Abstract
Bacillus subtilis produces a 35-kDa cell wall hydrolase, CwlF, during vegetative growth. The CwlF protein was extracted from B. subtilis cwlB sigD mutant cells and separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. N-terminal amino acid sequencing revealed that its sequence is completely identical to that of the internal region of the papQ gene product. Disruption of the papQ gene in the B. subtilis chromosome led to the complete loss of CwlF, indicating that papQ is identical to cwlF. CwlF exhibits high sequence similarity to the p60 proteins of Listeria species, NlpC proteins of Escherichia coli and Haemophilus influenzae, and Enp2 protein of Bacillus sphaericus. The β-galactosidase activity of the cwlF-lacZ transcriptional fusion and Northern blot analysis of the cwlF gene indicated that the gene is expressed as a monocistronic operon during the exponential growth phase, and primer extension analysis suggested that the cwlF gene is transcribed mainly by EςA RNA polymerase and weakly by EςH RNA polymerase. While the cells of the cwlF-deficient mutant were about twice as long as those of the wild-type strain, the cwlF sigD double mutant cells exhibited extraordinary microfiber formation, in contrast to the filamentation of the sigD mutant. The CwlF production was not affected by the pleiotropic mutations flaD1 and degU32(Hy), which endow cells with the ability of extensive filamentation.
Bacillus subtilis produces a complement set of enzymes capable of hydrolyzing the shape-maintaining and stress-bearing peptidoglycan layer of its own cell wall (8, 43, 50). Some of these peptidoglycan hydrolases can trigger cell lysis; therefore, they can truly be called autolysins or suicide enzymes (43). Autolysins have been implicated in several important cellular processes, such as cell wall turnover, cell separation, competence, and flagellation (motility), in addition to cell lysis, and they act as pacemaker and space-maker enzymes for cell wall growth (3, 9, 10, 38, 43). Therefore, fine-tuning of autolysin activity through efficient and strict regulation is a must for bacterial survival (17).
Two major vegetative-phase autolysins (a 50-kDa N-acetylmuramoyl-l-alanine amidase [amidase], CwlB [LytC], and a 90-kDa endo-β-N-acetylglucosaminidase [glucosaminidase], CwlG [LytD]) were initially purified and characterized from B. subtilis (16, 44). The cwlB gene is part of an operon containing sequences encoding a putative lipoprotein and a modifier protein and containing the cwlB gene, in that order (25, 29). Transcription of this operon proceeds from a distal ςA-type promoter and a proximal ςD-type one, the latter transcript being predominant in the exponential growth phase (23, 26). The cwlG gene has also been cloned by two groups (33, 41), and it is transcribed mainly, as a monocistronic operon, by EςD RNA polymerase (33, 41). A study on the physiological functions of CwlB and CwlG revealed that CwlB is responsible for cell lysis in the stationary phase (25) and after cold shock treatment (58) and that both proteins, but only in concert, are required for the motility function.
Several other amidase genes and their homologs have been cloned for the genus Bacillus. From B. subtilis, two prophage-borne amidase genes (cwlA and xlyA) (12, 24, 31), a sporulation-specific amidase gene (cwlC) (13, 22), a cortex maturation-specific and deduced amidase gene (cwlD) (48), and a germination-specific and deduced amidase gene (sleB) (35) have been cloned and studied, in addition to cwlB (25). Two amidase genes (cwlM and cwlL) from Bacillus licheniformis, a cell wall hydrolase (probably an amidase) gene from Bacillus species, and an amidase gene (sleB) from Bacillus cereus have been cloned and studied (27, 32, 36, 37, 39). Recently, the cwlB cwlC double mutant was found to be resistant to mother cell lysis during the late stage of sporulation (51). Evidence that many of these amidases are composed of a cell wall-specific and binding domain and a catalytic domain has accumulated (27). On the basis of the amino acid sequence similarity in their catalytic domains, these amidases can be classified into three groups. Class I includes CwlA, CwlL, XlyA, and Bacillus sp. amidase, class II includes CwlB, CwlC, CwlD, and CwlM, and class III includes the SleB proteins of B. subtilis and B. cereus. The cell wall-specific and binding domains of these amidases contain several (usually two or three) tandem repeated sequences. Interestingly, three tandem repeated sequences have also been observed in the noncatalytic cell wall-binding proteins CwbA (LytB) and WapA (14, 23).
The genome sequencing project on B. subtilis has revealed many cell wall hydrolase gene homologs (47). One of the homologs is the papQ gene, whose product was submitted to protein databases, as a phosphatase-associated and cell wall turnover-related protein precursor, by Whalen and Piggot (55). The papQ gene encodes a 334-amino-acid polypeptide having a molecular mass of 35,455 Da.
In this study, we identified papQ as a new cell wall hydrolase gene, cwlF, during the vegetative growth phase of B. subtilis, characterized the gene expression, and determined the cell morphology of the cwlF and cwlF sigD mutants.
(Preliminary data were presented at the 9th International Conference on Bacilli [Lausanne, Switzerland, 15 to 19 July 1997].)
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains of B. subtilis and Escherichia coli and the plasmids used in this study are described in Table 1. B. subtilis was grown on nutrient agar medium (Difco) at 30°C for 10 to 12 h and then inoculated into DSM (Schaeffer) medium (46), followed by a shake culture at 37°C. If necessary, tetracycline, chloramphenicol, and erythromycin were added to the medium to final concentrations of 15, 8, and 0.3 μg/ml, respectively. E. coli was grown in Luria-Bertani (LB) medium (45) at 37°C. If necessary, ampicillin was added to a final concentration of 50 or 100 μg/ml.
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Genotype | Source or reference |
|---|---|---|
| B. subtilis | ||
| 168 | trpC2 | D. Ehrlich |
| 1A165 | trpC2 degU32(Hy) | BGSCa |
| AC327 | purB his-1 smo-1 | 1 |
| AC334 | purB his-1 smo-1 flaD1(sinR) | 26 |
| 327SD1 | purB his-1 smo-1 sigD::tet | 42 |
| AN8SD1 | purB his-1 smo-1 sigD::tet cwlB::cat | 42 |
| EFD | trpC2 papQ(cwlF)::pM2cF | This study |
| 327FD | purB his-1 smo-1 papQ(cwlF)::pM2cF | This study |
| SD1F | purB his-1 smo-1 sigD::tet papQ(cwlF):: pM2cF | This study |
| ABDF | purB his-1 smo-1 sigD::tet cwlB::cat papQ(cwlF)::pM2cF | This study |
| 327UH | his-1 smo-1 degU32(Hy) | 53 |
| E. coli | ||
| JM109 | recA1 Δ(lac-proAB) endA1 gyrA96 thi-1 hsdR17 relA1 supE44 [F′:traD36 proAB+lacIqlacZΔM15] | Takara |
| C600 | supE44 hsdR thi-1 thr-1 leuB6 lacY1 tonA21 | Laboratory stock |
| Plasmids | ||
| pMUTin2 | lacZ lacI bla erm | D. Ehrlich |
| pM2cF | lacZ lacI bla erm ΔpapQ(cwlF) | This study |
| pGEM-3Zf(+) | lacZ bla | Promega |
| pGEMΔES | lacZ bla | This study |
| pGEMΔES-cwlF | lacZ bla | This study |
BGSC, Bacillus Genetic Stock Center, Ohio State University.
Preparation of the CwlF protein.
B. subtilis cells were harvested at various times, followed by washing with T buffer (25 mM Tris-HCl, pH 7.2, containing 1 mM phenylmethylsulfonyl fluoride). Then the cells were suspended in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer (28), followed by boiling for 10 min. The suspensions were centrifuged at 11,000 × g for 5 min, and the supernatants (extract S [42]) were used as samples for SDS-PAGE and zymography.
Preparation of cell wall.
Cell wall of B. subtilis 168S was prepared essentially as described previously (10, 24).
SDS-PAGE and zymography.
SDS-PAGE of proteins was performed in 10 or 12% (wt/vol) polyacrylamide gels as described by Laemmli (28). Zymography was performed essentially as described by Leclerc and Asselin (30), using SDS-polyacrylamide gels containing 0.1% (wt/vol) B. subtilis cell wall as a substrate.
N-terminal amino acid sequence.
After SDS-PAGE, peptides in the gel were transferred to a polyvinylidene difluoride membrane (Millipore) and then the gel was stained with Coomassie brilliant blue as described previously (25). The N-terminal amino acid sequence of the CwlF protein was determined with an automatic protein sequencer (model LF-3000; Beckman).
Plasmid construction.
To remove extra cloning sites, pGEM-3Zf(+) was digested with EcoRI and SmaI, blunt ended with mung bean nuclease, and then self-ligated. The resultant plasmid was designated pGEMΔES. To construct a B. subtilis cwlF mutant, an internal fragment in the cwlF gene was amplified by PCR with two primers, cFHF forward primer (5′-GCCGAAGCTTC15ATTACAGCTACGACAGC32; the internal sequence of the cwlF region is italicized, the numbers are with respect to the first A of the translational start codon of cwlF, and the HindIII site is underlined) and cFBR reverse primer (5′-GCGCGGATCCA250GGAAGAAGAACTGCTGC233; the sequence complementary to the internal region of cwlF is italicized and the BamHI site is underlined), and B. subtilis 168 DNA as a template. The PCR fragment was digested with HindIII and BamHI. Plasmids pMUTin2 and pGEMΔES were digested with HindIII and BamHI and then ligated into the digested PCR fragment. Transformation of E. coli C600 and JM109 was performed with the former and latter ligated solutions, respectively. The resulting pM2cF DNA was used for the transformation of B. subtilis strains, and the resulting pGEMΔES-cwlF DNA was used for Northern blot analysis. Insertion of the PCR fragment into pMUTin2 was confirmed by PCR with the primer PM-FK (5′-CGGGGTACCG−113TGTGGAATTGTGAGCG−97; the pMUTin2 sequence is italicized, the numbers are with respect to the first G of the translational start codon of lacZ, and the KpnI site is underlined) and primer PM-RX2 (5′-CCGCTCGAGG64ATTAAGTTGGGTAACGC47; the numbers are with respect to the complementary base of the first G of the translational start codon of lacZ, and the XhoI site is underlined).
Mutant construction.
A cwlF-deficient mutant, EFD, was constructed by transformation of B. subtilis 168 with pM2cF. Disruption of the cwlF gene by means of Campbell-type recombination was confirmed by Southern hybridization analysis (45) with an RNA probe. To construct isogenic disruptants, DNA from the EFD strain was used for the transformation of AC327, 327SD1, and AN8SD1, and transformants were selected on LB agar plates containing erythromycin. Disruption of the cwlF gene in the resultant disruptants, 327FD, SD1F, and ABDF, was confirmed by Southern hybridization analysis.
Transformation of E. coli and B. subtilis.
E. coli transformation was performed as described by Sambrook et al. (45), and B. subtilis transformation was performed by the competent-cell method (2).
β-Galactosidase assay.
The β-galactosidase assay was performed basically as described by Shimotsu and Henner (49). One unit of β-galactosidase activity was defined as the amount of enzyme necessary to release 1 nmol of 2-nitrophenol from o-nitrophenyl-β-d-galactopyranoside (ONPG) in 1 min.
Northern blot and primer extension analyses.
B. subtilis AC327 cells (optical density at 600 nm [OD600] of 15) cultured in DSM medium were harvested and then suspended in 1 ml of chilled killing buffer (20 mM Tris-HCl, pH 7.5, containing 5 mM MgCl2 and 20 mM Na2N3) (54). After centrifugation at 11,000 × g for 1 min at 4°C, the pellet was suspended in 1 ml of SET buffer containing lysozyme (final concentration, 6 mg/ml) (26). After incubation for 10 min at 0°C, the suspension was centrifuged at 11,000 × g for 1 min at 4°C. The pellet was used for RNA preparation with Isogen (Nippon Gene) according to the manufacturer’s instructions. Agarose-formaldehyde gel electrophoresis was performed as described by Sambrook et al. (45). The transfer of RNAs onto a nylon membrane (Magnagraph; Micron Separations) was performed with a vacuum blotter (model BE-600; BIOCRAFT). The DNA fragment used for preparing an RNA probe was amplified by PCR with M13(−21) and M13RV (Takara) as primers and pGEMΔES-cwlF DNA, containing the internal region of cwlF, as a template. The amplified fragment was digested with HindIII, and then fragments were purified by phenol and chloroform treatments followed by precipitation with ethanol. The RNA probe was prepared with a digoxigenin RNA labeling kit (Boehringer Mannheim), and Northern (RNA) hybridization was performed according to the manufacturer’s instructions. Primer extension analysis was performed as described previously (48), using the PEX-cF2 primer (5′-AGATCCCATAACGTGTCG; the 5′ and 3′ ends correspond to the complementary nucleotides at positions 116 and 99 with respect to the 5′ end of the cwlF gene).
Microscopic observation and determination of cell density.
Cells were shake cultured in a test tube (17-mm diameter) containing 5 ml of LB medium at 37°C. After 4, 6, and 8 h, 5-μl samples were mixed with 5 μl of 2% agarose on glass slides, and then the cell morphology was observed by phase-contrast microscopy. The OD600 was measured after strong vortexing of samples. In the case of the sigD cwlF mutant cells after 6 h of incubation, a small amount of lysozyme was added to the samples just before vortexing.
RESULTS AND DISCUSSION
Production of a vegetative cell wall hydrolase, CwlF.
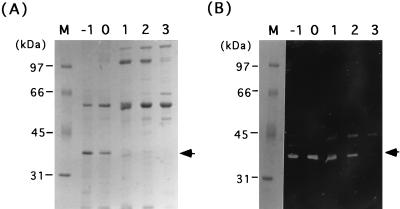
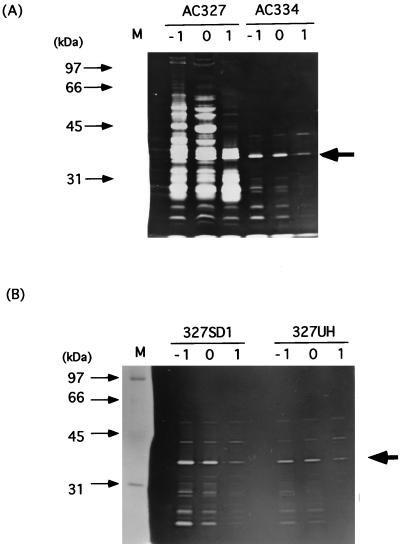
Cell wall extracts of B. subtilis AC327 in the exponential growth phase in modified Spizizen medium gave two strong cell wall-hydrolyzing bands during zymography after electrophoresis on an SDS-polyacrylamide gel containing B. subtilis cell wall (42). One of the bands corresponded to the major autolysin, CwlB (50 kDa), which overlapped with that of a new cell wall hydrolase, CwlE. The new cell wall hydrolase, CwlF, has a molecular mass of 35 kDa, as judged by SDS-PAGE. A ςD-deficient strain lost the ability to produce CwlE but not CwlF (42). Since it was expected that the cwlB sigD-deficient mutant would exhibit a reduction in the total amount of cell wall binding proteins other than CwlF, extract S of a sigD cwlB-deficient mutant, AN8SD1, was applied to an SDS–10% polyacrylamide gel containing 0.1% (wt/vol) B. subtilis cell wall. Figure 1 shows SDS-PAGE of the cell wall proteins. The 35-kDa protein band in panel A, corresponding to the cell wall-hydrolyzing band in panel B, was well separated from those of other proteins (Fig. 1A). Therefore, we prepared a sample at t−1 (1 h before the onset of sporulation) which was transferred to a polyvinylidene difluoride membrane. Then the N-terminal amino acid sequence of CwlF was determined to be QSIKVKKGDTLWDLSRKYDT.
FIG. 1.
Time course of production of CwlF. SDS-PAGE (A) and zymography (B) of protein extracts of B. subtilis AN8SD1 (sigD cwlF) cultured in DSM medium at 37°C are shown. Electrophoresis was performed in SDS–10% polyacrylamide gels, and the zymographic gel (B) contained 0.1% (wt/vol) B. subtilis cell wall as the substrate. Extracts S were prepared, and protein amounts equivalent to 4 OD600 units of cell growth were applied to each lane. Lane M contained protein standards (Bio-Rad), the molecular masses of which are shown on the left. Lanes −1 to 3 correspond to t−1, t0, t1, t2, and t3, respectively. The arrowheads indicate the CwlF protein.
Identity of CwlF to the papQ gene product.
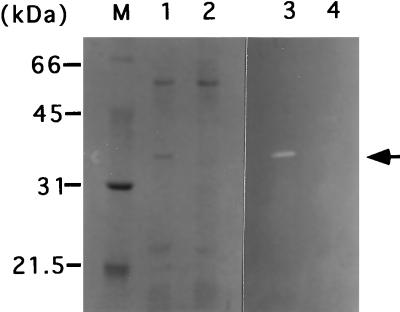
Comparison of the N-terminal amino acid sequence with those of proteins in a nonredundant protein database revealed that the sequence is completely identical to the internal 20-amino-acid sequence starting at position 26 (with respect to the N-terminal amino acid) of PapQ. The papQ gene encodes a 334-amino-acid polypeptide with a molecular mass of 35,455 Da (55). PapQ has two positively charged amino acids, K2 and K3 at the N terminus, followed by a hydrophobic core (from A11 to A23) and a deduced signal peptidase cleavage site (A23SA↓Q26 [the arrow indicates the cleavage site]). These results strongly suggest that the CwlF protein is identical to the PapQ protein. Therefore, the 236-bp internal region of the papQ gene was amplified by PCR with primers cFHF and cFBR and 168 DNA and then inserted into pMUTin2. The resultant plasmid, pM2cF, was introduced into strain 168, resulting in the EFD strain, and then the isogenic strains, AC327, 327SD1, and AN8SD1, were transformed with DNA of the EFD strain. Figure 2 shows SDS-PAGE and zymography of extract S proteins from the papQ-proficient AN8SD1 strain and papQ-deficient ABDF strain. The 35-kDa protein, having cell wall hydrolase activity, was completely lacking in the ABDF strain (Fig. 2, lanes 2 and 4). Moreover, the introduction of a plasmid containing the entire papQ gene into E. coli led to the appearance of a 35-kDa cell wall-hydrolytic band in zymography (19). These results definitely indicate that CwlF is identical to PapQ.
FIG. 2.
SDS-PAGE and zymography of B. subtilis ABDF (sigD cwlB cwlF) and AN8SD1 (sigD cwlB). Electrophoresis was performed in an SDS–12% polyacrylamide gel. Proteins were stained with Coomassie brilliant blue (lanes 1 and 2) or subjected to zymography (lanes 3 and 4). Samples were prepared as described in the legend to Fig. 1. Lane M contained protein standards (Bio-Rad), the molecular masses of which are shown on the left. Lanes 1 and 3, extract S of AN8SD1; lanes 2 and 4, extract S of ABDF. The arrowhead indicates the CwlF protein.
Amino acid sequence similarity of CwlF (PapQ) with other proteins.
The CwlF (PapQ) protein comprises a 25-amino-acid signal peptide region, three tandem repeated regions with three polyserine regions, and a C-terminal domain (Fig. 3). The C-terminal domain consists of 119 amino acid residues and exhibits 40.3% identity over 119 amino acids with that of p60 protein (Iap) of Listeria monocytogenes (21). Moreover, there is significant similarity in their N-terminal regions (25.1% identity over 227 amino acids). The p60 protein is associated with virulence in the mouse model of infection and possesses murein hydrolase activity (56). The C-terminal region of the CwlF protein also exhibits a high degree of sequence similarity with the C-terminal regions of p60 proteins from the different Listeria species (5). E. coli NlpC, comprising 154 amino acid residues, and Haemophilus influenzae NlpC, comprising 183 amino acid residues, also exhibit high degrees of sequence similarity (40.7 and 38.3% over 108 and 115 amino acid residues, respectively) with the C-terminal domain of CwlF (Fig. 3) (11, 20). NlpCs contain lipoprotein motif. Moreover, Bacillus sphaericus endopeptidase, Enp2, comprising 271 amino acids, exhibits a high degree of sequence similarity (31.3% over 112 amino acids) with the C terminus of CwlF (18). On the other hand, the repeated sequence in the N-terminal region of CwlF exhibits similarity with the repeated ones in the C-terminal regions of Lactococcus lactis muramidase AcmA (6), Streptococcus faecalis autolysin (4), and Enterococcus hirae muramidase-2 (7). These three cell wall hydrolases contain regions with high degrees of sequence similarity in their N termini, which encompass the active-site regions (6). Therefore, CwlF is not an AcmA-type muramidase. The amino acid sequence of CwlF indicates that it is a novel type of cell wall hydrolase in B. subtilis.
FIG. 3.
Alignment of the amino acid sequences of cell wall hydrolases B. subtilis CwlF (CwlF_BACSU; 334 amino acids) (55), L. monocytogenes p60 (p60_LISMO; 484 amino acids) (21), and B. sphaericus Enp2 (ENP2_BACSH; 271 amino acids) (18) and the deduced lipoproteins E. coli NlpC (NLPC_ECOLI; 154 amino acids) (20) and H. influenzae NlpC (NLPC_HAEIN; 183 amino acids) (11). Amino acid identities are indicated by two types of shading, and amino acids identical among the five proteins are indicated by asterisks. Amino acids are numbered from the N termini of the proteins. Dashes indicate the introduction of gaps in the alignment, and arrows and double overlines indicate tandem repeated regions and polyserine regions in CwlF, respectively. The arrowhead indicates a deduced signal peptidase cleavage site.
Regulation of the cwlF gene.
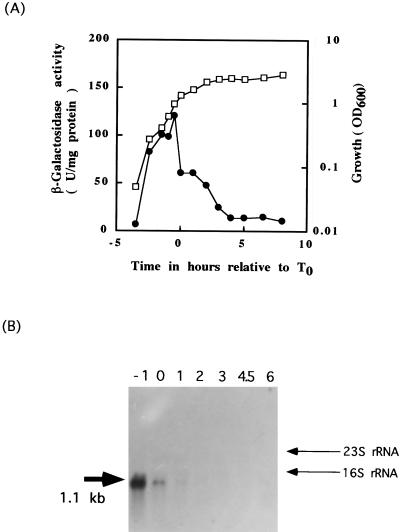
To study the expression of the cwlF gene in vivo, a cwlF-lacZ transcriptional fusion gene was constructed in the AC327 strain as described in Materials and Methods. Figure 4A shows the time courses of growth and expression of the cwlF-lacZ transcriptional fusion gene of the 327FD strain. The fusion gene expression started from the early growth phase, reaching a maximum at t−1, and then sharply decreased. The parent strain, B. subtilis AC327, exhibited β-galactosidase activities of less than 10 U/mg of protein during the exponential phase and less than 15 U/mg of protein during the sporulation phase (19). The Northern blot analysis in Fig. 5B shows that one transcript hybridized to a probe containing the internal region of the cwlF gene. This transcript, estimated to be 1.1 kb, was detected at t−1 to t1 but not after t2. Since the intensity of the signal was highest at t−1, cwlF is expressed as a monocistronic operon and may be transcribed by EςA RNA polymerase.
FIG. 4.
(A) β-Galactosidase activity of the cwlF-lacZ transcriptional fusion strain 327FD. Open symbols indicate growth; closed symbols indicate β-galactosidase activity. t0 is defined as the onset of sporulation. (B) Northern blot analysis with a cwlF RNA probe. Each lane contained 10 μg of total RNA isolated from B. subtilis AC327 at t−1, t0, t1, t2, t3, t4.5, and t6 (lanes −1 to 6, respectively). The 23S and 16S rRNAs are 2.8 and 1.4 kb, respectively. The calculated size of the detected transcript is shown on the left.
FIG. 5.
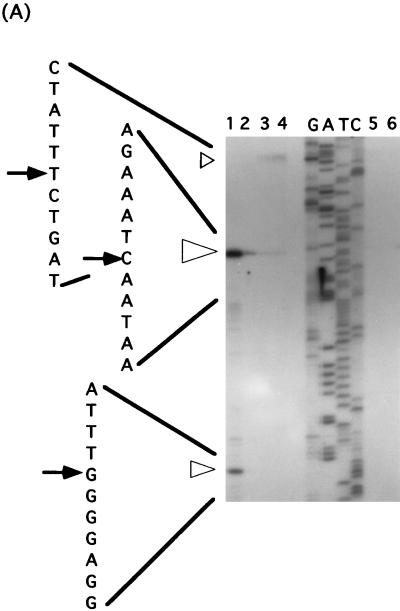
(A) Determination of the 5′ end of the cwlF transcript, by primer extension analysis (25 μg), isolated from the wild-type strain, AC327, at t−1 (lane 1), t0 (lane 2), t1 (lane 3), t2 (lane 4), t3 (lane 5), and t6 (lane 6). Open arrowheads indicate the three apparent transcriptional start sites. The dideoxy DNA sequencing reaction mixtures with the same primer (PEX-cF2) were electrophoresed in parallel (lanes G, A, T, and C). The positions of the products are indicated by arrows, and the sizes of arrowheads show the proportions of the activities of the three transcriptional start sites. (B) Nucleotide sequence of the putative promoter region of the cwlF gene. The nucleotides are numbered with respect to the translational start point (+1) of cwlF. PA−35 and PA−10 represent the −35 and −10 regions of a ςA-like promoter, and PH−35 and PH−10 represent the −35 and −10 regions of a ςH promoter. Arrowheads indicate the transcriptional start sites, and the sizes of the arrowheads indicate the proportions as described above. Dots indicate the N-terminal amino acid sequence of CwlF prepared from the culture. PEX-cF2 was the primer used for primer extension analysis. RBS, ribosome binding site. ρ-independent terminators are indicated by opposing arrows.
Determination of the 5′ end of cwlF RNA.
The cwlF gene is located near the phoA region, and the gene order is phoA-cwlF (papQ)-citR. phoA and citR are transcribed and translated in the same direction, but cwlF is transcribed and translated in a different direction (55). Two deduced ρ-independent terminators (ΔG = −10.2 and −10.8 kcal/mol) are located between phoA and cwlF, and one terminator (ΔG = −24.3 kcal/mol) is located between cwlF and citR. From the sequence information and the results of Northern blot analysis, it seemed likely that the 5′ end of cwlF RNA would be located upstream of cwlF and between phoA and cwlF. Primer extension analysis was performed with an oligonucleotide primer (PEX-cF2) that is complementary to the 5′ region of cwlF (bases 116 to 99) (Fig. 5A). A strong transcriptional signal starting at C−62 (the nucleotide is numbered with respect to the translational start point [+1] of cwlF) was observed with RNA from cells at t−1 (Fig. 5A, lane 1), t0 (lane 2), and t1 (lane 3). A signal with medium intensity, starting at G−12, was observed at t−1. Interestingly, a very weak but still significant signal, starting at T−93, was observed at t1 and t2. From the similarities in length and in the timing of the appearance of transcripts, the strong primer extension product seemed to correspond to the 5′ end of the 1.1-kb RNA. The −35 region (TTCTGA) and −10 region (TATAAT), with a spacing of 14 bp, were similar to those of the ςA consensus sequence (TTGACA for the −35 region and TATAAT for the −10 region, with a spacing of 17 bp; the underlined nucleotides are highly conserved) (Fig. 5B) (15, 34). The transcript, starting at G−12, is estimated to be 1.05 kb in length, if the same ρ-independent terminator located between cwlF and citR is used. The −35 region (AAAGAGATA) and −10 region (AGAGT), with a spacing of 10 bp, were very similar to those of the ςH consensus sequence (RWAGGAXXT for the −35 region and HGAAT for the −10 region, with a spacing of 14 bp; R = A or G; W = A, G, or C; X = A or T; and H = A or C) (15). The transcription of dual promoters was also found for those of the major autolysin gene, cwlB (ςD and ςA) (26, 29), and a cortex maturation gene, cwlD (ςE and ςG) (48).
Cell morphology of the cwlF and cwlF sigD disruptants.
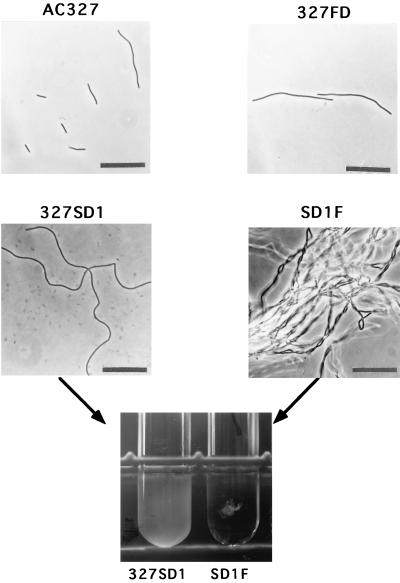
B. subtilis mutant cells which have deficiencies in the major autolysin gene (cwlB) and/or the glucosaminidase gene (cwlG) are rod shaped, while the sigD mutant forms filamentous cells, especially during exponential growth (25, 40, 42). Both autolysin genes were mainly transcribed by EςD RNA polymerase, and the sigD mutation led to less than 13% of the wild-type cwlB expression (26) and less than 8% of the wild-type cwlG expression (41). These results suggest that an unknown gene regulated by SigD is important for cell morphology. Although the cwlF gene is not regulated by SigD, we compared the morphology of the cwlF mutant 327FD with that of the wild type, AC327. The cwlF mutant cells were about twice as long as the wild-type cells (10.7 ± 8.7 μm and 6.5 ± 1.9 μm, respectively) (Fig. 6). However, in the case of the p60 protein of L. monocytogenes, a reduction in the amount of p60 leads to the formation of long cell chains (5, 56). Since these morphological differences may depend on the complement set of cell wall hydrolases in B. subtilis, we constructed a cwlB cwlF mutant and a sigD cwlF mutant. While the former double mutant showed a cell morphology similar to that of the cwlF mutant (19), the latter one showed extraordinary, dense microfiber formation (Fig. 6). The sigD mutant grew as a turbid suspension in a test tube culture, but SD1F grew like cotton waste in a transparent culture (Fig. 6). Recent studies have identified cell wall hydrolases (AcmA, p60, and Atl) involved in cell separation in L. lactis, L. monocytogenes, and Staphylococcus aureus, respectively (6, 56, 57). Atl is a bifunctional protein which has an amidase domain and a glucosaminidase domain and undergoes proteolytic processing to generate two extracellular cell wall hydrolases (amidase and glucosaminidase). These enzymes synergistically act on cell separation (52). Therefore, a combination of cell wall hydrolases may be important for cell separation in B. subtilis.
FIG. 6.
Phase-contrast microscopy of AC327 (wild-type), 327FD (cwlF), 327SD1 (sigD), and SD1F (cwlF sigD) cells and a picture of the test tube cultures of 327SD1 and SD1F. The pictures of AC327, 327FD, 327SD1, and SD1F were taken at OD600s of 0.263, 0.117, 0.146, and 0.205, respectively. Bar, 25 μm. To measure the OD600 of the SD1F strain, a small amount of lysozyme was added to the samples just before vigorous vortexing.
CwlF production is not affected by the pleiotropic genes flaD1 and degU32(Hy).
Since it is known that the pleiotropic mutations flaD1 and degU32(Hy) lead to extensive filamentation (10, 53), we measured CwlF productivity in organisms carrying these mutations. Figure 7 shows zymography of extracts S of the wild-type (AC327) strain and the sigD (327SD1), flaD1 (AC334), and degU32(Hy) (327UH) mutants. Compared with the wild-type cells, the mutants exhibited an extensive reduction in cell wall hydrolases. However, the 35-kDa CwlF protein was not greatly affected by these mutations.
FIG. 7.
Comparison of CwlF production by AC334 (flaD1), 327SD1 (sigD), 327UH [degU32(Hy)], and a parent strain, AC327 (wild type), by zymography. Extracts S at t−1 (lanes −1), t0 (lanes 0), and t1 (lanes 1) were prepared, and a protein amount equivalent to 3.6 OD600 units of cell growth was applied to each lane. Lane M contained protein standards (Bio-Rad), the molecular masses of which are shown on the left. The arrows indicate the CwlF protein.
This study indicated that CwlF is a new cell-separating enzyme. The gene is suggested to be transcribed mainly by EςA RNA polymerase and weakly by EςH RNA polymerase. The extraordinary microfiber formation by organisms carrying the sigD cwlF double mutation suggests that a SigD-dependent factor(s) and CwlF cooperatively play an important role in cell separation during the vegetative growth phase. Many cwlF homologs were recently found in the B. subtilis genome (47), and it is predicted that the cwlF group will become a big one among cell wall hydrolase genes. Research on the combination of cell wall hydrolases should reveal their cellular functions. The substrate specificity of CwlF has not been determined, and thus our research is now directed toward the purification and characterization of this enzyme.
REFERENCES
- 1.Akamatsu T, Sekiguchi J. Genetic mapping and properties of filamentous mutations in Bacillus subtilis. Agric Biol Chem. 1987;51:2901–2909. [Google Scholar]
- 2.Anagnostopoulos C, Spizizen J. Requirement for transformation in Bacillus subtilis. J Bacteriol. 1961;81:741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ayusawa D, Yoneda Y, Yamane K, Maruo B. Pleiotropic phenomena in autolytic enzyme(s) content, flagellation, and simultaneous hyperproduction of extracellular α-amylase and protease in a Bacillus subtilis mutant. J Bacteriol. 1975;124:459–469. doi: 10.1128/jb.124.1.459-469.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Béliveau C, Potvin C, Trudel J, Asselin A, Bellemare G. Cloning, sequencing, and expression in Escherichia coli of a Streptococcus faecalis autolysin. J Bacteriol. 1991;173:5619–5623. doi: 10.1128/jb.173.18.5619-5623.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bubert A, Kuhn M, Goebel W, Köhler S. Structural and functional properties of the p60 proteins from different Listeria species. J Bacteriol. 1992;174:8166–8171. doi: 10.1128/jb.174.24.8166-8171.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buist G, Kok J, Leenhouts K J, Dabrowska M, Venema G, Haandrikman A J. Molecular cloning and nucleotide sequence of the gene encoding the major peptidoglycan hydrolase of Lactococcus lactis, a muramidase needed for cell separation. J Bacteriol. 1995;177:1554–1563. doi: 10.1128/jb.177.6.1554-1563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chu C-P, Kariyama R, Daneo-Moore L, Shockman G D. Cloning and sequence analysis of the muramidase-2 gene from Enterococcus hirae. J Bacteriol. 1992;174:1619–1625. doi: 10.1128/jb.174.5.1619-1625.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Doyle R J, Koch A L. The functions of autolysins in the growth and division of Bacillus subtilis. Crit Rev Microbiol. 1987;15:169–222. doi: 10.3109/10408418709104457. [DOI] [PubMed] [Google Scholar]
- 9.Fein J E. Possible involvement of bacterial autolysin enzymes in flagellar morphogenesis. J Bacteriol. 1979;137:933–946. doi: 10.1128/jb.137.2.933-946.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fein J E, Rogers H J. Autolytic enzyme-deficient mutants of Bacillus subtilis 168. J Bacteriol. 1976;127:1427–1442. doi: 10.1128/jb.127.3.1427-1442.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fleischmann R D, Adams M D, White O, Clayton R A, Kirkness E F, Kerlavage A R, Bult C J, Tomb J-F, Dougherty B A, Merrick J M, McKenney K, Sutton G, FitzHugh W, Fields C, Gocayne J D, Scott J, Shirley R, Liu L-I, Glodek A, Kelley J M, Weidman J F, Phillips C A, Spriggs T, Hedblom E, Cotton M D, Utterback T R, Hanna M C, Nguyen D T, Saudek D M, Brandon R C, Fine L D, Fritchman J L, Fuhrmann J L, Geoghagen N S M, Gnehm C L, McDonald L A, Small K V, Fraser C M, Smith H O, Venter J C. Whole-genome random sequencing and assembly of Haemophilus influenzae Rd. Science. 1995;269:496–512. doi: 10.1126/science.7542800. [DOI] [PubMed] [Google Scholar]
- 12.Foster S J. Cloning, expression, sequence analysis and biochemical characterization of an autolytic amidase of Bacillus subtilis 168 trpC2. J Gen Microbiol. 1991;137:1987–1998. doi: 10.1099/00221287-137-8-1987. [DOI] [PubMed] [Google Scholar]
- 13.Foster S J. Analysis of the autolysins of Bacillus subtilis 168 during vegetative growth and differentiation by using renaturing polyacrylamide gel electrophoresis. J Bacteriol. 1992;174:464–470. doi: 10.1128/jb.174.2.464-470.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Foster S J. Molecular analysis of three major wall-associated proteins of Bacillus subtilis 168: evidence for processing of the product of a gene encoding a 258 kDa precursor two-domain ligand-binding protein. Mol Microbiol. 1992;8:299–310. doi: 10.1111/j.1365-2958.1993.tb01574.x. [DOI] [PubMed] [Google Scholar]
- 15.Haldenwang W G. The sigma factors of Bacillus subtilis. Microbiol Rev. 1995;59:1–30. doi: 10.1128/mr.59.1.1-30.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Herbold D R, Glaser L. Bacillus subtilis N-acetylmuramic acid l-alanine amidase. J Biol Chem. 1975;250:1676–1682. [PubMed] [Google Scholar]
- 17.Höltje J-V, Tuomanen E I. The murein hydrolases of Escherichia coli: properties, functions and impact on the course of infections in vivo. J Gen Microbiol. 1991;137:441–454. doi: 10.1099/00221287-137-3-441. [DOI] [PubMed] [Google Scholar]
- 18.Hourdou M-L, Duez C, Joris B, Vacheron M-J, Guinand M, Michel G, Ghuysen J-M. Cloning and nucleotide sequence of the gene encoding the γ-d-glutamyl-l-diamino acid endopeptidase II of Bacillus sphaericus. FEMS Microbiol Lett. 1992;91:165–170. doi: 10.1111/j.1574-6968.1992.tb05203.x. [DOI] [PubMed] [Google Scholar]
- 19.Ishikawa, S., Y. Hara, R. Ohnishi, and J. Sekiguchi. Unpublished data.
- 20.Kadner, R. J. 1989. Swiss-Prot database NLPC_ECOLI.
- 21.Köhler S, Leimeister-Wächter M, Chakraborty T, Lottspeich F, Goebel W. The gene coding for protein p60 of Listeria monocytogenes and its use as a specific probe for Listeria monocytogenes. Infect Immun. 1990;58:1943–1950. doi: 10.1128/iai.58.6.1943-1950.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuroda A, Asami Y, Sekiguchi J. Molecular cloning of a sporulation-specific cell wall hydrolase gene of Bacillus subtilis. J Bacteriol. 1993;175:6260–6268. doi: 10.1128/jb.175.19.6260-6268.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kuroda A, Rashid M H, Sekiguchi J. Molecular cloning and sequencing of the upstream region of the major Bacillus subtilis autolysin gene: a modifier protein exhibiting sequence homology to the major autolysin and the spoIID product. J Gen Microbiol. 1992;138:1067–1076. doi: 10.1099/00221287-138-6-1067. [DOI] [PubMed] [Google Scholar]
- 24.Kuroda A, Sekiguchi J. Cloning, sequencing and genetic mapping of a Bacillus subtilis cell wall hydrolase gene. J Gen Microbiol. 1990;136:2209–2216. doi: 10.1099/00221287-136-11-2209. [DOI] [PubMed] [Google Scholar]
- 25.Kuroda A, Sekiguchi J. Molecular cloning and sequencing of a major Bacillus subtilis autolysin gene. J Bacteriol. 1991;173:7304–7312. doi: 10.1128/jb.173.22.7304-7312.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kuroda A, Sekiguchi J. High-level transcription of the major Bacillus subtilis autolysin operon depends on expression of the sigma D gene and is affected by a sin (flaD) mutation. J Bacteriol. 1993;175:795–801. doi: 10.1128/jb.175.3.795-801.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kuroda A, Sugimoto Y, Funahashi T, Sekiguchi J. Genetic structure, isolation and characterization of a Bacillus licheniformis cell wall hydrolase. Mol Gen Genet. 1992;234:129–137. doi: 10.1007/BF00272354. [DOI] [PubMed] [Google Scholar]
- 28.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 29.Lazarevic V, Margot P, Soldo B, Karamata D. Sequencing and analysis of the Bacillus subtilis lytRABC divergon: a regulatory unit encompassing the structural genes of the N-acetylmuramoyl-l-alanine amidase and its modifier. J Gen Microbiol. 1992;138:1949–1961. doi: 10.1099/00221287-138-9-1949. [DOI] [PubMed] [Google Scholar]
- 30.Leclerc D, Asselin A. Detection of bacterial cell wall hydrolases after denaturating polyacrylamide gel electrophoresis. Can J Microbiol. 1989;35:749–753. doi: 10.1139/m89-125. [DOI] [PubMed] [Google Scholar]
- 31.Longchamp P F, Mauël C, Karamata D. Lytic enzymes associated with defective prophages of Bacillus subtilis: sequencing and characterization of the region comprising the N-acetylmuramoyl-l-alanine amidase gene of prophage PBSX. Microbiology. 1994;140:1855–1867. doi: 10.1099/13500872-140-8-1855. [DOI] [PubMed] [Google Scholar]
- 32.Makino S, Ito N, Inoue T, Miyata S, Moriyama R. A spore-lytic enzyme released from Bacillus cereus spores during germination. Microbiology. 1994;140:1403–1410. doi: 10.1099/00221287-140-6-1403. [DOI] [PubMed] [Google Scholar]
- 33.Margot P, Mauël C, Karamata D. The gene of the N-acetylglucosaminidase, a Bacillus subtilis cell wall hydrolase not involved in vegetative cell autolysis. Mol Microbiol. 1994;12:535–545. doi: 10.1111/j.1365-2958.1994.tb01040.x. [DOI] [PubMed] [Google Scholar]
- 34.Moran C P., Jr . RNA polymerase and transcription factors. In: Sonenshein A L, Hoch J A, Losick R, editors. Bacillus subtilis and other gram-positive bacteria: biochemistry, physiology, and molecular genetics. Washington, D.C: American Society for Microbiology; 1993. pp. 653–667. [Google Scholar]
- 35.Moriyama R, Hattori A, Miyata S, Kudoh S, Makino S. A gene (sleB) encoding a spore cortex-lytic enzyme from Bacillus subtilis and response of the enzyme to l-alanine-mediated germination. J Bacteriol. 1996;178:6059–6063. doi: 10.1128/jb.178.20.6059-6063.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moriyama R, Kudoh S, Miyata S, Nonobe S, Hattori A, Makino S. A germination-specific spore cortex-lytic enzyme from Bacillus cereus spores: cloning and sequencing of the gene and molecular characterization of the enzyme. J Bacteriol. 1996;178:5330–5332. doi: 10.1128/jb.178.17.5330-5332.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Oda Y, Nakayama R, Kuroda A, Sekiguchi J. Molecular cloning, sequence analysis, and characterization of a new cell wall hydrolase, CwlL, of Bacillus licheniformis. Mol Gen Genet. 1993;241:380–388. doi: 10.1007/BF00284691. [DOI] [PubMed] [Google Scholar]
- 38.Pooley H M, Karamata D. Genetic analysis of autolysin-deficient and flagellaless mutants of Bacillus subtilis. J Bacteriol. 1984;160:1123–1129. doi: 10.1128/jb.160.3.1123-1129.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Potvin C, Leclerc D, Tremblay G, Asselin A, Bellemare G. Cloning, sequencing and expression of a Bacillus bacteriolytic enzyme in Escherichia coli. Mol Gen Genet. 1988;214:241–248. doi: 10.1007/BF00337717. [DOI] [PubMed] [Google Scholar]
- 40.Rashid M H, Kuroda A, Sekiguchi J. Bacillus subtilis mutant deficient in the major autolytic amidase and glucosaminidase is impaired in motility. FEMS Microbiol Lett. 1993;112:135–140. doi: 10.1111/j.1574-6968.1993.tb06438.x. [DOI] [PubMed] [Google Scholar]
- 41.Rashid M H, Mori M, Sekiguchi J. Glucosaminidase of Bacillus subtilis: cloning, regulation, primary structure and biochemical characterization. Microbiology. 1995;141:2391–2404. doi: 10.1099/13500872-141-10-2391. [DOI] [PubMed] [Google Scholar]
- 42.Rashid M H, Sato N, Sekiguchi J. Analysis of the minor autolysins of Bacillus subtilis during vegetative growth by zymography. FEMS Microbiol Lett. 1995;132:131–137. [Google Scholar]
- 43.Rogers H J, Perkins H R, Ward J B. Microbial cell walls and membranes. London, England: Chapman and Hall; 1980. [Google Scholar]
- 44.Rogers H J, Taylor C, Rayter S, Ward J B. Purification and properties of autolytic endo-β-N-acetylglucosaminidase and the N-acetylmuramoyl-l-alanine amidase from Bacillus subtilis strain 168. J Gen Microbiol. 1984;130:2395–2402. doi: 10.1099/00221287-130-9-2395. [DOI] [PubMed] [Google Scholar]
- 45.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 46.Schaeffer P, Millet J, Aubert J P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci USA. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sekiguchi, J. Unpublished data.
- 48.Sekiguchi J, Akeo K, Yamamoto H, Khasanov F K, Alonso J C, Kuroda A. Nucleotide sequence and regulation of a new putative cell wall hydrolase gene, cwlD, which affects germination in Bacillus subtilis. J Bacteriol. 1995;177:5582–5589. doi: 10.1128/jb.177.19.5582-5589.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shimotsu H, Henner D J. Modulation of Bacillus subtilis levansucrase gene expression by sucrose and regulation of the steady-state mRNA level by sacU and sacQ genes. J Bacteriol. 1986;168:380–388. doi: 10.1128/jb.168.1.380-388.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shockman G D, Höltje J-V. Microbial peptidoglycan (murein) hydrolases. In: Ghuysen J-M, Hakenbeck R, editors. Bacterial cell wall. Amsterdam, The Netherlands: Elsevier; 1994. pp. 131–166. [Google Scholar]
- 51.Smith T J, Foster S J. Characterization of the involvement of two compensatory autolysins in mother cell lysis during sporulation of Bacillus subtilis 168. J Bacteriol. 1995;177:3855–3862. doi: 10.1128/jb.177.13.3855-3862.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sugai M, Komatsuzawa H, Akiyama T, Hong Y-M, Oshida T, Miyake Y, Yamaguchi T, Suginaka H. Identification of endo-β-N-acetylglucosaminidase and N-acetylmuramoyl-l-alanine amidase as cluster-dispersing enzymes in Staphylococcus aureus. J Bacteriol. 1995;177:1491–1496. doi: 10.1128/jb.177.6.1491-1496.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tokunaga T, Rashid M H, Kuroda A, Sekiguchi J. Effect of degS-degU mutations on the expression of sigD, encoding an alternative sigma factor, and autolysin operon of Bacillus subtilis. J Bacteriol. 1994;176:5177–5180. doi: 10.1128/jb.176.16.5177-5180.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Völker U, Engelmann S, Maul B, Riethdorf S, Völker A, Schmid R, Mach H, Hecker M. Analysis of the induction of general stress proteins of Bacillus subtilis. Microbiology. 1994;140:741–752. doi: 10.1099/00221287-140-4-741. [DOI] [PubMed] [Google Scholar]
- 55.Whalen, M. B., and P. J. Piggot. 1996. Swiss-Prot database PAPQ_BACSU.
- 56.Wuenscher M D, Köhler S, Bubert A, Gerike U, Goebel W. The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, p60, has bacteriolytic activity. J Bacteriol. 1993;175:3491–3501. doi: 10.1128/jb.175.11.3491-3501.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamada S, Sugai M, Komatsuzawa H, Nakashima S, Oshida T, Matsumoto A, Suginaka H. An autolytic ring associated with cell separation of Staphylococcus aureus. J Bacteriol. 1996;178:1565–1571. doi: 10.1128/jb.178.6.1565-1571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yamanaka K, Araki J, Takano M, Sekiguchi J. Characterization of Bacillus subtilis mutants resistant to cold shock-induced autolysis. FEMS Microbiol Lett. 1997;150:267–275. doi: 10.1111/j.1574-6968.1997.tb10380.x. [DOI] [PubMed] [Google Scholar]