26497–26504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (45).Balchin D; Wallace L; Dirr HW S-nitrosation of glutathione transferase P1–1 Is controlled by the conformation of a dynamic active site helix. J. Biol. Chem. 2013, 288, 14973–14984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (46).Parker LJ; Bocedi A; Ascher DB; Aitken JB; Harris HH; Lo Bello M; Ricci G; Morton CJ; Parker MW Glutathione transferase P1–1 as an arsenic drug-sequestering enzyme. Protein Sci. 2017, 26, 317–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (47).De Luca A; Parker LJ; Ang WH; Rodolfo C; Gabbarini V; Hancock NC; Palone F; Mazzetti AP; Menin L; Morton CJ; Parker MW; Lo Bello M; Dyson PJ A structure-based mechanism of cisplatin resistance mediated by glutathione transferase P1–1. Proc. Natl. Acad. Sci. U.S.A. 2019, 116, 13943–13951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (48).Vega MC; Walsh SB; Mantle TJ; Coll M. The three-dimensional structure of Cys-47-modified mouse liver glutathione Stransferase P1–1. Carboxymethylation dramatically decreases the affinity for glutathione and is associated with a loss of electron density in the alphaB-310B region. J. Biol. Chem. 1998, 273, 2844–2850. [DOI] [PubMed] [Google Scholar]
- (49).Lu KP; Zhou XZ The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat. Rev. Mol. Cell Biol. 2007, 8, 904–916. [DOI] [PubMed] [Google Scholar]
- (50).Chen Y; Wu YR; Yang HY; Li XZ; Jie MM; Hu CJ; Wu YY; Yang SM; Yang YB Prolyl isomerase Pin1: a promoter of cancer and a target for therapy. Cell Death Dis. 2018, 9, No. 883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (51).Pinch BJ; Doctor ZM; Nabet B; Browne CM; Seo HS; Mohardt ML; Kozono S; Lian X; Manz TD; Chun Y; Kibe S; Zaidman D; Daitchman D; Yeoh ZC; Vangos NE; Geffken EA; Tan L; Ficarro SB; London N; Marto JA; Buratowski S; Dhe-Paganon S; Zhou XZ; Lu KP; Gray NS Identification of a potent and selective covalent Pin1 inhibitor. Nat. Chem. Biol. 2020, 16, 979–987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (52).Dubiella C; Pinch BJ; Koikawa K; Zaidman D; Poon E; Manz TD; Nabet B; He SN; Resnick E; Rogel A; Langer EM; Daniel CJ; Seo HS; Chen Y; Adelmant G; Sharifzadeh S; Ficarro SB; Jamin Y; da Costa BM; Zimmerman MW; Lian XL; Kibe S; Kozono S; Doctor ZM; Browne CM; Yang AN; Stoler-Barak L; Shah RB; Vangos NE; Geffken EA; Oren R; Koide E; Sidi S; Shulman Z; Wang C; Marto JA; Dhe-Paganon S; Look T; Zhou XZ; Lu KP; Sears RC; Chesler L; Gray NS; London N. Sulfopin is a covalent inhibitor of Pin1 that blocks Myc-driven tumors in vivo. Nat. Chem. Biol. 2021, 17, 954–963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (53).Yaffe MB; Schutkowski M; Shen M; Zhou XZ; Stukenberg PT; Rahfeld JU; Xu J; Kuang J; Kirschner MW; Fischer G; Cantley LC; Lu KP Sequence-specific and phosphorylation-dependent proline isomerization: a potential mitotic regulatory mechanism. Science 1997, 278, 1957–1960. [DOI] [PubMed] [Google Scholar]
- (54).Dymecki SM; Niederhuber JE; Desiderio SV Specific expression of a tyrosine kinase gene, blk, in B lymphoid cells. Science 1990, 247, 332–336. [DOI] [PubMed] [Google Scholar]
- (55).Lanning BR; Whitby LR; Dix MM; Douhan J; Gilbert AM; Hett EC; Johnson TO; Joslyn C; Kath JC; Niessen S; Roberts LR; Schnute ME; Wang C; Hulce JJ; Wei B; Whiteley LO; Hayward MM; Cravatt BF A road map to evaluate the proteome-wide selectivity of covalent kinase inhibitors. Nat. Chem. Biol. 2014, 10, 760–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (56).Klaeger S; Heinzlmeir S; Wilhelm M; Polzer H; Vick B; Koenig PA; Reinecke M; Ruprecht B; Petzoldt S; Meng C; Zecha J; Reiter K; Qiao H; Helm D; Koch H; Schoof M; Canevari G; Casale E; Depaolini SR; Feuchtinger A; Wu Z; Schmidt T; Rueckert L; Becker W; Huenges J; Garz AK; Gohlke BO; Zolg DP; Kayser G; Vooder T; Preissner R; Hahne H; Tonisson N; Kramer K; Gotze K; Bassermann F; Schlegl J; Ehrlich HC; Aiche S; Walch A; Greif PA; Schneider S; Felder ER; Ruland J; Medard G; Jeremias I; Spiekermann K; Kuster B. The target landscape of clinical kinase drugs. Science 2017, 358, No. eaan4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (57).Cameron F; Sanford M. Ibrutinib: first global approval. Drugs 2014, 74, 263–271. [DOI] [PubMed] [Google Scholar]
- (58).Li X; Zuo Y; Tang G; Wang Y; Zhou Y; Wang X; Guo T; Xia M; Ding N; Pan Z. Discovery of a series of 2,5-diaminopyrimidine covalent irreversible inhibitors of Bruton’s tyrosine kinase with in vivo antitumor activity. J. Med. Chem. 2014, 57, 5112–5128. [DOI] [PubMed] [Google Scholar]
- (59).Bender AT; Gardberg A; Pereira A; Johnson T; Wu Y; Grenningloh R; Head J; Morandi F; Haselmayer P; Liu-Bujalski L. Ability of Bruton’s tyrosine kinase inhibitors to sequester Y551 and prevent phosphorylation determines potency for inhibition of Fc receptor but not B-cell receptor signaling. Mol. Pharmacol. 2017, 91, 208–219. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.