Abstract
The yeast YCC5 gene encodes a putative amino acid permease and is homologous to GNP1 (encoding a high-affinity glutamine permease). Using strains with disruptions in the genes for multiple permeases, we demonstrated that Ycc5 (which we have renamed Agp1) is involved in the transport of asparagine and glutamine, performed a kinetic analysis of this activity, and showed that AGP1 expression is subject to nitrogen repression.
The yeast Saccharomyces cerevisiae can utilize a wide variety of compounds as nitrogen sources for growth. The permeases responsible for amino acid transport in yeast form a family of proteins with conserved sequences and structural features (1, 17). These integral membrane proteins are approximately 600 amino acids long and have 12 transmembrane domains in the central portion of the molecule. Most of the known amino acid permeases are specific for a group of structurally related amino acids, transporting various members of the family with different affinities. For example, the permease Bap2 transports leucine, isoleucine, and valine but not other nonpolar amino acids (10). One permease which behaves differently is the general amino acid permease (Gap1), which transports all 20 naturally occurring l-amino acids, as well as their d-isomers and many structurally related compounds, with high efficiency (5, 13).
The Yeast Genome Project has identified several open reading frames (ORFs) which, based on known permease genes, probably encode amino acid permeases (1, 17). One of these is the ORF YCL025c (encoding a sequence 633 amino acids long; also called YCC5 [18]) on chromosome III, which is most similar to the high-affinity glutamine permease gene GNP1 (27). We isolated the YCC5 region by PCR, constructed a ycc5::HIS3 plasmid, and integrated the disrupted gene into the yeast genome. The resultant disruption strains were used to study the substrate specificity of this permease.
Strains and growth conditions.
The wild-type strain SP1 is MATa ura3-52 leu2-3,112 his3Δ1 trp1-289 can1 ade8. JGY50 (MATa ura3-52 his3Δ1 trp1-289 can1 ade8 gap1::LEU2) and JGY51 (MATa his3Δ1 trp1-289 can1 ade8 gap1::LEU2 gnp1::URA3) were described previously (27), and the construction of JSY1 (MATa ura3-52 leu2-3,112 trp1-289 can1 ade8 ycc5::HIS3) and JGY52 (MATa trp1-289 can1 ade8 gap1::LEU2 gnp1::URA3 ycc5::HIS3) is described below. A set of Σ1278b-based strains, 23344c (MATα ura3-52), 30505b (MATα ura3-52 gln3Δ), SBS21 (MATα ura3-52 nil1Δ::KanMX2), 50027c (MATα ura3-52 leu2 gln3 nil1Δ::KanMX2), and 26854 (MATa ura3-52 ure2 [gdhCR]), were obtained from B. André (22). Yeast strains were grown by routine methods in standard yeast media (20, 21). Minimal medium (25) contained the following supplements unless indicated otherwise: l-histidine (20 mg/liter), l-leucine (30 mg/liter), l-tryptophan (20 mg/liter), l-lysine (30 mg/liter), adenine (20 mg/liter), and uracil (20 mg/liter). For the determination of growth rates on different nitrogen sources, the amino acid acting as the principal nitrogen source was added at 0.01% (wt/vol) and the supplements (adenine, histidine, lysine, and tryptophan) were added at 0.1 times the normal concentration.
Isolation of the YCC5 gene.
Standard protocols were used for all DNA manipulations in yeast and Escherichia coli (19, 21). PCR amplification of a 2,690-bp fragment containing the YCC5 ORF was achieved with primers (5′-CAGCGGATCCCTGCTCCTTAGTAGTCC and 5′-CTCGGATCCATTTCCATCACGCAATCG, obtained from Gibco BRL) which generate BamHI sites at both ends of the amplified YCC5 fragment (12). The BamHI sites were used to place the fragment in pUC18 to create a plasmid called pJAK2. This plasmid was opened at the unique BglII site in YCC5, and a BamHI fragment containing the HIS3 gene, flanked by multiple stop codons (1,100-bp fragment of YDp-H [2]), was inserted to disrupt the YCC5 ORF. The 3,500-bp BamHI ycc5::HIS3 fragment was used to transform the desired haploid yeast strain. Disruption of the chromosomal YCC5 gene was checked by PCR and restriction digest analysis of the PCR product (12). A CEN-based plasmid containing a wild-type YCC5 gene was constructed by inserting the BamHI YCC5 PCR fragment into the plasmid YCp410 (14), which contains TRP1 as a selectable marker.
Disruption of YCC5 confers resistance to toxic analogs of asparagine and glutamine.
In preliminary studies to determine the substrate(s) of the Ycc5 permease, the rates of growth of strains with (SP1) and without (JSY1) permease were compared with each amino acid as the principal nitrogen source and on plates containing various amino acid analogs. No differences in growth were observed under any of these conditions. Because yeasts have multiple permeases capable of taking up each amino acid, we reasoned that the phenotype of the YCC5 disruption was being masked by the activity of related permeases. The growth of strains with disruptions in gap1 (JGY50), gap1 gnp1 (JGY51), and gap1 gnp1 ycc5 (JGY52) was then compared with that of the parental strain on minimal ammonia plates (auxotrophic-requirement supplements were added at 0.1 times the normal concentration) containing toxic levels of various amino acid analogs. Disruption of YCC5 did not alter the growth of gap1 gnp1 yeast on plates containing l-canavanine (10 μg/ml), cycloleucine (500 μg/ml), dl-cycloserine (30 μg/ml), dl-ethionine (25 μg/ml), parafluorophenylalanine (120 μg/ml), trifluoroleucine (120 μg/ml), or β-2-thienylalanine (30 μg/ml) (data not shown). However, JGY52 was completely resistant to 400 μg of β-hydroxyaspartate (an analog of asparagine)/ml, whereas the wild-type and gap1 and gap1 gnp1 disruption strains were sensitive to it (data not shown). Disruption of YCC5 also conferred a growth advantage on cells growing on 400 μg of γ-hydroxyglutamate (an analog of glutamine)/ml and conferred a very slight growth advantage on cells growing on 200 μg of norleucine/ml (data not shown). Growth of yeast on γ-hydroxyglutamate has been shown to be affected by deletion of the high-affinity glutamine permease gene GNP1 (27). gap1 gnp1 strains are partially resistant to this analog of glutamine but are not resistant to the asparagine analog (data not shown). Transformation of JGY52 (gap1 gnp1 ycc5) with a wild-type copy of YCC5 cloned in the CEN plasmid YCp410 restores the sensitivity of JGY52 to β-hydroxyaspartate (data not shown) and returns all phenotypes of the ycc5::HIS3 strains described below to those of the parent strain. This shows that the resistance of JGY52 yeast to the toxic asparagine analog, and other asparagine- and glutamine-linked phenotypes described below, is a result of disruption of the YCC5 gene. These results demonstrate that the permease encoded by YCC5 transports both asparagine and glutamine, and we therefore have renamed the YCC5 gene AGP1.
Kinetic analysis of asparagine and glutamine transport by Agp1.
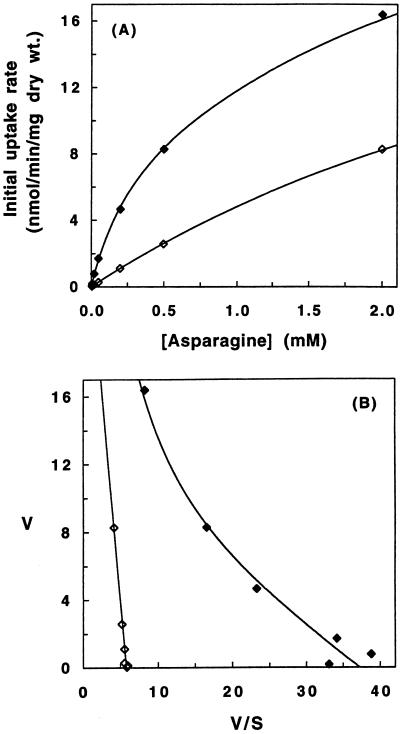
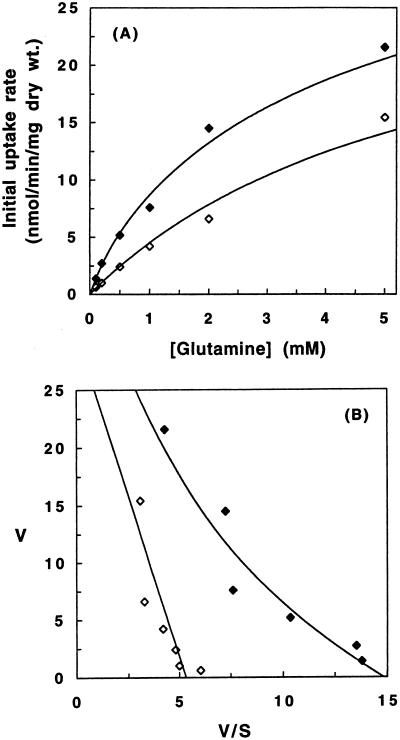
In order to determine the contribution of Agp1 to asparagine and glutamine uptake, the transport of these amino acids was compared in the ammonia-grown strains JGY51 and JGY52 (Fig. 1A and 2A). Amino acid uptake was assayed as described previously (20, 26). Uptake was measured at asparagine concentrations between 0.005 and 2.0 mM (specific activity between 140 and 0.36 μCi/μmol) and glutamine concentrations between 0.1 and 5.0 mM (specific activity between 5.0 and 0.1 μCi/μmol). The Eadie-Hofstee plots of asparagine and glutamine uptake by JGY52 (Fig. 1B and 2B) were approximately linear, indicating that only one transport system for these amino acids remained in this triple disruption strain and that this system displayed Michaelis-Menten kinetics (11). The Solver program of Microsoft Excel was used to simultaneously fit the data and obtain the kinetic parameters of this transport. This transporter had very low affinity for both amino acids (Kmapp > 5.0 mM) and may represent the nonspecific activity of another amino acid permease at these very high substrate concentrations or could be the result of a facilitated diffusion system. The Eadie-Hofstee plots of asparagine and glutamine uptake in the double disruption strain JGY51 were clearly biphasic. Uptake by JGY51 was analyzed by assuming the participation of two transport systems: the low-affinity system identified in JGY52 and the permease encoded by AGP1. The Solver program was used to simultaneously fit the JGY51 data with the parameters obtained from the JGY52 data for the low-affinity system and by adjusting the Agp1 parameters to obtain the best fit. The apparent Km values of Agp1 were found to be 0.29 mM for asparagine and 0.79 mM for glutamine. The Vmax values were 9.0 and 7.5 nmol/min/mg (dry weight) for asparagine and glutamine, respectively.
FIG. 1.
Kinetics of asparagine uptake in JGY51 (gap1 gnp1) and JGY52 (gap1 gnp1 agp1). The rate of [14C]asparagine uptake was determined in JGY51 (filled symbols) and JGY52 (open symbols) as described previously (20, 26). The data was plotted as Michaelis-Menten (A) and Eadie-Hofstee (B) graphs. The Michaelis-Menten graphs were fitted to curves assuming two permeases (JGY51) and one permease (JGY52) with the Solver program of Microsoft Excel. Each data point is the average of at least three measurements, with a standard error of <10%. The deduced Kmapp for asparagine of Agp1 is 0.29 mM, and the Vmax is 9.0 nmol/min/mg (dry weight).
FIG. 2.
Kinetics of glutamine uptake in JGY51 (gap1 gnp1) and JGY52 (gap1 gnp1 agp1). The rate of [14C]glutamine uptake was determined as described in the legend for Fig. 1. The data were plotted as Michaelis-Menten (A) and Eadie-Hofstee (B) graphs. Each data point is the average of at least two measurements, with a standard error of <15%. The deduced Kmapp for glutamine of Agp1 is 0.79 mM, and the Vmax is 7.5 nmol/min/mg (dry weight).
AGP1 expression is subject to nitrogen repression.
Many genes involved in the uptake and metabolism of nitrogenous compounds are coregulated by a system called nitrogen repression (15). Cells growing on “poor” nitrogen sources, such as proline or urea, activate the transcription of a set of genes, including those encoding the permeases Gap1 and Can1, by the action of transcription factors, e.g., Gln3 and Nil1, at GATAA sites found upstream of these genes (4, 7, 23, 24). In the preferred nitrogen sources, asparagine and glutamine, cells repress the transcription of these genes through the action of Ure2, which inactivates some of the positive transcription factors (3, 6, 22).
In order to test the transcriptional regulation of AGP1, an AGP1-lacZ fusion plasmid was made. A 1,157-bp fragment composed of 1,124 bp of the AGP1 promoter region and the coding sequence for the first nine amino acids of Agp1 was synthesized by PCR. The primers were designed to contain EcoRI and BamHI sites at the termini so that the fragment could then be inserted directly into the yeast vector pSEY101 (9) to create an in-frame fusion between AGP1 and lacZ. This construct was used to assay the level of AGP1 expression of yeast growing in ammonia, glutamate, glutamine, and proline in the Σ1278b strain 23344, a strain subject to nitrogen repression (22). Cells were grown in minimal media to an optical density at 550 nm of between 0.10 and 0.40, and β-galactosidase activity was assayed in disruptant cells (16).
The highest levels of AGP1-directed β-galactosidase activity were found in cells grown in glutamate and the lowest levels of activity were found in cells grown on glutamine (Table 1). This is consistent with previous studies on GAP1 expression (15, 22). The relative levels of expression of AGP1 and GAP1 were also similar in ammonia and proline media. The levels of AGP1 expression in cells growing in these four media were also determined by Northern analysis, which showed the same pattern of AGP1 regulation by the nitrogen source (data not shown). There are six GATAA sequences in the upstream region of the AGP1 promoter which are potential upstream activator sequences for binding the nitrogen-regulated transcription factors Gln3 and Nil1. In order to determine if AGP1 is regulated by Gln3 and Nil1 in a manner similar to GAP1, AGP1-driven β-galactosidase production was measured in the gln3, nil1, and gln3 nil1 strains (Table 1). The results show that both Gln3 and Nil1 have a role in the activation of AGP1 expression. The pattern of AGP1 expression is very similar to that of GAP1 expression except that Gln3 appears to play a more important role in activating AGP1 expression in ammonia- and proline-grown cells. The repression of AGP1 expression in glutamine-grown cells was relieved in cells with mutations in the regulatory protein Ure2 (wild type, 295 U; ure2 strain, 3,320 U), also indicating that nitrogen repression controls both AGP1 and GAP1. One major difference in the control of expression of the two permeases was noticed in asparagine-grown cells. The AGP1 level in these cells was relatively high (approximately 2,000 U), whereas asparagine has routinely been used as a repressing nitrogen source in studies of GAP1 expression (8). This may be explained by the fact that asparagine is a primary substrate for Agp1 permease.
TABLE 1.
Nitrogen regulation of AGP1 expressiona
| Nitrogen source | Relative β-galactosidase activity of strain
|
|||
|---|---|---|---|---|
| WT | gln3 strain | nil1 strain | gln3 nil1 strain | |
| Glutamate | 100 (100) | 12 (19) | 72 (85) | <0.1 (<0.1) |
| Proline | 70 (89) | 23 (87) | 31 (64) | <0.1 (<0.1) |
| Ammonia | 35 (47) | 5 (137) | 22 (11) | <0.1 (<0.1) |
| Glutamine | 3.2 (<2) | <0.1 (ND) | 1.3 (ND) | <0.1 (ND) |
Strains containing the AGP1-lacZ fusion plasmid were grown in minimal glucose medium containing the indicated nitrogen source (amino acids, 0.1%; ammonium sulfate, 0.2%) and 30 mg of leucine/liter (necessary to supplement the leucine auxotrophy of the 50027c strain [gln3 nil1]). Exponentially growing cells were harvested and their β-galactosidase activities were measured as described by Miller (16) at least three times for each value reported. The activity of the wild-type (WT) strain grown in glutamate medium (5,760 U) was set as 100% and the other relative values were calculated. The numbers in parentheses are the equivalent values for β-galactosidase expression from the GAP1 promoter as published previously (24) and are included for comparison. ND, not determined (24).
Agp1 has a relatively broad substrate range.
Agp1 has been demonstrated to transport asparagine and glutamine with intermediate affinity (Fig. 1 and 2). However, these amino acids may not be the only substrates for this permease. In order to determine if other amino acids are transported by Agp1, the growth rates of JGY51 and JGY52 were determined with different amino acids (approximately 1 mM) as the principal nitrogen source. As expected, the loss of Agp1 activity resulted in slower growth of yeast on asparagine (doubling times: JGY51, 2.1 h; JGY52, 4.1 h) or glutamine (doubling times: JGY51, 2.3 h; JGY52, 4.2 h). The two strains grew equally well on some nitrogen sources, e.g., arginine, glutamic acid, and proline. However, the doubling times of yeast utilizing leucine, isoleucine, methionine, phenylalanine, serine, threonine, tryptophan, tyrosine, and valine were increased (in most cases approximately doubled) by the loss of Agp1 activity (data not shown). This suggests that Agp1 permease can transport these amino acids when they are present at high concentrations in the medium. Furthermore, inhibition studies of [14C]asparagine (0.2 mM) uptake by competing amino acids (added at 1.0 mM) showed that aspartic acid, glutamic acid, isoleucine, leucine, methionine, serine, and threonine all significantly (>30%) decreased asparagine uptake. However, when a range of amino acids (arginine, asparagine, glutamine, glutamate, leucine, proline, tryptophan, and tyrosine) were tested in the 14C-amino acid uptake assay (substrate at 0.2 mM) in JGY51 (gap1 gnp1), JGY52 (gap1 gnp1 agp1), JGY51p410, and JGY51pAGP1 (AGP1 cloned into p410), only the uptake of asparagine and glutamine differed among the strains. JGY52 showed significantly decreased transport (>30%) of asparagine and glutamine, and the overexpression of AGP1 resulted in increased uptake of both amino acids (approximately 200%) whereas the uptake of the other amino acids tested was unchanged in both JGY52 and JGY51 and its derivatives. The results suggest that asparagine and glutamine are the primary substrates of Agp1, with Kmapp values of <1.0 mM; however, other amino acids can be taken up by this permease if they are present in millimolar concentrations. One other amino acid which may be a major substrate of this permease is serine. A recent analysis of the kinetics of serine uptake has shown that asparagine is an efficient competitive inhibitor of serine transport (18a).
The permease encoded by YCC5 has been identified as an asparagine and glutamine permease and renamed Agp1 to reflect its substrate specificity. The substrate specificity of Agp1 was first determined from the increased resistance of ycc5::HIS3 strains to toxic analogs of asparagine and glutamine. Its role in the transport of asparagine and glutamine was confirmed by both the decreased growth rate of agp1 strains using these amino acids as nitrogen sources and the decreased rate of [14C]asparagine and [14C]glutamine uptake in these strains. The results of the growth rate experiments and the asparagine uptake inhibition studies suggest that Agp1 can act as a low-affinity broad-substrate-range permease and is involved in the uptake of amino acids for catabolism in a manner similar to Gap1 (5). Expression of AGP1 was shown to be subject to nitrogen repression by Gln3 and Nil1, which is consistent with a role for this permease in supplying amino acids as nitrogen sources for cell growth.
Acknowledgments
This work was supported by NIH grant R15GM54280 to J.M.G. J.K.S. was supported by Hamilton College’s senior research program.
We thank B. André for strains and T. Michaeli and M. Brandriss for helpful discussions.
REFERENCES
- 1.André B. An overview of membrane transport proteins in Saccharomyces cerevisiae. Yeast. 1995;11:1575–1611. doi: 10.1002/yea.320111605. [DOI] [PubMed] [Google Scholar]
- 2.Berben G, Dumont J, Gilliquet V, Bolle P, Hilger F. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- 3.Coffman J A, El Berry H M, Cooper T G. The URE2 protein regulates nitrogen catabolic gene expression through the GATAA-containing UASNTR element in Saccharomyces cerevisiae. J Bacteriol. 1994;176:7476–7483. doi: 10.1128/jb.176.24.7476-7483.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Coffman J A, Rai R, Cooper T G. Genetic evidence for Gln3p-independent, nitrogen catabolite repression-sensitive gene expression in Saccharomyces cerevisiae. J Bacteriol. 1995;177:6910–6918. doi: 10.1128/jb.177.23.6910-6918.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper T G. Nitrogen metabolism in Saccharomyces cerevisiae. In: Strathern J N, Jones E W, Broach J, editors. The molecular biology of the yeast Saccharomyces: metabolism and gene expression. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. pp. 39–99. [Google Scholar]
- 6.Coschigano P W, Magasanik B. The URE2 gene product of Saccharomyces cerevisiae plays an important role in the cellular response to the nitrogen source and has homology to glutathione S-transferases. Mol Cell Biol. 1991;11:822–832. doi: 10.1128/mcb.11.2.822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Courchesne W E, Magasanik B. Regulation of nitrogen assimilation in Saccharomyces cerevisiae: roles of the URE2 and GLN3 genes. J Bacteriol. 1988;170:708–713. doi: 10.1128/jb.170.2.708-713.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daugherty J R, Rai R, El Berry H M, Cooper T G. Regulatory circuit for responses of nitrogen catabolic gene expression to the GLN3 and DAL80 proteins and nitrogen catabolite repression in Saccharomyces cerevisiae. J Bacteriol. 1993;175:64–73. doi: 10.1128/jb.175.1.64-73.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Emr S E, Vassarotti A, Garrett J M, Geller B L, Takeda M, Douglas M G. The amino terminus of the yeast F1-ATPase β-subunit precursor functions as a mitochondrial import signal. J Cell Biol. 1986;102:523–533. doi: 10.1083/jcb.102.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grauslund M, Didion T, Kielland-Brandt M C, Anderson H A. BAP2, a gene encoding a permease for branched-chain amino acids in Saccharomyces cerevisiae. Biochim Biophys Acta. 1995;1269:275–280. doi: 10.1016/0167-4889(95)00138-8. [DOI] [PubMed] [Google Scholar]
- 11.Hofstee B H J. On the evaluation of the constants Vm and KM in enzyme reactions. Science. 1952;116:329–331. doi: 10.1126/science.116.3013.329. [DOI] [PubMed] [Google Scholar]
- 12.Innis M A, Gelfand D H, Sninsky J J, White T J, editors. PCR protocols: a guide to methods and applications. San Diego, Calif: Academic Press; 1990. [Google Scholar]
- 13.Jauniaux J-C, Grenson M. GAP1, the general amino acid permease gene of Saccharomyces cerevisiae. Eur J Biochem. 1990;190:39–44. doi: 10.1111/j.1432-1033.1990.tb15542.x. [DOI] [PubMed] [Google Scholar]
- 14.Ma H, Kunes S, Schatz P J, Botstein D. Plasmid construction by homologous recombination in yeast. Gene. 1987;58:201–216. doi: 10.1016/0378-1119(87)90376-3. [DOI] [PubMed] [Google Scholar]
- 15.Magasanik B. Regulation of nitrogen utilization. In: Strathern J N, Jones E W, Broach J, editors. The molecular biology of the yeast Saccharomyces: metabolism and gene expression. II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. pp. 283–317. [Google Scholar]
- 16.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. pp. 352–355. [Google Scholar]
- 17.Nelissen B, Mordant P, Jonniaux J-L, De Wachter R, Goffeau A. Phylogenetic classification of the major superfamily of membrane transport facilitators, as deduced from genome sequencing. FEBS Lett. 1995;377:232–236. doi: 10.1016/0014-5793(95)01380-6. [DOI] [PubMed] [Google Scholar]
- 18.Oliver S G, et al. The complete DNA sequence of yeast chromosome III. Nature. 1992;357:38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- 18a.Regenberg, B. Personal communication.
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 20.Schreve J, Garrett J M. The branched-chain amino acid permease gene of Saccharomyces cerevisiae, BAP2, encodes the high-affinity leucine permease (S1) Yeast. 1997;13:435–439. doi: 10.1002/(SICI)1097-0061(199704)13:5<435::AID-YEA95>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 21.Sherman F, Fink G R, Hicks J B. Methods in yeast genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- 22.Soussi-Boukedou S, Vissers S, Urrestarazu A, Jauniaux J-C, André B. Gzf3p, a fourth GATA factor involved in nitrogen-regulated transcription in Saccharomyces cerevisiae. Mol Microbiol. 1997;23:1157–1168. doi: 10.1046/j.1365-2958.1997.3021665.x. [DOI] [PubMed] [Google Scholar]
- 23.Stanbrough M, Magasanik B. Two transcription factors, Gln3p and Nil1p, use the same GATAAG sites to activate the expression of GAP1 of Saccharomyces cerevisiae. J Bacteriol. 1996;178:2465–2468. doi: 10.1128/jb.178.8.2465-2468.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanbrough M, Rowen D W, Magasanik B. Role of the GATA factors Gln3p and Nil1p of Saccharomyces cerevisiae in the expression of nitrogen-regulated genes. Proc Natl Acad Sci USA. 1995;92:9450–9454. doi: 10.1073/pnas.92.21.9450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wickerham L J. A critical evaluation of the nitrogen assimilation tests commonly used in the classification of yeasts. J Bacteriol. 1946;52:293–301. doi: 10.1128/JB.52.3.293-301.1946. [DOI] [PubMed] [Google Scholar]
- 26.Woodward J R, Cirillo V P. Amino acid transport and metabolism in nitrogen-starved cells of Saccharomyces cerevisiae. J Bacteriol. 1977;130:714–723. doi: 10.1128/jb.130.2.714-723.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu X, Garrett J M, Schreve J, Michaeli T. GNP1, a glutamine permease whose overproduction induces growth defects in the yeast Saccharomyces cerevisiae. Curr Genet. 1996;30:107–114. doi: 10.1007/s002940050108. [DOI] [PubMed] [Google Scholar]