Abstract
Holins are small phage-encoded cytoplasmic membrane proteins, remarkable for their ability to make membranes permeable in a temporally regulated manner. The purification of S105, the λ holin, and one of the two products of gene S is described. Because the wild-type S105 holin could be only partially purified from membrane extracts by ion-exchange chromatography, an oligohistidine tag was added internally to the S105 sequence for use in immobilized metal affinity chromatography. An acceptable site for the tag was found between residues 94 and 95 in the highly charged C-terminal domain of S. This allele, designated S105H94, had normal lysis timing under physiological expression conditions. The S105H94 protein was overproduced, purified, and characterized by circular dichroism spectroscopy, which revealed approximately 40% alpha-helix conformation, consistent with the presence of two transmembrane helices. The purified protein was then used to achieve release of fluorescent dye loaded in liposomes in vitro, whereas protein from an isogenic construct carrying an S mutation known to abolish hole formation was inactive in this assay. These results suggest that S is a bitopic membrane protein capable of forming aqueous holes in bilayers.
Bacteriophage holins are small membrane proteins required for host cell lysis. For most phages, a bacteriolytic activity, or endolysin, is elaborated in the cytoplasm during the vegetative phase. Endolysins with a variety of enzyme activities have been reported, including true lysozymes (17, 20), transglycosylases (3), endopeptidases (24), and amidases (15, 22). In general, endolysins have no secretory signal sequence and thus accumulate as fully folded enzymes in the cytoplasm (46). The holin functions to effect release of the endolysin across the cytoplasmic membrane at a precisely scheduled time. There appears to be no specific interaction between the holin and endolysin, because unrelated endolysins with different enzymatic activities can act with the same holin to effect lysis (7, 13, 25, 33, 42). More than 60 different genes have been identified as holins by function or postulated to be holins by sequence analysis (38, 46, 47).
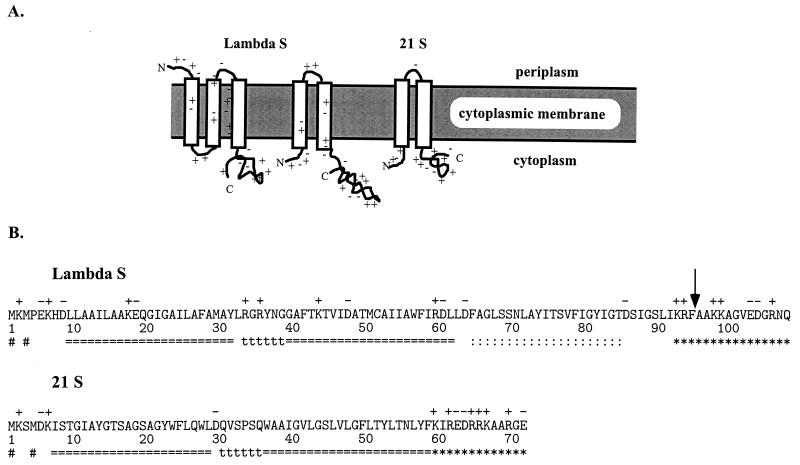
On the basis of primary sequence, holins appear to fall into two size classes: class I holins, the prototype for which is the protein S from phage λ, are 90 residues or more in length; class II holins, the prototype for which is the S protein from the lambdoid phage 21, are usually less than 75 residues (Fig. 1A) (7, 46, 47). Membrane topology seems obvious for the class II holins, which have two putative transmembrane domains (TMDs) and almost certainly are oriented with the N and C termini in the cytoplasm (Fig. 1B) (46, 47). The topology of S as the prototype class I is not settled, because there is a third uncharged although relatively hydrophilic domain which could be a transmembrane alpha-helix (Fig. 1B). Analysis of the distribution of positively charged residues according to the von Heijne “positive-inside” rule favors the three-domain model, with the N terminus outside (Fig. 1B) (31, 44).
FIG. 1.
(A) Topological models for class I and II holins. Class I holins may have three TMDs, but class II holins are limited by their size to two TMDs. In both cases, the longer form (λ S107 and 21 S71) is depicted. The C and N termini and charges are indicated. (B) Amino acid sequences of the class I holin of λ and the class II holin of 21. Charged residues are denoted above the sequence, while putative TMDs (===), turn regions (ttt), and highly charged C-terminal regions (∗∗∗) are indicated below the sequence. The ambiguous uncharged domain in λ S (::::) is indicated. The # signs indicate the dual starts demonstrated for λ S (5, 31) and suggested for 21 S (7). The vertical arrow indicates the site of the inserted G2H6G2 sequence in the S105H94 protein (see text).
Genetic analysis of S has been fruitful. Because S is small and lethal, it has been possible to select for a large number of missense mutations. During a screen for dominant mutations, it was found that some alleles with changes in the 5′ flanking region of S had dominant character (31). This led to the identification of a secondary structure, called sdi (for structure-directed initiation), which directs translational initiation events not only to codon Met1 but also to the Met3 codon (Fig. 1A and 2). S thus produces two distinct proteins, called S107 and S105, reflecting the number of amino acid residues in the products resulting from initiation at codons 1 and 3, respectively (5). Remarkably, the two proteins have opposing functions in vivo, with the shorter product S105 acting as the lethal lysis effector, whereas the S107 protein functions as an inhibitor of S105 function (30, 31). The inhibitory function of S107 was shown to be due to the positively charged residue at codon 2 (4). Thus, it was apparent that one level of the temporal regulation of S-mediated lysis was the partition of S expression between S105 and S107 products. This “dual-start” mode of regulation appears to be conserved in lambdoid and other holins, including both class I and class II holins (6, 47). The conservation of this motif implies that the functional regulation of the two classes of holins is the same. This supports the notion that the N termini of both proteins are cytoplasmically disposed, which in turn requires that λ S have two TMDs like the phage 21 class II holin (Fig. 1B).
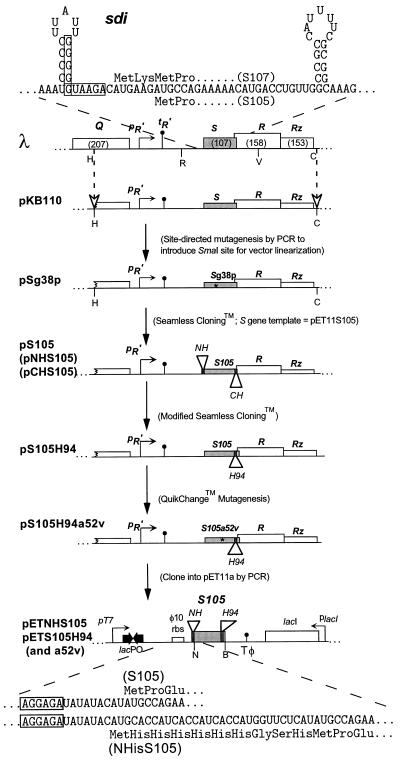
FIG. 2.
Map of the λ lysis gene region and plasmid derivatives. The sdi mRNA and N-terminal amino acid sequences of the S gene products are depicted. The boxed sequences in the mRNA indicate the Shine-Dalgarno sequences for the dual translational starts of the S gene (5). sdi is the RNA stem-loop structure in which single-base changes are known to cause dominant-negative lysis defects (31). The pR′ and tR′ promoter and terminator for the λ late transcriptional unit are shown upstream of the EcoRI site. Restriction sites are indicated (H, HindIII; R, EcoRI; V, EcoRV; C, ClaI; N, NdeI; B, BamHI). pKB110 contains a HindIII/ClaI fragment corresponding to the wild-type “lysis cassette” of λ (46). N- and C-terminal oligohistidine tags were inserted in frame into the S105 gene by Seamless Cloning. An isogenic trans activation plasmid, pS105, bearing the M1L mutation was also made. A derivative of this plasmid, pS105H94, encoding an oligohistidine tag between codons 94 and 95, was made by modified Seamless Cloning. The plasmid pS105H94 was subjected to QuikChange mutagenesis to introduce the A52V lysis-deficient allele, creating the plasmid pS105H94a52v. The S105 alleles of the internally tagged oligohistidine trans activation plasmids were subcloned into pET11a by standard PCR. In the diagram, oligohistidine tags in S at the N terminus or between codons 94 and 95 are mutually exclusive. The N-terminal coding regions of pETNHS105 and pETS105 and their translations are illustrated. The boxes indicate the Shine-Dalgarno sequences. rbs, ribosome binding site.
Genetic analysis further revealed that the structure of S contains timing information independent of the N terminus. A missense mutant was isolated with the Ala-to-Gly change at position 52 (A52G) in the second TMD. This mutant had an unconditional defect in plaque formation resulting from host lysis which occurs so early that, on average, the first virion has not been assembled (23). This result demonstrates that holins have two essential functions: not only must holes be formed to give the endolysin access to the peptidoglycan but also these lesions must form only after a period adequate to support unabated intracellular production of the progeny virions. Quantitative analysis suggests that the λ holin accumulates in the membrane to approximately 103 molecules per cell before hole formation is triggered to terminate the infective cycle (10, 39, 48).
In this light, it was doubtful that purification of the S holin was going to be feasible, given the fact that this molecule evolved to kill the host cell at very low concentrations and is a small membrane protein without enzyme activity. However, we have found that, although hyperexpression of the S gene results in loss of more than 5 orders of magnitude of viability in the induced culture within 5 min, the induced cells are metabolically active for 10 to 15 min, allowing accumulation of S protein in the cytoplasmic membrane to levels 100-fold in excess of the normal triggering levels (39, 40). At this level of expression, milligram quantities of this small membrane protein could, in principle, be obtained from liter cultures. Here we report successful efforts to achieve purification of biochemically useful quantities of the λ holin by using an inserted oligohistidine tag. The results are discussed in terms of the sensitivity of S and membrane proteins to multiple-residue insertions and the potential for developing an in vitro system for investigating the molecular basis of holin function and regulation.
MATERIALS AND METHODS
Strains, phages, plasmids, oligonucleotides and standard DNA manipulations and sequencing.
Relevant Escherichia coli strains, bacteriophages, plasmids, and oligonucleotides used in this study are listed in Tables 1 and 2. Oligonucleotides were purchased from the Gene Technologies Laboratory in the Department of Biology at Texas A&M University. Luria-Bertani (LB), LB-AMP (LB medium supplemented with 100 μg of ampicillin per ml), TB, and TB-maltose (TBM) media were prepared by the methods of Miller (26). The pET11a plasmid vector was acquired from Novagen (Madison, Wis.). Octylglucoside (n-octyl-β-d-galactopyranoside) was purchased from ICN Biomedicals Inc. (Costa Mesa, Calif.). Egg yolk lecithin (l-α-phosphatidylcholine [PC]) was obtained from Avanti Polar Lipids Inc. (Alabaster, Ala.). Unless otherwise indicated, chemical compounds were obtained from Sigma (St. Louis, Mo.).
TABLE 1.
Bacterial strains, bacteriophages, and plasmids used in this study
| Strain, phage, or plasmid | Genotype and relevant features | Source or reference |
|---|---|---|
| Bacterial strains | ||
| BL21(DE3) | E. coli B dcm hsdS(rB− mB−) gal(λimm21nin5 int::lacI-lac puv5-T7 gene1) | 43 |
| MC4100 | E. coli K-12 F− araD139 Δ(argF-lac)U169 rpsL150 relA1 flbB5301 deoC1 ptsF25 rbsR | 37 |
| XL1-Blue | E. coli K-12 recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lac [F′ proAB lacIqZΔM15::Tn10 (Tetr)] | Stratagene |
| Phages | ||
| λS+ | cI857 | Laboratory stock |
| λ Cmr Δ(SR) | stf::cat::tfa cI857 Δ(SR); replacement of stf and tfa genes (λ nt 19996–22220) with cat gene (36); Δ(SR); loss of λ nt 45136–45815 (31) | Laboratory stock |
| Plasmids | ||
| pOR19 | Derivative of pBR322; EcoRI site inactivated by blunt ending and ligation | 34 |
| pKB1 | Derivative of pOR19; λ lysis gene region (Sam7) cloned as HindIII/ClaI fragment (λ nt 44141–46440) | 26a |
| pKB110 | Derivative of pOR19; λ lysis gene region cloned as HindIII/ClaI fragment (λ nt 44141–46440) | 9 |
| pSg38p | Site-directed mutagenesis derivative of pKB110; S Gly38 mutated to Pro to create a SmaI site in codons 38 and 39 | This work |
| pET11a | Expression vector containing hybrid T7-lacPO promoter, φ10 ribosome binding site, and lacI gene (pBR322 derivative) | Novagen |
| pETNH | N-terminal oligohistidine tag (MH6GSH)-coding region inserted into pET11a as NdeI/BglII fragment | This study |
| pETNHS105 | Same as pETNHS107, but encodes NHS105 | This study |
| pS105 | Seamless Cloning derivative of pSg38p; Met1Leu mutation; encodes S105 only | This study |
| pNHS105 | Seamless Cloning derivative of pSg38p; oligohistidine tag inserted at the N terminus of S105 | This study |
| pCHS105 | Seamless Cloning derivative of pSg38p; oligohistidine tag inserted at the C terminus of S105 | This study |
| pS105H94 | Modified Seamless Cloning derivative of pS105; oligohistidine tag (G2H6G2) inserted behind Phe94 | This study |
| pS105H94a52v | QuikChange mutagenesis derivative of pS105H94 with Ala52Val allele | This study |
| pETS105H94 | Derivative of pET11a; S105H94 allele inserted as Nde/BamHI fragment | This study |
| pETS105H94a52v | Derivative of pET11a; S105H94a52v allele inserted as NdeI/BamHI fragment | This study |
TABLE 2.
Oligonucleotides and primers used in this study
| Oligonucleotide or primer | Sequencea | Relevant features |
|---|---|---|
| Oligonucleotides | ||
| ForSG38P | GATATAATccCGGgGCGTTTACAAAAACAG | Binds λ nt 45289–45418; mutates Gly 38 to Pro; creates a SmaI site |
| RevSG38P | GTAAACGCcCCGggATTATATCTGCCGCG | Reverse primer used in conjunction with ForSG38P |
| PETHis | ggaattccatatgagaaccatggtgatggtgatggtgcatgtaTATATCTCCTTCTTAAAGTTAAAC | Anneals downstream of NdeI site in pET11a and primes backward toward the T7 promoter; destroys the original NdeI site; used to create pETNH |
| RevT7 | cggaatTCATAAGTGCGGCGACG | Anneals to pET11a lacI flanking region; primes into lacI gene; used to create pETNH |
| Seamless Cloning primers | ||
| ForpKBEam | agttactcttcATAATCAACGTAAGGCGTTCC | Forward primer for generating vector fragments of transactivation plasmids |
| RevpKBEam | agttactcttcaGCATCTTCAGGTCTTACCCCC | Reverse primer for generating vector fragments of trans activation plasmids |
| ForS105Eam | agttactcttcaTATGCCAGAAAAACATGACCTG | Forward primer for generating S105 alleles with wild-type N termini |
| RevSEam | agttactcttcaATTATTGATTTCTACCATCTTCTAC | Reverse primer for generating S alleles with wild-type C termini |
| ForNHS105Eam | agttactcttcaCATGCACCATCACCATCACC | Forward primer for generating N-terminally tagged S105 alleles |
| RevCHSEam | agttactcttcaCTTAGTGATGGTGATGGTGATG | Reverse primer for generating C-terminally tagged S alleles |
| ForPhe94His | agttactcttctatcaccatcacggcggcGCTGCTAAAAAAGCCGGAG | Forward primer for generating an oligohistidine tag behind codon 94 |
| RevPhe94His | agttactcttctgatgatggtggccgccGAAGCGTTTGATAAGCGAACC | Reverse primer to be used in conjunction with ForPhe94His |
Lowercase letters represent either mismatches or insertions, and new restriction sites are indicated by bold type.
Standard DNA manipulations were done as previously described (39). The method of Chung et al. (12) was used to produce E. coli competent cells for transformation and long-term storage at −80°C.
All PCRs were done as previously described (39), except that for 1,100-bp products, the 72°C segment was extended for 5 min. Sequencing reactions were performed as previously described (39) and were run on an ABI 373A DNA Sequencer (Perkin-Elmer, Foster City, Calif.) by the Gene Technologies Laboratory in the Department of Biology at Texas A&M University, and sequence data were analyzed by using the ABI Sequencer software program.
Site-directed mutagenesis of S by PCR.
PCR was used to create a unique SmaI site in codons 38 and 39 of S in the plasmid pKB110 (Table 1) (Fig. 2). Two mutagenic oligonucleotides, ForSG38P and RevSG38P, were annealed to covalently closed circular pKB110 plasmid DNA under standard PCR conditions and subjected to 30 cycles of the following thermal cycler program: 95°C for 1 min, 50°C for 1 min, and 72°C for 12 min. The resultant PCR product was gel purified, digested with SmaI, ligated, and transformed as previously described. Plasmid DNA was obtained from transformant colonies and screened by restriction with SmaI. One SmaI-sensitive plasmid, pSg38p, was sequenced to confirm the mutagenized sequence.
Mutagenesis of S using Seamless Cloning.
All manipulations of S were done using the reading frame for S105, beginning with codon 3 and terminating at codon 107. For simplicity, this reading frame will be designated S105 throughout this work. Seamless Cloning (Stratagene, La Jolla, Calif.) was used to place the NHS105 allele under λ transcriptional and translational control by substituting it for the S+ allele of pKB110. This plasmid and its derivatives are used as functional test vectors for all S alleles by introduction into a host carrying λ Cmr Δ(SR). Induction of the lysis-defective prophage results in trans activation of the pR′ promoter and thus transcription of the lysis genes on the plasmid (Fig. 2). S105 alleles with the oligohistidine tags are designated as follows: NHS105 has the sequence MH6GSH at the N terminus, CHS105 has the sequence G2H6 at the C terminus, and S105H94 has the sequence G2H6G2 inserted between residues 94 and 95. The manufacturer’s protocol was followed exactly. A vector fragment was synthesized by using the primers ForpKBEam and RevpKBEam with the SmaI-linearized pSg38p template. The S105 insert was synthesized with ForS105Eam and RevSEam. The NHS105 insert was amplified with ForNHS105Eam and RevSEam, while the CHS105 insert was amplified with ForS105Eam and RevCHSEam.
Modified Seamless Cloning.
An oligohistidine tag was inserted between codons 94 and 95 without creating a new restriction site in the resulting plasmid (Fig. 2). The Seamless Cloning (Stratagene) protocol was modified in two respects. First, only two oligonucleotides or primers, ForPhe94His and RevPhe94His, were used, encoding the Eam1104I restriction site, half of the oligohistidine tag, and 19 to 21 nucleotides (nt) of homology with the S gene (Table 2). Second, the two primers were annealed to a covalently closed circular plasmid (pS105) and extended completely around the plasmid under the following conditions: 1 cycle of 94°C for 3 min, 50°C for 1 min, and 72°C for 12 min and then 10 cycles, with 1 cycle consisting of 94°C for 1 min, 50°C for 1 min, and 72°C for 12 min. 5-Methyl-dCTP was then added to the sample exactly as specified in the Seamless Cloning protocol, and five cycles were performed, with one cycle consisting of 94°C for 1 min, 50°C for 1 min, and 72°C for 12 min. The reaction mixtures were digested, ligated, and transformed, and transformants were screened exactly as described above for the Seamless Cloning technique.
QuikChange mutagenesis.
Specific alleles were introduced into the plasmids pS105 and pS105H94 by site-directed mutagenesis by using the QuikChange kit from Stratagene (Fig. 2). Manufacturer’s instructions were specifically followed. The plasmids pS105 and pS105H94 were used as template DNA (Table 1). Constructs were confirmed by diagnostic PCR and sequencing as previously described (39).
Induction of pET11 clones of the S gene.
Cells carrying the T7 vector plasmid, pET11a (Novagen), or any of its derivatives (Table 1) were grown and induced as previously described (39, 40).
Induction of functional test vectors harboring the S gene.
In each case, MC4100 [λ Cmr Δ(SR)] harboring the trans activation plasmid derived from pKB110 was induced as previously described (10), except that the period of aeration at 42°C was decreased from 20 to 15 min.
Membrane protein sample preparation and analysis.
Detergent-solubilized preparations of inner membrane proteins were obtained, resolved, immunoblotted, and analyzed as described previously (10, 39, 40).
Immobilized metal affinity chromatography (IMAC).
Ni-NTA (nickel-nitrilotriacetic acid) (Qiagen, Chatsworth, Calif.) and cobalt-based Talon affinity resin (CLONTECH Laboratories, Inc., Palo Alto, Calif.) were purchased as 50:50 slurries. Columns (0.5- to 1.0-cm diameter) with bed volumes of 0.5 to 1.0 ml were poured and preserved in 30% ethanol. Prior to use, columns were equilibrated with 20 column volumes of a solution consisting of 1% Triton X-100, 20 mM Tris-HCl, and 10% glycerol (TTG) plus 0.5 M NaCl (TTGN), pH 8.0. Columns were developed at a flow rate of no more than 4 column volumes per h. Detergent-solubilized membrane extracts were loaded based on estimates of the extracts’ S protein content and on an optimized 10-mg/ml binding capacity of the resin for oligohistidine-tagged protein. Columns were then washed with 10 column volumes each of the following: (i) TTGN at pH 8.0, (ii) TPGN (1% Triton X-100, 10% glycerol, 0.5 M NaCl, 100 mM sodium phosphate) at pH 6.5, and (iii) TPGN at pH 4.5. Columns were finally eluted with a solution consisting of 1% octylglucoside, 10% glycerol, and 100 mM sodium phosphate using a linear gradient from pH 4.5 to 2.5. Column fractions were analyzed for protein purity by sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis.
Determination of protein concentration.
Estimates of protein concentration of various samples were determined by DC (detergent-compatible) Bio-Rad Protein Assay (Bio-Rad Laboratories, Hercules, Calif.) according to the manufacturer’s recommendations. Bovine serum albumin was used as a standard. Concentrations of purified samples of S protein were determined by the method of Pace et al. (28) after complete dialysis against 6 M guanidine hydrochloride (GdnHCl) in 20 mM Tris-HCl–150 mM NaCl (pH 7.6) (Tris-buffered saline [TBS]), yielding an ɛ280 of 11,460 M−1 cm−1.
CD.
Purified samples of S protein were analyzed on an Aviv model 62DS circular dichroism (CD) spectrometer (Aviv Associates Inc., Lakewood, N.J.) at wavelengths from 195 to 320 nm in a 0.1-cm pathlength cuvette at a final concentration of 15 to 25 μM at room temperature (23°C). Scans were measured in millidegrees and converted to mean residue ellipticity. Alpha-helical content was estimated from the molar ellipticity signal at 222 nm.
Preparation of calcein-loaded liposomes.
First, 350 μl of a solution of PC dissolved in hexane-ethanol (20 mg of PC per ml) was mixed with 143 μl of cholesterol dissolved in chloroform (7 mg/ml) and dried under air. After removal of residual organic solvents under a vacuum for 15 min, the lipids were resuspended in 1 ml of 0.2 M calcein in TBS with vigorous vortexing. The Liposofast apparatus (Avestin, Inc., Ottawa, Ontario, Canada) was fitted with polycarbonate membranes (100-nm pore size), and the multilamellar lipid suspension was extruded through the membranes 19 times. The nominally unilamellar liposomes loaded with calcein were separated from unincorporated calcein by gel filtration chromatography on a 150-ml Sephadex G50 resin column equilibrated in TBS. We estimate that 1012 liposomes were added to each assay based on the lipid content of E. coli (27).
Dye release assay.
Dye release assays were performed in an SLM-Aminco 8000 or 8100 fluorescence spectrophotometer (Spectronic Instruments Inc., Rochester, N.Y.) in 2.5-ml glass cuvettes with all four sides polished. Two milliliters of TBS was added to a cuvette with 40 μl of liposomes. The samples were excited at a wavelength of 490 nm, and emission was monitored at 520 nm. Baseline, or 0% relative fluorescence, was defined as the signal acquired upon addition of a volume of buffer (6 M GdnHCl in TBS) equal to the volume of protein added. Total release, and therefore 100% relative fluorescence, was obtained by adding 10 μl of 10% Triton X-100. Protein (2 to 15 μg in a volume of 5 to 40 μl) was added to initiate the assay. Alpha-hemolysin (9 to 18 μg in a volume of 5 to 10 μl) was used as a control to establish the quality and fraction of unilamellar vesicles (2, 14). Relative fluorescence was plotted versus time.
RESULTS
Preliminary purification of unmodified S105 and S107 proteins.
No holin has been purified in biochemically useful quantities nor has an in vitro assay been devised for crude preparations. Gene fusion approaches in which easily purifiable domains (i.e., β-galactosidase or ubiquitin) were fused with proteolytically cleavable linkers to the N- or C-terminal end of the S protein were unsuccessful, because in every case cleavage of the hybrid purified in detergent was at best inefficient (11). Taking advantage of the recent achievement of hyperexpression of S (39), we undertook to obtain enriched fractions of S holin by ion-exchange chromatography and preparative-scale isoelectric focusing of inner membrane extracts in detergent. The S holin could be purified to a state where only one contaminant was detectable, but the available concentration of S holin in such preparations was limited by its tendency to precipitate near its isoelectric point (38). Consequently, purification of S protein by standard chromatographic methods was abandoned.
Oligohistidine tags at the N and C termini of S.
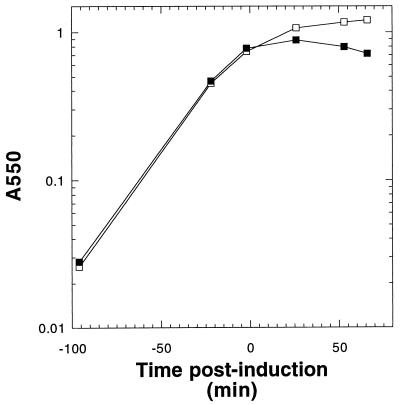
The potential for single-step purification of S from detergent extracts by IMAC led us to create oligohistidine-tagged versions of S (21). The reading frame encoding S105 was fused to an N-terminal oligohistidine tag sequence under the translational control of the consensus T7φ10 ribosome binding site and the T7 promoter of pET11a (Novagen) (Fig. 2). Induction of this construct, designated as pETNHS105, resulted in a sudden halt in culture growth after about 10 min, followed by a gradual loss of turbidity (Fig. 3), which correlated with the accumulation of translucent, vacuolized cells as determined by phase-contrast microscopy (data not shown). These growth characteristics were very similar to those observed when the normal S105 reading frame, lacking an oligohistidine tag, is fused to the same gene expression signals (39), suggesting that the addition of the N-terminal oligohistidine tag did not grossly affect the toxicity of the S105 sequence, at least at these hyperexpression levels. Coomassie blue-detectable protein species of the expected sizes accumulated in membrane extracts (Fig. 4A, lane 1), and immunoblot analysis using the C-terminal antibody confirmed that the accumulated protein was S (Fig. 4B, lane 1).
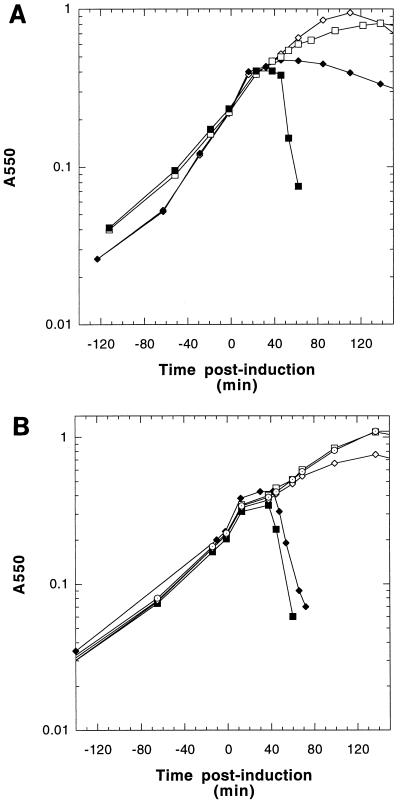
FIG. 3.
Profiles of BL21(DE3) pETNH induction. Cells harboring plasmid pETNH (□) or pETNHS105 (▪) were induced at time zero and followed by monitoring A550.
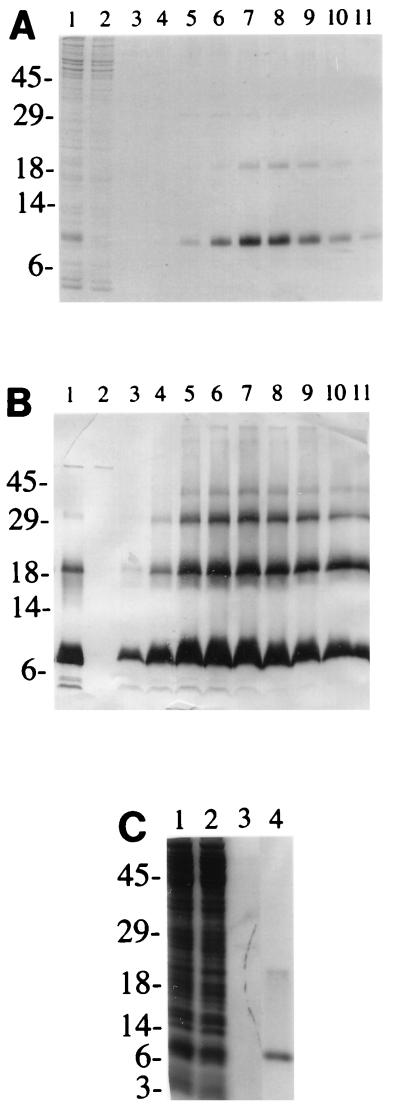
FIG. 4.
NHS105 accumulates in inner membrane and can be purified by metal affinity chromatography. (A) Coomassie blue-stained SDS-polyacrylamide gel revealing hyperexpression of NHS105 and its extraction from the inner membrane (lane 1), column fractions from purification (flowthrough [lane 2], pH 4.5 wash [lane 3], and pH 4.5 to 2.5 gradient fractions [lanes 4 to 11]). S protein dimers are visible in the peak fractions from the gradient. (B) Western immunoblot of identical fractions from the purification revealing oligomers up to tetramers. Note that some NHS105 is detected in the pH 4.5 wash. (C) Purification of S105H94. The fractions are inner membrane extract (lane 1), metal affinity column flowthrough (lane 2), pH 4.5 wash (lane 3), and pooled fractions from the pH 4.5 to 2.5 gradient (lane 4). The positions of molecular size markers (in kilodaltons) are shown to the left of the gels.
The protein could be purified to apparent homogeneity by binding the Triton-solubilized membrane fraction to an IMAC resin and eluting at low pH in the dialyzable detergent octylglucoside (Fig. 4, lanes 4 to 11). Isogenic constructs carrying S− missense alleles were also constructed (Fig. 2) and induced, and the resultant membranes were used for S purification to supply material for negative controls in development of an in vitro assay (data not shown). These purified NHS proteins were subjected to analysis by CD spectroscopy. The results showed that the NHS protein has approximately 40% alpha-helical content (Fig. 5), which would correspond to about 45 residues. These results support the model for S protein in which there are two TMDs, as opposed to three (6, 31), at least in detergent. However, repeated attempts to detect pore formation in vitro using liposome or “black lipid membrane” systems were unsuccessful with these purified protein preparations (8).
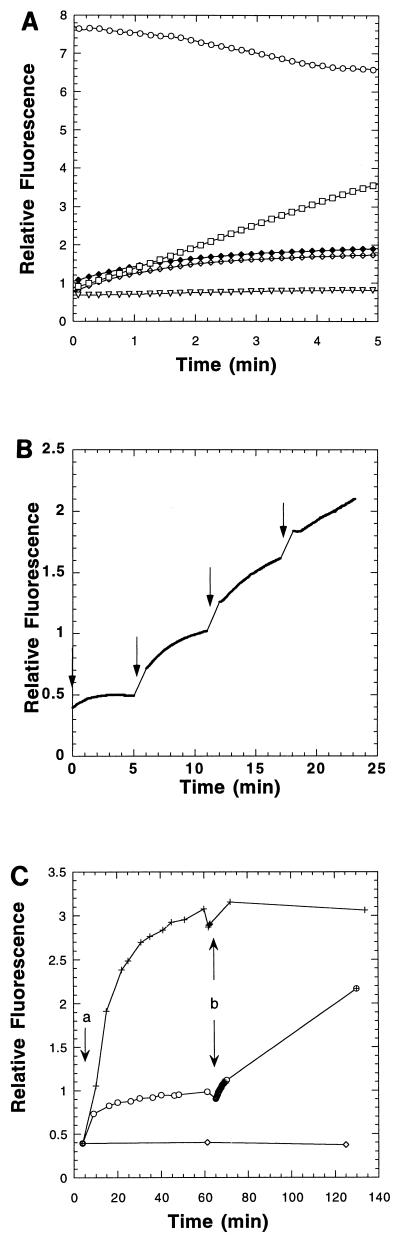
FIG. 5.
CD spectra of NHS105 (□) and S105H94 (▪) proteins. See Materials and Methods for conditions of measurement.
In vivo functional assay for oligohistidine-tagged S alleles.
Our inability to detect pore formation in vitro by a variety of methods prompted a more rigorous examination of the biological function of the NHS105 protein. Although the physiological effects of inducing the pETNHS105 plasmid closely resembled those obtained from inducing the isogenic constructs without the oligohistidine tag, the abnormal physiology derived in these T7 polymerase expression systems precludes useful phenotypic analysis. The S gene is normally subject to sophisticated and complex transcriptional and translational controls, upon which the timing of lysis, and thus the ultimate productivity of the vegetative cycle, is dependent (6, 46). To assess biological function, it was necessary to reinsert the NHS105 allele into the normal transcriptional and translational context of the phage.
Using a mutagenesis system which allows insertion of DNA sequences at any site without regard to restriction site, a construct was made with the oligohistidine-tagged S105 reading frame placed under the normal wild-type transcriptional and translational control, in cis with the R and Rz genes (Fig. 2, plasmid pNHS105). Lysis supported by the NHS105 allele, when trans-activated by induction of a lysis-defective prophage, was severely delayed (Fig. 6A), resembling the delayed lysis typical of induced prophages bearing the Sm3l allele (producing only S107) (31). This result suggested that the tag sequence at the N terminus was providing an effect similar to the Met-Lys… sequence which differentiates S107 from S105 (4, 31). We thus concluded that the NHS105 protein would not be a suitable protein for developing an in vitro hole-forming system. Moreover, it was clear that no tagged S allele could be tested for functionality outside of the normal transcriptional and translational context of the S gene, presumably because the timing and function of S depend critically on the kinetics of its accumulation in the cytoplasmic membrane.
FIG. 6.
Profiles of MC4100 [λ Cmr Δ(SR)] induction. (A) Cells harboring pKB1 (□), pS105 (▪), pNHS105 (◊), or pCHS105 (⧫) were induced at time zero and monitored for 1.5 h. (B) Cells harboring pKB1 (□), pS105 (▪), pS105H94 (⧫), pS105H94a52v (◊), or pS107H94m3l (⊙) allele.
The next construction, pCHS105, in which an oligohistidine tag is attached to the C terminus of S, was done directly in the context of the trans activation plasmid vector to facilitate immediate assessment of whether the allele was sufficiently normal in lysis and lysis scheduling (Fig. 2). Although the C-terminally tagged allele showed nearly normal scheduling of the onset of lysis, the rate of lysis after triggering was much more gradual than with the wild type, suggesting a partial defect in the membrane lesion through which the endolysin must pass (Fig. 6A). It was clear that CHS105 protein would also not be an acceptable protein to develop an in vitro system for hole formation.
Construction of a functional internally tagged S105 allele.
Despite the fact that the S105 constructs with oligohistidine tags at the N and C termini had proven to have aberrant lysis behavior in terms of timing or efficiency of endolysin release, the ease with which the tagged S protein could be purified from membrane extracts prompted us to look for internal sites in S where the tag would not seriously affect S-holin function. Constructs containing the sequence G2H6G2 inserted in different locations within S105 were made by using a modified version of Seamless Cloning, and each was tested for S function at physiologically relevant levels of S expression (41). One allele, in which the tag was inserted between codons 94 and 95, exhibited a lysis phenotype sufficiently similar to wild-type S to warrant its use as a source of S protein (Fig. 6B). This allele, designated S105H94, was inserted into the pET11a hyperexpression vector, generating pETS105H94 (Fig. 2). Induction of this plasmid resulted in accumulation of S protein at levels indistinguishable from that obtained with pETS105 (Fig. 4C). As a control for in vitro hole formation assays, S105H94a52v, an isogenic allele bearing the A52V lysis-defective missense change (30), was constructed (Fig. 2). This allele was tested in the trans activation plasmid context and found to have the expected lysis-defective phenotype (Fig. 6B). Induction of an isogenic pET11a construct of this defective allele also yielded Coomassie blue-detectable quantities of S protein, at approximately twofold-higher levels than for the S105H94 lysis-proficient allele, presumably because mass accumulation continues a few minutes longer after induction (data not shown). Purification of the S105H94 (and S105H94a52v [data not shown]) protein was accomplished in the same manner as described above (Fig. 4C). All the purified S protein preparations had CD spectra indistinguishable from that observed for the original purified N-tagged S protein, with approximately 40% alpha-helical conformation in 1% octylglucoside detergent solution (Fig. 5).
In vitro assay for S function.
The purified S105H94 protein, solubilized in octylglucoside, exhibited a marked tendency to aggregate in insoluble masses, which were readily resolubilized by resuspension in 6 M GdnHCl in TBS (data not shown). After replacing the chaotrope with detergent by dialysis, the S protein refolded to a conformation indistinguishable from that of S eluted from the IMAC column in octylglucoside, as determined by CD spectroscopy (data not shown). We reasoned that this renaturability might allow denatured S to insert into a target membrane in vitro. To test this idea, calcein-loaded PC-cholesterol liposomes were created by extrusion of lipid suspensions through polycarbonate membranes. At the concentrations used, the dye is self-quenched, and only when it is released from the liposomes and diluted into the assay solution does it become highly fluorescent (1). For a positive control, the alpha-hemolysin from Staphylococcus aureus, a soluble hole-forming toxin (2, 19, 45), was used to demonstrate that most of the liposomes could be made permeable efficiently by attack from the external aqueous environment and were thus not multilamellar (Fig. 7A). When S105H94 was added to the suspension of calcein-loaded liposomes, a fluorescent signal was obtained, indicating that a fraction of the dye was released, whereas the S105H94a52v protein was essentially inactive under these conditions (Fig. 7A). We reasoned that the S protein could have several fates at the instant of dilution into the aqueous suspension of liposomes (Fig. 8). The desired pathway, in which S inserted into the membrane bilayer, would compete with various unproductive pathways, including the formation of insoluble aggregates and the accretion of S protein onto the surfaces of the liposomes. The latter pathways might be favored by higher concentrations of S, and consequently the assay was repeated by adding the same total amount of S in a series of smaller sequential aliquots. Using this protocol, more-efficient calcein release was indeed observed, although the final yield of dye released never exceeded approximately 50% of that obtained using the alpha-hemolysin (Fig. 7B). Interestingly, each addition of S protein resulted in dye release with slow kinetics on the order of minutes. Because the partition between membrane insertion and aggregation is likely to be extremely rapid after dilution out of detergent or chaotrope, we presume that the slow kinetics reflects the time required for the productive formation of holes within the membrane. In any case, because this dye release was obtained with purified S protein and protein-free artificial membranes, we conclude that S protein is necessary and sufficient to make a bilayer permeable, at least sufficiently to allow passage of a fluorescein dye. The fact that the isogenic A52V protein is incapable of the same reaction strongly suggests that this permeabilization capacity is at least related, if not identical, to the “hole formation” which allows passage of endolysin through the cytoplasmic membrane in phage lysis. Moreover, we observed that prior treatment of the liposomes with S105H94a52v followed by treatment with S105H94 resulted in a more gradual dye release (Fig. 7C), suggesting that the preinserted defective holins interfered with the hole-forming activity of the active holin. This result is consistent with the partial dominant character of the A52V allele when it is present on a multicopy plasmid in trans to the parental S105 allele on an induced prophage (41).
FIG. 7.
Permeabilization of calcein-loaded liposomes by purified S protein. (A) Kinetics of dye release by holin and alpha-hemolysin. Total release is effected by adding 0.05% Triton X-100 (○). Relative fluorescence was monitored continuously for the first 5 min. Single point measurements were taken between 10 and 20 min postaddition of protein. Amounts of protein added to initiate the assay were as follows: 8.75 μg of alpha-hemolysin (□), 15.2 μg of S105H94 (⧫), 3.8 μg of S105H94 (◊), and 17.6 μg of S105H94a52v (▿). (B) Sequential amounts (1.9 μg) of S105H94 were added to calcein-loaded liposomes, as indicated by the vertical arrows. Fluorescence was monitored continuously. (C) Pretreatment of calcein-loaded liposomes with a S105H94a52v. Maximum release of dye was established by addition of 0.05% Triton X-100 (⧫). A baseline signal was established by adding an equal volume of 6 M GdnHCl in TBS (◊). At point a, 7.6 μg of S105H94 protein (+) or 8.8 μg of S105H94a52v protein (○) was mixed separately with calcein-loaded liposomes, and fluorescence was monitored for 1 h. At point b, 7.6 μg of S105H94 was then added to both S105 (+) and S105H94 (⊕) samples, and fluorescence was monitored.
FIG. 8.
Model for permeabilization of calcein-loaded liposomes by purified S protein. Purified S protein is diluted out of 6 M GdnHCl into the assay. Two immediate fates are portrayed: (i) insoluble aggregation and (ii) association with liposome membrane bilayers. S protein association with the bilayer is depicted as having three basic outcomes: (i) association with the bilayer in a nonintegral fashion, (ii) spontaneous integration into the bilayer, and/or (iii) oligomerization and hole formation. A successful hole formation event would be detected as an increase in fluorescence, as the dye goes from a self-quenched state (filled circles) to a nonquenched state (open circles) due to release and dilution into the assay buffer.
DISCUSSION
Holins constitute one of the largest functional groups of proteins known, with more than 60 reported holin sequences from more than 30 nonorthologous groups (40, 46, 47). This work reports the first purification, the first biophysical characterization, and the first in vitro assay of a holin, the S protein of bacteriophage lambda.
Positioning of the oligohistidine tag in the S protein.
Oligohistidine tags are frequently added to proteins to facilitate purification by IMAC (21), and in most cases, the tag is placed at the amino or carboxy terminus of the polypeptide chain. The initial choice of the N-terminal histidine tag position for S was based on the fact that our best antibody was raised against a C-terminal epitope (10). N-tagged S protein was stable and straightforward to purify in milligram quantities from detergent-solubilized membranes. Under overexpression conditions, NHS105 affected cellular morphology and culture growth very similarly to the untagged protein. Moreover, the CD spectrum of detergent-solubilized NHS105 was exactly as expected from genetic and modeling considerations. However, in the exact transcriptional and translational context of the S gene in lambda, NHS105 was found to function much more like the inhibitor form of S, S107, than the effector form, S105. This serves as a cautionary note for the future analysis of other holins. The holin protein is designed to cause hole formation and thus terminate the vegetative cycle, at a precisely defined time, which corresponds to a certain concentration in the membrane (10, 39). It is thus unproductive to assess the function of a holin variant, such as an oligohistidine-tagged form, under conditions where the rate or amount of holin production is significantly different from the normal context of the lytic cycle. The inhibitor character of the NHS105 allele has been verified by demonstrating its ability to inhibit S function in trans (41). Assuming that this inhibitory property is based on the same requirement for N-terminal positive charge found for the S107 and P22 gp13108 inhibitor forms, this phenotype suggests that significant positive charge exists on the side chains of the hexahistidine sequence in vivo.
The C-terminal tagged protein appears to have essentially normal timing for the onset of the lytic event, but the lysis profile after onset is shallow, suggesting that endolysin release is inefficient compared to the parental S105 protein. It should be noted that fusions of lacZ to the extreme 3′ end of the S gene result in stable, membrane-bound S–β-galactosidase hybrid proteins which have no lytic function (9, 29, 48). These observations suggest that alterations in the C-terminal domain may occlude the “hole” through which the endolysin proteins must pass.
Purification and characterization of the holin.
The use of IMAC for the purification of the holin was successful, achieving apparent homogeneity in a single step from solubilized inner membrane material. This is important because holins are small membrane proteins which have evolved to kill E. coli and thus cannot be synthesized in vivo at levels exceeding approximately 1 mg per liter of culture (39). Multistep purifications would thus be likely to reduce useable yields to unacceptable levels. However, it should be noted that the site used (between residues 94 and 95 in the middle of the highly charged C terminus) was one of only two positions of 13 tested for the oligohistidine tag in the S105 reading frame which left the timing and lysis functions of the holin largely unaffected (41). Assuming that a similar site for the oligohistidine tag would be effective in other holins, these findings should allow rapid purification of other holins of interest, including the prototypical class II holin S21 and the nonorthologous class I holins of phages P1 and P2 (7, 35, 49).
The single-step purification was notable for lack of copurifying proteins, which might have been evidence for interactions of S with host proteins in the membrane. Although multiple genetic selections have failed in identifying host loci which could give rise to S-resistant mutations (16, 29), there was heretofore no direct evidence that S alone was required to form holes. The fact that SDS-resistant oligomers of appropriate molecular mass were observed in Western blots of concentrated purified S protein (Fig. 4B) is evidence that S forms a homo-oligomer, consistent with the patterns of oligomeric species found after chemical cross-linking and immunoblot analysis of bacterial membranes containing S protein (7, 48). Whether the single cysteine within the second TMD is involved in disulfide links in any of these oligomeric species is unknown, although a covalent function for this residue is unlikely, considering the fact that only one other of the 63 known holin sequences (φ29 P15) contains a cysteine residue in the putative TMDs (47). The prominence of the oligomeric ladder up to tetramers suggests that tetrameric S may be a functional unit of the hole assembly, as suggested previously (6).
The CD spectrum of S clearly shows that, in detergent, approximately 40% of the residues are in alpha-helical conformation (Fig. 5). Based on the 115-residue length of S105H94, this would correspond to about 45 residues, consistent with two alpha-helical TMDs, but not three. The topology of S in the membrane has not been directly determined. The C termini of all holins have numerous charged residues, with an excess of basic residues (38, 46, 47). Based on sequence considerations alone, this domain is thus almost certainly cytoplasmic, a fact which has been confirmed by protease accessibility experiments on inverted membrane vesicles and spheroplasts (11) and by the normal enzymatic specific activity and membrane localization of C-terminal S–β-galactosidase hybrids (32). Thus, evidence for two alpha-helical TMDs can be taken as support for the cytoplasmic location of the N terminus (Fig. 1B). The two TMD model for S would also unify the topologies of type I and II holins, because the type II holins are too small to support formation of more than two alpha-helical TMDs (Fig. 1). Such unified topology is desirable because the dual-start motif is apparently conserved in both type I and II lambdoid holins, and it is difficult to imagine that such a conserved regulatory motif could function from opposite sides of the cytoplasmic membrane.
An in vitro assay for holin function.
This work demonstrates that purified S protein, when diluted out of a chaotropic solution, is capable of forming holes in artificial liposomes. This is a major step in the effort to understand how holins achieve the hole-forming function. It is not yet clear that the holes responsible for the release of the dye are the same as those formed in vivo. However, it is unlikely that the A52V missense change would adversely affect the ability of the S protein to enter the membrane, since it actually increases hydrophobic character. Thus, the inability of S protein carrying this missense change, which abolishes hole formation in vivo, to form these holes is support for the notion that the channel formation being measured by the release of the encapsulated carboxyfluorescein is related, if not identical, to the lethal biological lesion. Moreover, each addition of S protein was followed by a definable kinetics of fluorescein release over a period of minutes (Fig. 7B). This is a tantalizing result which may reflect the other required biological function of S: not just to form a hole but to do so slowly to allow a macroscopic vegetative phase on the order of many minutes (23). Nevertheless, in its present form, this in vitro assay for holin function is still far from an optimal tool for detailed structure-function analysis of this unusual class of membrane proteins. Most S protein added to the assays ends up as insoluble aggregates, and very little S protein actually ends up associated with the liposomes, as determined by Western blot analysis of the liposome and aqueous fractions after addition of the holin (data not shown). Moreover, the assay appears limited to the release of only a fraction of the total dye entrapped in the liposome preparation, approximately 50% by the sequential method of S addition (Fig. 7B). One interpretation of this limitation is that significant amounts of S become trapped on the surface of the liposome and thus block entry of further S molecules (Fig. 8). Some of the material which is bound to the membranes is apparently bound nonproductively. In any case, substantial effort will now be required to optimize this assay. The use of altered ionic and osmotic conditions, different phospholipid mixtures, and soluble chaperones to help prevent excessive aggregation of S may be indicated. It should be noted that the lysis protein L of the RNA phage MS2 has previously been studied using similar technology (18). In that case, a fragment of the lysis protein was synthesized chemically and tested for its ability to liberate calcein from liposomes. An important difference is that L is not a holin and does not function by allowing endolysin to penetrate the cytoplasmic membrane and thus gain access to the peptidoglycan. Moreover, there is no evidence that the fragment of L used functions in the same way as the L protein does in vivo. That is, there are no mutant forms of L which have been demonstrated to fail in the same biochemical assay. In any case, experiments to optimize the dye release assay are now under way in this laboratory.
ACKNOWLEDGMENTS
Alpha-hemolysin was a kind gift from Hagan Bayley and Orit Braha. We express our appreciation to Art Johnson and David Geidroc for the use of their fluorescence spectrophotometers. We are also indebted to the members of the Young lab past and present for help and encouragement and to Sharyll Pressley for competent clerical assistance. In particular, we thank Rudi Martinez for help with the purification of NHS105 proteins and Matilda Powers and George Han for plasmid construction and sequencing and for inductions. Special thanks to Bill Roof for insight and encouragement.
This work was funded by grant GM27099 from the National Institute of General Medical Sciences, National Institutes of Health, to R.Y. and by funds from the College of Agriculture at Texas A&M University.
REFERENCES
- 1.Allen T M, Cleland L G. Serum-induced leakage of liposome contents. Biochim Biophys Acta. 1980;597:418–426. doi: 10.1016/0005-2736(80)90118-2. [DOI] [PubMed] [Google Scholar]
- 2.Bhakdi S, Tranum-Jensen J. Alpha-toxin of Staphylococcus aureus. Microbiol Rev. 1991;55:733–751. doi: 10.1128/mr.55.4.733-751.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bienkowska-Szewczyk K, Lipinska B, Taylor A. The R gene product of bacteriophage λ is the murein transglycosylase. Mol Gen Genet. 1981;184:111–114. doi: 10.1007/BF00271205. [DOI] [PubMed] [Google Scholar]
- 4.Bläsi U, Chang C-Y, Zagotta M T, Nam K, Young R. The lethal λ S gene encodes its own inhibitor. EMBO J. 1990;9:981–989. doi: 10.1002/j.1460-2075.1990.tb08200.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bläsi U, Nam K, Hartz D, Gold L, Young R. Dual translational initiation sites control function of the λ S gene. EMBO J. 1989;8:3501–3510. doi: 10.1002/j.1460-2075.1989.tb08515.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bläsi U, Young R. Two beginnings for a single purpose: the dual-start holins in the regulation of phage lysis. Mol Microbiol. 1996;21:675–682. doi: 10.1046/j.1365-2958.1996.331395.x. [DOI] [PubMed] [Google Scholar]
- 7.Bonovich M T, Young R. Dual start motif in two lambdoid S genes unrelated to λ S. J Bacteriol. 1991;173:2897–2905. doi: 10.1128/jb.173.9.2897-2905.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braha, O. Personal communication.
- 9.Chang C-Y. Synthesis, function and regulation of the lambda holin. Ph.D. dissertation. College Station: Texas A&M University; 1994. [Google Scholar]
- 10.Chang C-Y, Nam K, Young R. S gene expression and the timing of lysis by bacteriophage λ. J Bacteriol. 1995;177:3283–3294. doi: 10.1128/jb.177.11.3283-3294.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang, C.-Y., and R. Young. Unpublished data.
- 12.Chung C T, Niemela S L, Miller R H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA. 1989;86:2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Diaz E, Munthali M, Lünsdorf H, Höltje J-V, Timmis K N. The two-step lysis system of pneumococcal bacteriophage EJ-1 is functional in Gram-negative bacteria: triggering of the major pneumococcal autolysin in Escherichia coli. Mol Microbiol. 1996;19:667–681. doi: 10.1046/j.1365-2958.1996.399929.x. [DOI] [PubMed] [Google Scholar]
- 14.Fiechtner M, Wong M, Bieniarz C, Shipchandler M T. Hydrophilic fluorescein derivatives: useful reagents for liposome immunolytic assays. Anal Biochem. 1989;180:140–146. doi: 10.1016/0003-2697(89)90102-4. [DOI] [PubMed] [Google Scholar]
- 15.Garcia P, Mendez E, Garcia E, Ronda C, Lopez R. Biochemical characterization of a murine hydrolase induced by bacteriophage Dp-1 in Streptococcus pneumoniae: comparative study between bacteriophage-associated lysin and the host amidase. J Bacteriol. 1984;159:793–796. doi: 10.1128/jb.159.2.793-796.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garrett J M. The mechanism and control of host cell lysis by bacteriophage lambda. Ph.D. dissertation. College Station: Texas A&M University; 1982. [Google Scholar]
- 17.Garvey K J, Saedi M S, Ito J. Nucleotide sequence of Bacillus phage φ29 genes 14 and 15: homology of gene 15 with other phage lysozymes. Nucleic Acids Res. 1986;14:10001–10008. doi: 10.1093/nar/14.24.10001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goessens W H F, Driessen A J M, Wilschut J, van Duin J. A synthetic peptide corresponding to the C-terminal 25 residues of phage MS2 coded lysis protein dissipates the proton motive force in Escherichia coli membrane vesicles by generating hydrophilic pores. EMBO J. 1988;7:867–873. doi: 10.1002/j.1460-2075.1988.tb02886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gouaux J E, Braha O, Hobaugh M R, Song L, Cheley S, Shustak C, Bayley H. Subunit stoichiometry of staphylococcal alpha-hemolysin in crystals and on membranes: a heptameric transmembrane pore. Proc Natl Acad Sci USA. 1994;91:12828–12831. doi: 10.1073/pnas.91.26.12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gutter M G, Weaver L H, Gray T M, Matthews B W. Structure, function, and evolution of the lysozyme from bacteriophage T4. In: Mathews C K, Kutter E M, Mosig G, Berget P B, editors. Bacteriophage T4. 1st ed. Washington, D.C: American Society for Microbiology; 1983. pp. 356–360. [Google Scholar]
- 21.Hochuli E, Bannwarth W, Döbeli R, Gentz R, Stüber D. Genetic approach to facilitate purification of recombinant proteins with a novel metal chelate adsorbent. Bio/Technology. 1988;6:1321–1325. [Google Scholar]
- 22.Inouye M, Arnheim N, Sternglanz R. Bacteriophage T7 lysozyme is an N-acetylmuramyl-L-alanine amidase. J Biol Chem. 1973;248:7247–7252. [PubMed] [Google Scholar]
- 23.Johnson-Boaz R, Chang C-Y, Young R. A dominant mutation in the bacteriophage lambda S gene causes premature lysis and an absolute defective plating phenotype. Mol Microbiol. 1994;13:495–504. doi: 10.1111/j.1365-2958.1994.tb00444.x. [DOI] [PubMed] [Google Scholar]
- 24.Loessner M J, Wendlinger G, Scherer S. Heterogeneous endolysins in Listeria monocytogenes bacteriophages: a new class of enzymes and evidence of conserved holin genes within the siphoviral lysis cassettes. Mol Microbiol. 1995;16:1231–1241. doi: 10.1111/j.1365-2958.1995.tb02345.x. [DOI] [PubMed] [Google Scholar]
- 25.Lu M-J, Henning U. Lysis protein T of bacteriophage T4. Mol Gen Genet. 1992;235:253–258. doi: 10.1007/BF00279368. [DOI] [PubMed] [Google Scholar]
- 26.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 26a.Nam K. Translational regulation of the S gene of bacteriophage λ. Ph.D. dissertation. College Station: Texas A&M University; 1991. [Google Scholar]
- 27.Neidhardt F G, Ingraham J L, Schaechter M. Physiology of the bacterial cell: a molecular approach. Sunderland, Mass: Sinauer Associates, Inc.; 1990. pp. 1–506. [Google Scholar]
- 28.Pace C N, Vajdos F, Fee L, Grimsley G, Gray T. How to measure and predict the molar absorption coefficient of a protein. Protein Sci. 1995;4:2411–2423. doi: 10.1002/pro.5560041120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raab R. The structure, function and regulation of the S gene of bacteriophage lambda. Ph.D. dissertation. College Station: Texas A&M University; 1988. [Google Scholar]
- 30.Raab R, Neal G, Garrett J, Grimaila R, Fusselman R, Young R. Mutational analysis of bacteriophage lambda lysis gene S. J Bacteriol. 1986;167:1035–1042. doi: 10.1128/jb.167.3.1035-1042.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Raab R, Neal G, Sohaskey C, Smith J, Young R. Dominance in lambda S mutations and evidence for translational control. J Mol Biol. 1988;199:95–105. doi: 10.1016/0022-2836(88)90381-6. [DOI] [PubMed] [Google Scholar]
- 32.Raab, R., and R. Young. Unpublished data.
- 33.Rennell D, Poteete A R. Phage P22 lysis genes: nucleotide sequences and functional relationships with T4 and λ genes. Virology. 1985;143:280–289. doi: 10.1016/0042-6822(85)90115-1. [DOI] [PubMed] [Google Scholar]
- 34.Rossetti O L, Altman R, Young R. Kinetics of Tn5 transposition. Gene. 1984;32:91–98. doi: 10.1016/0378-1119(84)90036-2. [DOI] [PubMed] [Google Scholar]
- 35.Schmidt C, Velleman M, Arber W. Three functions of bacteriophage P1 involved in cell lysis. J Bacteriol. 1996;178:1099–1104. doi: 10.1128/jb.178.4.1099-1104.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schweizer H P. The pUC18CM plasmids: a chloramphenicol resistance gene cassette for site-directed insertion and deletion mutagenesis in Escherichia coli. BioTechniques. 1990;8:612–616. [PubMed] [Google Scholar]
- 37.Silhavy T J, Berman M L, Enquist L W. Bacterial strains. In: Silhavy T J, Berman M L, Enquist L W, editors. Experiments with gene fusions. 1st ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1984. pp. xi–xiii. [Google Scholar]
- 38.Smith D L. Purification and biochemical characterization of the bacteriophage λ holin. Ph.D. dissertation. College Station: Texas A&M University; 1998. [Google Scholar]
- 39.Smith D L, Chang C-Y, Young R. The λ holin accumulates beyond the lethal triggering concentration under hyper-expression conditions. Gene Expr. 1998;7:39–52. [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, D. L., and R. Young. Unpublished data.
- 41.Smith, D. L., and R. Young. Oligohistidine tag mutagenesis of the lambda holin gene. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 42.Steiner M, Lubitz W, Bläsi U. The missing link in phage lysis of gram-positive bacteria: gene 14 of Bacillus subtilis phage φ29 encodes the functional homolog of lambda S protein. J Bacteriol. 1993;175:1038–1042. doi: 10.1128/jb.175.4.1038-1042.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 44.von Heijne G. Membrane protein structure prediction hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 45.Walker B, Krishnasastry M, Zorn L, Kasianowicz J, Bayley H. Functional expression of the alpha-hemolysin of Staphylococcus aureus in intact Escherichia coli and in cell lysates. Deletion of five C-terminal amino acids selectively impairs hemolytic activity. J Biol Chem. 1992;267:10902–10909. [PubMed] [Google Scholar]
- 46.Young R. Bacteriophage lysis: mechanism and regulation. Microbiol Rev. 1992;56:430–481. doi: 10.1128/mr.56.3.430-481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Young R, Bläsi U. Holins: form and function in bacteriophage lysis. FEMS Microbiol Rev. 1995;17:191–205. doi: 10.1111/j.1574-6976.1995.tb00202.x. [DOI] [PubMed] [Google Scholar]
- 48.Zagotta M T, Wilson D B. Oligomerization of the bacteriophage lambda S protein in the inner membrane of Escherichia coli. J Bacteriol. 1990;172:912–921. doi: 10.1128/jb.172.2.912-921.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ziermann R, Bartlett B, Calendar R, Christie G E. Functions involved in bacteriophage P2-induced host cell lysis and identification of a new tail gene. J Bacteriol. 1994;176:4974–4984. doi: 10.1128/jb.176.16.4974-4984.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]