Abstract
Quantitative polymerase chain reaction (qPCR) is widely used in detection of nucleic acids, but existing methods either lack sequence-specific detection or are costly because they use chemically modified DNA probes. In this work, we apply a DNA aptamer and light-up dye-based chemistry for qPCR for nucleic acid quantification. In contrast to the conventional qPCR, in our method, we observe an exponential decrease in fluorescence upon DNA amplification. The qPCR method we developed produced consistent Ct vs log10 (DNA amount) standard curves, which have a linearfit with R2 value > 0.99. This qPCR technique was validated by quantifying gene targets from Streptococcus zooepidemicus (SzhasB) and Mycobacterium tuberculosis (MtrpoB). We show that our strategy is able to successfully detect DNA at as low as 800 copies/μL. To the best of our knowledge, this is the first study demonstrating the application of light-up dyes and DNA aptamers in qPCR.
Introduction
Quantitative PCR (qPCR) has revolutionized the specific detection and quantification of targeted nucleic acid sequences.1 It has been successfully applied in biomedical research, diagnostics, species identification, and nucleic acid quantification.2,3 Currently available qPCR methods to monitor amplification rely on modified-DNA hydrolysis probes or DNA-intercalating fluorescent dyes that bind to double-stranded DNA.2,4−7 The oligonucleotide hydrolysis probes are capable of multiplexed detection as they are sequence-specific but need to be custom designed and are expensive due to the chemical modifications.2,7 On the other hand, fluorescent dyes, which bind non-specifically to any DNA sequence being amplified are prone to false positives and cannot be multiplexed.2,7 Most advances in PCR-based nucleic acid detection strategies involve these two fundamental chemistries or their variations.8 Other technologies include quantification of PCR amplification using un-natural bases in the plexor technology,9 molecular beacons,10,11 scorpion probes,12 LNA-based probes,13 or other chemically modified technologies.7 However, all of these methods contain the limitations of current methods, requiring chemically modified primers or probes.
Recently, aptamers have been used for detection applications in diagnostics and therapeutics.14 Aptamers are short sequences of single-stranded DNA or RNA molecules that can form secondary and tertiary structures, which allows them to bind to their targets with high affinity.15 Aptamers have been highly successful due to their low cost of production, high stability, low batch-to-batch variability and ease of integration into complex genetic circuits for target detection.14 RNA-aptamers were initially used to develop GFP-mimics where aptamer binding enhances the fluorescence of a GFP fluorophore analogue DMHBI or other dyes significantly.10,16,17 Since then, light-up RNA aptamers have been used for a plethora of applications, including sensors for in vivo monitoring of biomolecules.18−20 A single DNA-based light-up dye system was developed in 2008 for Hoescht dye, but DNA-based systems were ignored due to a lack of their applications in vivo.21 Recently, various DNA-based light-up aptamer systems have been developed for multiple dyes, including dimethyl-indole red (DIR), berberine, thioflavin T, crystal violet, malachite green, DFHBI, and dapoxyl SEDA, for in vitro detection and diagnostic applications.20,22−28
In this work, we employed a novel chemistry for monitoring qPCR amplification based on light-up dye and DNA aptamers. Our method uses the previously described DIR dye and its DNA aptamer DIR2-1 for a proof-of-concept of our qPCR strategy.16,24 The aptamer–dye complex is sequence-specific, unlike fluorescent intercalating dyes, and does not require expensive chemical modifications, such as in hydrolysis probes.2,3 The DIR dye is less fluorescent in its free state, but when bound to its 42 nt DNA aptamer, its fluorescence increases 140-fold.24 It is designed to be specific for its aptamer target, with the bulky dimethyl-indole group preventing intercalation with bases and a negatively charged propylsulfonate group preventing nonspecific binding to nucleic acids.16 Thus, DIR and its aptamer DIR2-1 present an ideal system for testing our qPCR strategy. We demonstrate that our qPCR design detects in real-time, the amplification of DNA with every cycle, and compare our method to conventional DNA intercalating dye (TB green)-based qPCR. Our technique can be used for multiple gene targets using commonly available Taq DNA polymerase and generates linear Ct vs log10 (copy number/μL) plots. Our investigations show that this method is simple, quantitative, and compatible with current instruments and reagents. Further, this method has the potential to significantly bring down the costs of multiplexed qPCR detection for research and diagnostics.
Results and Discussion
Design of the Novel qPCR Strategy
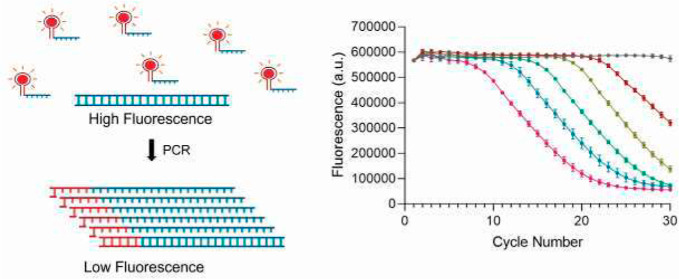
We placed the 42 nt DIR2-1 aptamer sequence upstream of our primer designed for the PCR reaction.24 The DIR dye is added at 400 nM as an additional component in the standard PCR reaction without any other modification. Before the reaction starts, the aptamer upstream of the primer is free to take its 3D conformation and bind specifically to the DIR dye. This bound state emits a high fluorescence. As the PCR reaction progresses, the primers are consumed and made double-stranded, resulting in the aptamer losing conformation and thereby its ability to bind to the DIR dye. This results in a significant decrease in fluorescence with consumption of the aptamer-containing free primers in each cycle (Figure 1a).
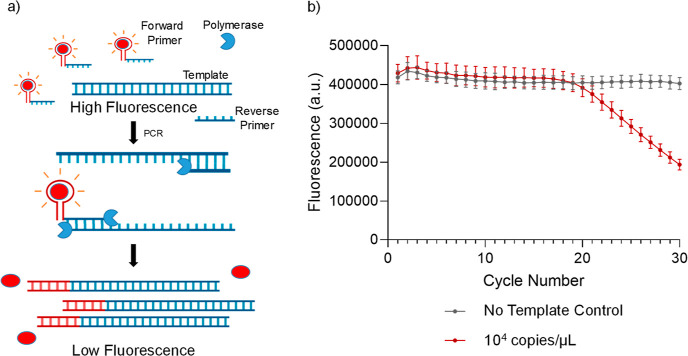
Figure 1.
Quantitative PCR using chemically un-modified DNA primer probes (a) strategy to use light-up DNA-aptamer pair for quantitation in PCR. A red sphere indicates the dye, the blue part of the primer is complementary to the template, and the red part is the dye-specific aptamer (b) DNA aptamer and its partner dye can be used to monitor the progress of a PCR reaction with a decrease in fluorescence. Data represents the mean and standard deviation of n = 3 replicates.
Calibration of DIR Dye-Aptamer Fluorescence on qPCR Instrument
For testing our qPCR strategy, we first calibrated DIR fluorescence in the presence of its aptamer, using 400 nM dye in the presence of 400 nM DIR2-1 aptamer, on the Applied Biosystems QuantStudio 7 Pro Dx real-time PCR system. The highest fluorescence signal on the instrument was obtained using the excitation filter x5:640 ± 10 nm and emission filter m5:682 ± 14 nm (Figure S1).
Detection of qPCR Amplification in Real-Time
Next, we performed qPCR to detect a 96 bp segment of the rpoB gene from Mycobacterium tuberculosis present at 104 copies/μL. We observed the expected decrease in fluorescence with increasing cycles (Figure 1b). This is typical of a quantitative PCR reaction and demonstrates that we can monitor DNA amplification with every cycle by following the decrease in fluorescence in real-time.
Light-up aptamers have been previously combined in different nucleic acid circuits for specific diagnostics and detection including for single nucleotide variants, pathogenic bacteria, and SARS-CoV-2 RNA.28−31 Generally, these methods rely on direct binding of the aptamer to a small amount of the target28 or require prior amplification of the target before detection.31,32 Both strategies lack the real-time quantification capabilities observed for qPCR. Our method combines the qPCR advantages with light-up dye and aptamer-based detection.
A previous study tried to incorporate light-up RNA aptamers for the detection of nucleic acids using PCR amplification but did not monitor the PCR reaction in real-time as the described method required in vitro transcription to generate the RNA aptamers for signal production.33 It involved multiple indirect steps and did not have a cost advantage over the existing technologies.33 Our technique uses light-up DNA aptamers that directly monitor the PCR reaction in real-time and does not require any significant modification to currently available reagents or qPCR instruments.
Standard Curves Generated Using Taq DNA Polymerase
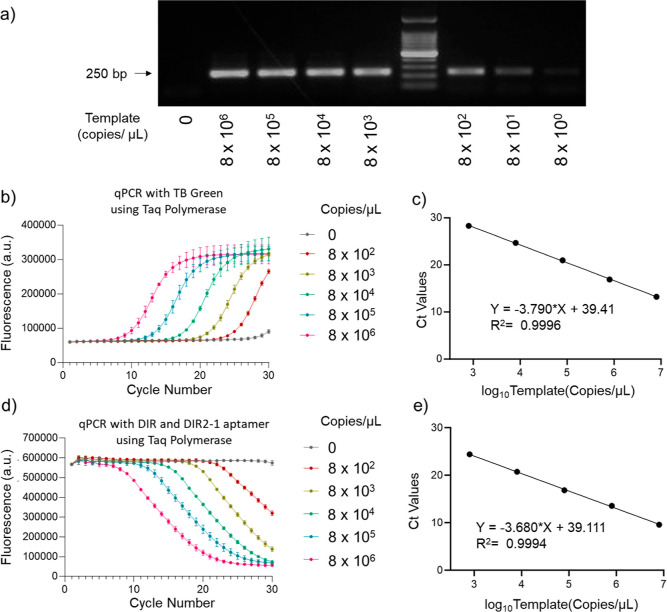
After confirming the proof-of-concept for DNA-aptamer-based nucleic acid detection, we generated a standard curve of Ct values for different sample concentrations.34 We used Taq polymerase to amplify a 208 bp sequence of the hasB gene fromStreptococcus zooepidemicus at different target concentrations. The agarose gel electrophoresis showed that our primers can detect as low as 800 copies/μL template within 30 cycles of PCR (Figure 2a). We tested if the light-up aptamer and DIR dye-based method can sensitively detect the template at the limit of detection and compared it simultaneously to qPCR based on the DNA intercalating dye, TB Green. We observed an exponential increase in fluorescence for TB green (Figure 2b) with a linear Ct value versus log10 (copy number/μL), with an R2 of 0.9996 (Figure 2c).
Figure 2.
qPCR using light-up dye-aptamer and conventional DNA intercalating dye-based method with Taq polymerase and its quantification using Ct values. (a) Agarose gel electrophoresis to determine the limit of detection using the custom designed primers. (b) Exponential increase in fluorescence with the progress of the PCR reaction using Taq polymerase and DNA intercalating dye, TB Green, n = 3 (c) Ct values were calculated from (b) and plotted against the log10 (copies/μL). The curve was fitted to a linear model and the fitted equation with the R2 value is shown. Each curve represents the mean and standard deviation of the replicates. (d) Exponential decrease in fluorescence with the progress of the PCR reaction using Taq polymerase with DIR dye and DIR2-1 aptamer fluorescence detection, n = 3. (e) Ct values were calculated from (d) and plotted against the log10 (copies/μL). The curve was fitted to a linear model and the fitted equation with the R2 value is shown. Each curve represents the mean and standard deviation of the replicates.
DIR and DIR2-1 aptamer-based qPCR chemistry performed similarly to the commercial TB green chemistry with an exponential decrease in fluorescence (Figure 2d). Like with TB green qPCR chemistry, we see a linear decrease (R2 = 0.9994) in the Ct value with an increase in log10 (copy number/μL) between 2.9 and 6.9 (Figure 2c,e). This demonstrates that our method can be easily incorporated with the commercially available PCR kits.
Quantification of Unknown DNA Sample
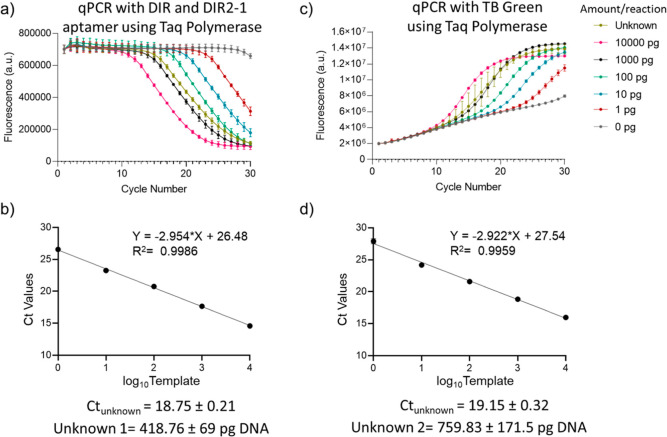
We further validated the method to quantify an unknown amount of DNA target using both the TB green and DIR2-1 aptamer and DIR dye-based chemistries. A standard curve of Ct vs log10 (amount of DNA) of our standards, serially diluted between 1 and 10000 pg/reaction, was plotted using the change in fluorescence during the PCR (Figure 3a–d). 0.1 pg target DNA/reaction corresponds to 800 copies/μL. The standard curve for DIR and DIR2-1 chemistry estimated the amount of DNA in the unknown sample to be 418.76 ± 69 pg, which is close to the prepared double-blinded sample containing 400 pg template (Figure 3b). The method performs equivalent to the TB green chemistry, which determined the unknown concentration as 759.83 ± 171.5 pg, when the sample was prepared double-blinded to contain 800 pg of DNA (Figure 3c,d). Thus, the proof-of-concept strategy described here can be used for the qPCR quantification of unknown nucleic acids in different samples and applied for environmental monitoring and diagnostics.
Figure 3.
Determination of the concentration of an unknown DNA sample with qPCR using conventional and light-up dye-aptamer detection chemistry. (a) Exponential decrease in DIR dye and DIR2-1 aptamer fluorescence with the progress of the PCR reaction using Taq polymerase. (b) Ct values were calculated from (a) and plotted against the log10 (DNA amount). The curve was fitted to a linear model and the fitted equation with the R2 value is shown. Each curves represents the mean and standard deviation of n = 4 replicates. (c) Exponential increase in fluorescence with the progress of the PCR reaction using Taq polymerase and DNA intercalating dye TB green, n = 4. (d) Ct values were calculated from (c) and plotted against the log10 (DNA amount). The curve was fitted to a linear model and the fitted equation with the R2 value is shown. Each curves represents the mean and standard deviation of n = 4 replicates.
Our method provides the first description of the application of light-up DNA aptamer and dye pair for the quantification of amplification in PCR. However, our current design strategy has the probe attached to the primer which could lead to a false positive in the scenario of nonspecific amplification or primer dimers. This could be prevented in the future work by detaching the aptamer probe from the primer and integrating DNA circuit principles including polymerase or toehold-mediated strand displacement, to monitor PCR amplification.35
Conclusions
In this study, we demonstrate that a dye-binding DNA-aptamer can be used for monitoring amplification in qPCR. We obtained linear Ct vs log10 (DNA amount) standard curves with R2 value > 0.99 using a Taq DNA polymerase. Our method can detect nucleic acids as low as 800 copies/μL. We successfully quantitated an unknown DNA sample using the standard curve. Our method is as highly sensitive as conventional methods, cost-effective, and does not require any modified oligonucleotide probes. In the future, this technique will be tested for its capability to multiplex the detection of multiple targets in a single tube using the available wavelength spectrum of DNA-aptamer light-up dyes. It also needs to be optimized for use in reverse-transcription qPCR to detect RNA.
Materials and Methods
Instruments and Reagents
qPCR was performed using Applied Biosystems Quant studio 7 Pro Dx or Quant studio 3. Custom DIR dye in the presence of its aptamer DIR2-1 was successfully calibrated on the qPCR instrument as per the manufacturer’s protocol (Figure S1). Plasmid isolation was done using Qiagen miniprep kit. Taq DNA polymerase was purchased from NEB. The DIR aptamer used in our study is the same as published before (H. Wang et al. 2017). All the oligonucleotides were obtained from Bioserve Biotechnologies, India.
Templates and Primers Used for PCR
A 208 bp fragment from the UDP-glucose 6-dehydrogenase (hasB) gene of S. zooepidemicus and a 96 bp fragment of the DNA-dependent RNA polymerase (RNAP) rpoB gene from M. tuberculosis were amplified using custom-designed primers (Table 1). The template plasmid for hasB gene amplification was pGJP2, a nisin-inducible plasmid with pNZ8148 backbone containing hasA and hasB genes.36 The plasmid template for rpoB gene amplification was a gift from Yaathum Biotech (IIT Madras Research Park).
Table 1. Primers Used in the qPCR Amplification of hasB (GenBank: CP065056.1) and rpoB (GenBank: L27989.1).
| target | primers (42 nt DIR2-1, bold) | nucleotides |
|---|---|---|
| hasB | 5′-atgggctcacaggaggctgag | 203647–203667 |
| hasB | 5′-gacgacgacgctaggaaggcgttggtgggcacgccggtcgtc cctttggcaggcaatagccgc | 203834–203854 |
| rpoB | 5′-tcacgtgacagaccgccg | 2461–2444 |
| rpoB | 5′-gacgacgacgctaggaaggcgttggtgggcacgccggtcgtc ccagctgagccaattcatggg | 2366–2386 |
qPCR with Taq Polymerase Using Light-Up Dye and Aptamer Chemistry
The 18 μL reaction contains 1× Taq buffer, 200 nM primers, 200 μM dNTPs, 400 nM DIR dye, 0.45 units of Taq polymerase (NEB no. M0273), and template DNA as indicated. Reaction conditions for the PCR of hasB or rpoB gene with Taq polymerase include an initial denaturation step at 95 °C for 5 min, followed by 30 cycles of denaturation at 95 °C for 15 s, annealing at 60 °C for 20 s, extension at 68 °C for 25 s, and fluorescence measurement at 25 °C for 25 s.
qPCR with Taq Polymerase Using TB Green, a DNA Intercalating Dye, Based Chemistry
A TB Green Premix Ex TaqII (Tli RNase H Plus) kit (Takara #RR82WR) was used according to the manufacturer’s protocol. In summary, the 20 μL reaction contains 1× TB Green Premix Ex TaqII (Tli RNaseH Plus) master mix, 200 μM primers, 1× ROX reference dye, and template DNA as indicated. Reaction conditions for the PCR of hasB include an initial denaturation step at 95 °C for 30 s, followed by 30 cycles of denaturation at 95 °C for 5 s and a combined annealing and extension step at 60 °C for 30 s.
Normalization of Fluorescence and Standard Curve Analysis
The normalization method used was first described by Higuchi et al., 1993. Briefly, the average of initial fluorescence of samples is calculated. A multiplication factor for each sample is calculated by dividing the initial fluorescence of each sample with the above average. This multiplication factor is used to normalize all of the fluorescence measurements of that sample. For the calculation of Ct value and generation of standard curve, an arbitrary threshold in the linear region of the exponential change in fluorescence is defined to find fractional Ct values, which is then plotted against log10 (amount of initial DNA).
Acknowledgments
We would like to thank Dr. Bruce Armitage from Carnegie Mellon University for generously providing us with Dimethylindole Red dye. We also thank IIT Madras Bioincubator for providing research facilities and space.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/acsomega.3c07599.
Fluorescence calibration of DIR dye with DIR2-1 aptamer (PDF)
Author Present Address
† Department of Molecular Biosciences, University of Texas at Austin, Austin, TX, USA
Author Contributions
S.G. conceptualized the project. S.G., P.S., S.S., A.B.M., and G.J. contributed to experimental design. S.G., P.S., S.S., and A.B.M. performed the experiments. The manuscript was written with contributions from all authors, and all authors have given approval to the final version of the manuscript.
Research in the laboratory was supported by Student Innovation Grant from the Indian Institute of Technology Madras.
The authors declare no competing financial interest.
Special Issue
Published as part of ACS Omegavirtual special issue “Nucleic Acids: A 70th Anniversary Celebration of DNA”.
Supplementary Material
References
- Higuchi R.; Fockler C.; Dollinger G.; Watson R. Kinetic PCR Analysis: Real-Time Monitoring of DNA Amplification Reactions. Nat. Biotechnol. 1993, 11 (9), 1026–1030. 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- Arya M.; Shergill I. S.; Williamson M.; Gommersall L.; Arya N.; Patel H. R. Basic Principles of Real-Time Quantitative PCR. Expert Rev. Mol. Diagn. 2005, 5 (2), 209–219. 10.1586/14737159.5.2.209. [DOI] [PubMed] [Google Scholar]
- Khodakov D.; Wang C.; Zhang D. Y. Diagnostics Based on Nucleic Acid Sequence Variant Profiling: PCR, Hybridization, and NGS Approaches. Adv. Drug Delivery Rev. 2016, 105, 3–19. 10.1016/j.addr.2016.04.005. [DOI] [PubMed] [Google Scholar]
- Schneeberger C.; Speiser P.; Kury F.; Zeillinger R. Quantitative Detection of Reverse Transcriptase-PCR Products by Means of a Novel and Sensitive DNA Stain. Genome Res. 1995, 4 (4), 234–238. 10.1101/gr.4.4.234. [DOI] [PubMed] [Google Scholar]
- Holland P. M.; Abramson R. D.; Watson R.; Gelfand D. H. Detection of Specific Polymerase Chain Reaction Product by Utilizing the 5′ → 3′ Exonuclease Activity of Thermus Aquaticus DNA Polymerase. Proc. Natl. Acad. Sci. U.S.A. 1991, 88 (16), 7276–7280. 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heid C. A.; Stevens J.; Livak K. J.; Williams P. M. Real Time Quantitative PCR. Genome Res. 1996, 6 (10), 986–994. 10.1101/gr.6.10.986. [DOI] [PubMed] [Google Scholar]
- Navarro E.; Serrano-Heras G.; Castaño M.; Solera J. Real-Time PCR Detection Chemistry. Clin. Chim. Acta 2015, 439, 231–250. 10.1016/j.cca.2014.10.017. [DOI] [PubMed] [Google Scholar]
- Marras S. A. E.; Tyagi S.; Kramer F. R. Real-Time Assays with Molecular Beacons and Other Fluorescent Nucleic Acid Hybridization Probes. Clin. Chim. Acta 2006, 363 (1–2), 48–60. 10.1016/j.cccn.2005.04.037. [DOI] [PubMed] [Google Scholar]
- Sherrill C. B.; Marshall D. J.; Moser M. J.; Larsen C. A.; Daudé-Snow L.; Prudent J. R. Nucleic Acid Analysis Using an Expanded Genetic Alphabet to Quench Fluorescence. J. Am. Chem. Soc. 2004, 126 (14), 4550–4556. 10.1021/ja0315558. [DOI] [PubMed] [Google Scholar]
- Paige J. S.; Wu K. Y.; Jaffrey S. R. RNA Mimics of Green Fluorescent Protein. Science 2011, 333 (6042), 642–646. 10.1126/science.1207339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nazarenko I. A.; Bhatnagar S. K.; Hohman R. J. A Closed Tube Format for Amplification and Detection of DNA Based on Energy Transfer. Nucleic Acids Res. 1997, 25 (12), 2516–2521. 10.1093/nar/25.12.2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitcombe D.; Theaker J.; Guy S. P.; Brown T.; Little S. Detection of PCR Products Using Self-Probing Amplicons and Fluorescence. Nat. Biotechnol. 1999, 17 (8), 804–807. 10.1038/11751. [DOI] [PubMed] [Google Scholar]
- Costa J. M.; Ernault P.; Olivi M.; Gaillon T.; Arar K. Chimeric LNA/DNA Probes as a Detection System for Real-Time PCR. Clin. Biochem. 2004, 37 (10), 930–932. 10.1016/j.clinbiochem.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Dunn M. R.; Jimenez R. M.; Chaput J. C. Analysis of Aptamer Discovery and Technology. Nat. Rev. Chem 2017, 110, 76. 10.1038/s41570-017-0076. [DOI] [Google Scholar]
- Adachi T.; Nakamura Y. Aptamers: A Review of Their Chemical Properties and Modifications for Therapeutic Application. Molecules 2019, 24 (23), 4229. 10.3390/molecules24234229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Constantin T. P.; Silva G. L.; Robertson K. L.; Hamilton T. P.; Fague K.; Waggoner A. S.; Armitage B. A. Synthesis of New Fluorogenic Cyanine Dyes and Incorporation into RNA Fluoromodules. Org. Lett. 2008, 10 (8), 1561–1564. 10.1021/ol702920e. [DOI] [PubMed] [Google Scholar]
- Babendure J. R.; Adams S. R.; Tsien R. Y. Aptamers Switch on Fluorescence of Triphenylmethane Dyes. J. Am. Chem. Soc. 2003, 125 (48), 14716–14717. 10.1021/ja037994o. [DOI] [PubMed] [Google Scholar]
- Ouellet J. RNA Fluorescence with Light-Up Aptamers. Front. Chem. 2016, 4, 29. 10.3389/fchem.2016.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Litke J. L.; Jaffrey S. R. Highly Efficient Expression of Circular RNA Aptamers in Cells Using Autocatalytic Transcripts. Nat. Biotechnol. 2019, 376, 667–675. 10.1038/s41587-019-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T.; Luo Y.; Li W.; Cao Y.; Pei R. Progress in the Isolation of Aptamers to Light-up the Dyes and the Applications. Analyst 2020, 145 (3), 701–718. 10.1039/C9AN01825E. [DOI] [PubMed] [Google Scholar]
- Sando S.; Narita A.; Aoyama Y. Light-Up Hoechst-DNA Aptamer Pair: Generation of an Aptamer-Selective Fluorophore from a Conventional DNA-Staining Dye. ChemBioChem 2007, 8 (15), 1795–1803. 10.1002/cbic.200700325. [DOI] [PubMed] [Google Scholar]
- Wang J.; Zhang Y.; Wang H.; Chen Y.; Xu L.; Gao T.; Pei R. Selection and Analysis of DNA Aptamers to Berberine to Develop a Label-Free Light-up Fluorescent Probe. New J. Chem. 2016, 40 (11), 9768–9773. 10.1039/C6NJ02290A. [DOI] [Google Scholar]
- Wang H.; Wang J.; Xu L.; Zhang Y.; Chen Y.; Chen H.; Pei R. Selection and Characterization of Thioflavin T Aptamers for the Development of Light-up Probes. Anal. Methods 2016, 8 (48), 8461–8465. 10.1039/C6AY02890J. [DOI] [Google Scholar]
- Wang H.; Wang J.; Wang Q.; Chen X.; Liu M.; Chen H.; Pei R. Selection and Characterization of Dimethylindole Red DNA Aptamers for the Development of Light-up Fluorescent Probes. Talanta 2017, 168, 217–221. 10.1016/j.talanta.2017.03.041. [DOI] [PubMed] [Google Scholar]
- Wang H.; Wang J.; Sun N.; Cheng H.; Chen H.; Pei R. Selection and Characterization of Malachite Green Aptamers for the Development of Light-up Probes. ChemistrySelect 2016, 1 (8), 1571–1574. 10.1002/slct.201600154. [DOI] [Google Scholar]
- Kato T.; Shimada I.; Kimura R.; Hyuga M. Light-up Fluorophore-DNA Aptamer Pair for Label-Free Turn-on Aptamer Sensors. Chem. Commun. 2016, 52 (21), 4041–4044. 10.1039/C5CC08816J. [DOI] [PubMed] [Google Scholar]
- Chen Y.; Wang J.; Zhang Y.; Xu L.; Gao T.; Wang B.; Pei R. Selection and Characterization of a DNA Aptamer to Crystal Violet. Photochem. Photobiol. Sci. 2018, 17 (6), 800–806. 10.1039/c7pp00457e. [DOI] [PubMed] [Google Scholar]
- VarnBuhler B. S.; Moon J.; Dey S. K.; Wu J.; Jaffrey S. R. Detection of SARS-CoV-2 RNA Using a DNA Aptamer Mimic of Green Fluorescent Protein. ACS Chem. Biol. 2022, 17 (4), 840–853. 10.1021/acschembio.1c00893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolpashchikov D. M.; Spelkov A. A. Binary (Split) Light-up Aptameric Sensors. Angew Chem. Int. Ed. Engl. 2021, 60 (10), 4988–4999. 10.1002/anie.201914919. [DOI] [PubMed] [Google Scholar]
- Zhang T.; Zhou W.; Lin X.; Khan M. R.; Deng S.; Zhou M.; He G.; Wu C.; Deng R.; He Q. Light-up RNA Aptamer Signaling-CRISPR-Cas13a-Based Mix-and-Read Assays for Profiling Viable Pathogenic Bacteria. Biosens. Bioelectron. 2021, 176, 112906. 10.1016/j.bios.2020.112906. [DOI] [PubMed] [Google Scholar]
- O’Steen M. R.; Kolpashchikov D. M. A Self-Assembling Split Aptamer Multiplex Assay for SARS-COVID19 and Miniaturization of a Malachite Green DNA-Based Aptamer. Sens. Actuators Rep. 2022, 4, 100125. 10.1016/J.SNR.2022.100125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuchi N.; Reed A.; Gerasimova Y. V.; Kolpashchikov D. M. Split Dapoxyl Aptamer for Sequence-Selective Analysis of Nucleic Acid Sequence Based Amplification Amplicons. Anal. Chem. 2019, 91 (4), 2667–2671. 10.1021/acs.analchem.8b03964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasmeen A.; Du F.; Zhao Y.; Dong J.; Chen H.; Huang X.; Cui X.; Tang Z. Sequence-Specific Biosensing of DNA Target through Relay PCR with Small-Molecule Fluorophore. ACS Chem. Biol. 2016, 11 (7), 1945–1951. 10.1021/acschembio.5b01081. [DOI] [PubMed] [Google Scholar]
- Bustin S. A.; Benes V.; Garson J. A.; Hellemans J.; Huggett J.; Kubista M.; Mueller R.; Nolan T.; Pfaffl M. W.; Shipley G. L.; Vandesompele J.; Wittwer C. T. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55 (4), 611–622. 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- Wang B.; Thachuk C.; Ellington A. D.; Winfree E.; Soloveichik D. Effective Design Principles for Leakless Strand Displacement Systems. Proc. Natl. Acad. Sci. U.S.A. 2018, 115 (52), E12182–E12191. 10.1073/pnas.1806859115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeeva P.; Shanmuga Doss S.; Sundaram V.; Jayaraman G. Production of Controlled Molecular Weight Hyaluronic Acid by Glucostat Strategy Using Recombinant Lactococcus Lactis Cultures. Appl. Microbiol. Biotechnol. 2019, 103 (11), 4363–4375. 10.1007/s00253-019-09769-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.