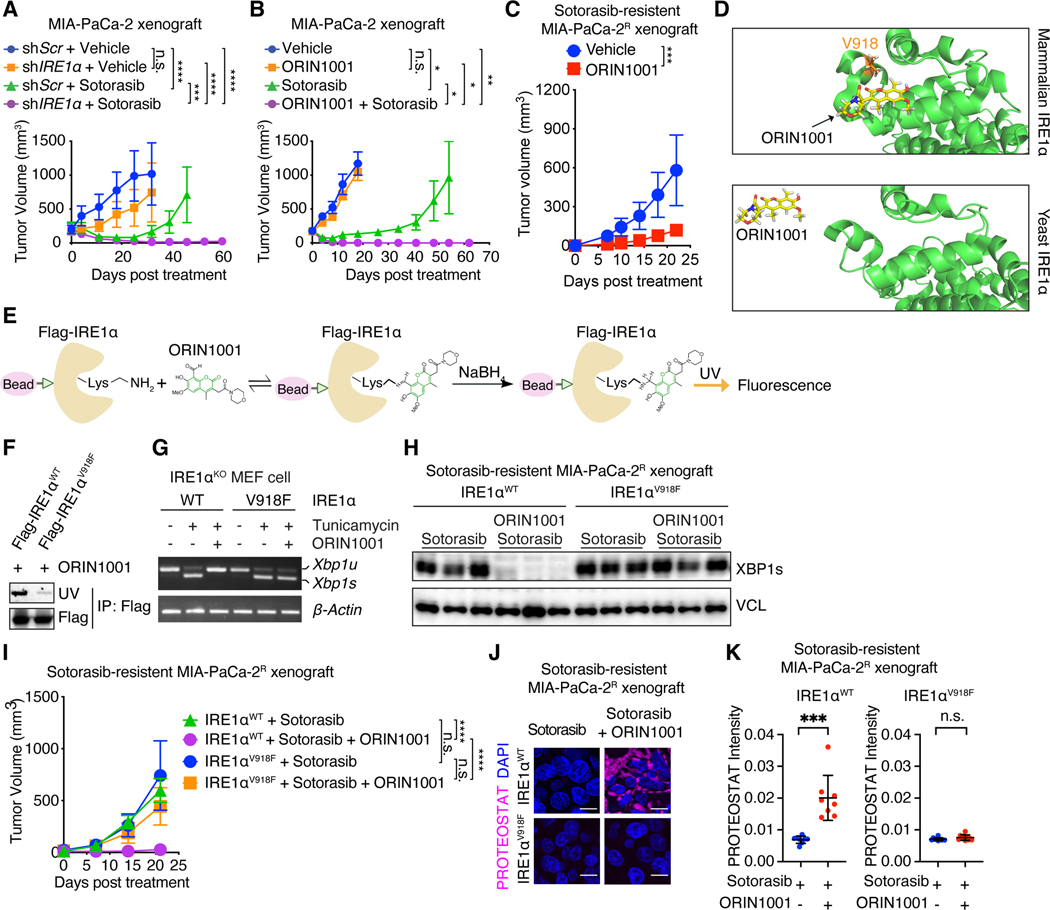
Fig. 7. IRE1α inhibition enhances tumor responses to sotorasib.
(A) Tumor volume of MIA-PaCa-2 tumors transduced with dox-inducible shScr or shIRE1α and treated with doxycycline water, vehicle (n=5) or sotorasib (n=6). (B) Tumor volume of MIA-PaCa-2 tumors treated with vehicle (n=5), ORIN1001 (n=4), sotorasib (n=4), or both (n=5). (C) Tumor volume of sotorasib-resistant MIA-PaCa-2R tumors treated with vehicle (n=4) or ORIN1001 (n=7). (D) Docking modeling of ORIN1001 with IRE1α. V918 is critical for the formation of the shallow pocket at mammalian IRE1α RNase-active site for ORIN1001 binding. (E) Biochemical fluorescence assay detecting the binding between ORIN1001 and IRE1α in vitro. (F) Equal amount of Flag-IRE1αWT or Flag-IRE1αV918F protein purified from 293T cells was used to pull down ORIN1001 in vitro. UV transmission was used to detect ORIN1001 that is covalently bound to IRE1α protein in the SDS-PAGE. (G) Xbp1 splicing in Ire1α-knock out MEF cells expressing IRE1αWT or IRE1αV918F and treated with tunicamycin (5 μg/mL) and/or ORIN1001 (5 μM) for 6 hours as indicated. (H) Immunoblot of XBP1s in shRNA-resistant IRE1αWT or IRE1αV918F-transduced, endogenous IRE1α-depleted MIA-PaCa-2R tumors treated as indicated. (I) Tumor volume of established shRNA-resistant IRE1αWT- or IRE1αV918F- expressing, endogenous IRE1α-depleted, sotorasib-resistant MIA-PaCa-2R tumors treated as indicated. n=10. (J-K) Representative images (J) and quantification (K) of PROTEOSTAT and DAPI staining in MIA-PaCa-2R tumors treated as in (I). Data represent average fluorescence intensity of PROTEOSTAT/cell from each image and are presented as mean ± SD from n>10 independent images. Scale bar: 20μm. Sotorasib: 100mg/kg. ORIN1001: 300mg/kg. Data are presented as mean ± SEM (A, B, C, and I). Two-way ANOVA test with Bonferroni’s multiple comparisons test (A, B, C, and I) or ordinary one-way ANOVA with Dunnett’s multiple comparisons test (K) was used to calculate P values. n.s., not significant, * P<0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.