Abstract
Major DNA binding proteins, designated Ssh7, were purified from the thermoacidophilic archaeon Sulfolobus shibatae. The Ssh7 proteins have an apparent molecular mass of 6.5 kDa and are similar to the 7-kDa DNA binding proteins from Sulfolobus acidocaldarius and Sulfolobus solfataricus in N-terminal amino acid sequence. The proteins constitute about 4.8% of the cellular protein. Upon binding to DNA, the Ssh7 proteins constrain negative supercoils. At the tested Ssh7/DNA mass ratios (0 to 1.65), one negative supercoil was taken up by approximately 20 Ssh7 molecules. Our results, together with the observation that the viral DNA isolated from S. shibatae is relaxed, suggest that regions of free DNA in the S. shibatae genome, if present, are highly positively supercoiled.
DNA supercoiling plays key roles in DNA replication, transcription, recombination, and compaction (23, 29, 30). While closed DNA molecules isolated from eukaryotes, bacteria, and mesophilic archaea are negatively supercoiled, those from thermophilic archaea are relaxed (6, 28). The unique DNA topology in thermophilic archaea appears to be consistent with the need of the organisms to maintain the integrity of their genome at high growth temperatures. Although DNA is under no net superhelical stress in thermophilic archaea, it may be bound by DNA binding proteins in such a way that local superhelical tension results. A few years ago, Musgrave et al. showed that HMf, a histone-like protein from the hyperthermophilic methanogen Methanothermus fervidus, is capable of constraining positive DNA supercoils (20). Binding by HMf would conceivably introduce compensatory negative superhelical tension into protein-free regions of the genome. By comparison, the eucaryal histones and Escherichia coli HU protein constrain DNA in negative supercoils, resulting in complete or partial relaxation, respectively, of the regions of free DNA in these organisms (25, 28). A number of DNA binding proteins have been isolated from other hyperthermophilic archaea, but they have not been tested for their effect on DNA topology. In this study, we sought to determine if DNA binding proteins from other hyperthermophilic archaea resemble HMf in constraining DNA supercoils. For this purpose, we isolated major DNA binding proteins, designated Ssh7, from the thermoacidophilic archaeon Sulfolobus shibatae, which harbors a virus containing DNA known to be relaxed or even slightly positively supercoiled (6, 21). The Ssh7 proteins appear to be sufficiently abundant to bind the entire genome of the organism. We show that the Ssh7 proteins are capable of constraining negative supercoils, a property that has not been shown for any other DNA binding protein from Sulfolobus.
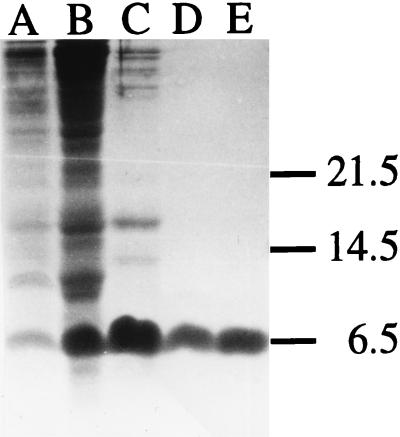
S. shibatae ATCC 51178 was obtained from the American Type Culture Collection. The organism was grown for 72 h at 75°C with mild mixing in 1 liter of Brock’s basal medium (5) supplemented with 0.2% tryptone and 0.1% sucrose in a 4-liter flask on a stirring hot plate. Cells were harvested by centrifugation (10,000 × g, 4°C, 10 min). About 1.5 g (wet weight) of cell paste was normally obtained from 1 liter of culture. Cells were lysed immediately after harvest by the method of Hüdepohl et al. (12) with modifications. The cell pellet was resuspended in 5 volumes of ice-cold lysis buffer (20 mM Tris-HCl [pH 7.6], 2 M KCl, 0.1 mM EDTA, 1 mM dithiothreitol [DTT]). Lysis was initiated by the addition of 10-fold-diluted Triton X-100 to the cell suspension to a final concentration of 0.1%. Following incubation for 30 min on ice, the lysate was cleared by centrifugation (30,000 × g, 4°C, 30 min). The supernatant was further centrifuged (150,000 × g, 4°C, 2.5 h) to yield a postribosomal fraction. All the subsequent purification steps were carried out at 0 to 4°C. A sample (50 ml) of the postribosomal fraction obtained from approximately 7 g of cell paste was dialyzed for 12 h against 25 mM HEPES/KOH (pH 7.6)–0.1 mM EDTA–0.1 mM DTT–5% (vol/vol) glycerol (buffer A). The dialyzed material was loaded onto an SP Sepharose column (30 ml; Pharmacia) equilibrated in buffer A plus 50 mM KCl. The column was washed with buffer A plus 50 mM KCl (90 ml), and bound proteins were eluted with a KCl gradient (0.05 to 1.0 M) in buffer A (400 ml). To identify DNA binding proteins, we employed a gel retardation assay. A sample of each fraction was mixed with plasmid pBluescript KS(−) (0.5 μg; Stratagene) in 20 mM Tris-HCl (pH 7.6)–10 mM MgCl2–1 mM DTT–100 μg of bovine serum albumin (BSA) per ml in a final volume of 20 μl. After incubation for 10 min at room temperature, a loading solution (4 μl) containing 0.25% bromophenol blue, 0.25% xylene cyanol FF, and 30% (vol/vol) glycerol was added to each mixture, and the samples were electrophoresed at room temperature on a 0.8% agarose gel at a constant voltage (4 V/cm) in 0.5× TBE (16). Fractions eluting from the SP Sepharose column at about 0.3 and 0.5 M KCl were capable of significant retardation of the plasmid in the gel and, thus, presumably contained DNA binding proteins. In the present study, we focused on the DNA binding activity in the 0.3 M KCl fraction (the purification and characterization of the DNA binding protein in the 0.5 M KCl fraction will be reported elsewhere). As judged by tricine-sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (26), the molecular mass of the predominant protein species in the 0.3 M KCl fraction is approximately 6.5 kDa (Fig. 1, lane C). It was later found that the 6.5-kDa species probably comprises two polypeptides that have nearly identical N-terminal amino acid sequences (see below) and behave indistinguishably on chromatographic columns used in this study. Fractions containing the 6.5-kDa proteins were pooled, dialyzed against 25 mM ethanolamine-HCl, pH 9.7, and applied to a PBE 94 chromatofocusing column (10 ml; Pharmacia) equilibrated in 25 mM ethanolamine-HCl, pH 9.7. The column was washed with the equilibration buffer. The 6.5-kDa proteins were found in the flowthrough, suggesting that the proteins are highly basic. Since most cellular proteins in S. shibatae presumably have isoelectric points below 9.7, the chromatofocusing step resulted in a significant purification of the 6.5-kDa proteins (Fig. 1, lane D). As expected, the maximum DNA binding activity coeluted with the 6.5-kDa proteins. To eliminate the remaining contaminants, the protein fractions in the flowthrough were pooled, dialyzed against 50 mM Tris-HCl (pH 7.6)–0.1 mM EDTA–0.1 mM DTT–5% (vol/vol) glycerol (buffer B), and applied to a phosphocellulose P11 column (10 ml; Whatman) equilibrated in buffer B plus 50 mM KCl. The column was washed with buffer B plus 50 mM KCl (30 ml), and proteins were eluted with a KCl gradient (0.05 to 1.5 M) in buffer B (300 ml). The 6.5-kDa proteins eluted at 0.5 M KCl. When this protein preparation was subjected to tricine-SDS-polyacrylamide gel electrophoresis and the gel was stained with Coomassie brilliant blue R-250, only a single protein band was observed (Fig. 1, lane E). Fractions containing the 6.5-kDa proteins were pooled, concentrated in an Amicon ultrafiltration unit using a YM-5 membrane, and stored at −20°C in 50 mM Tris-HCl (pH 7.6)–1 mM DTT–20% (vol/vol) glycerol. Approximately 2 mg of the pure 6.5-kDa proteins was obtained, representing a recovery of 20%.
FIG. 1.
Tricine-SDS-polyacrylamide gel electrophoresis of samples taken at various stages during the purification of Ssh7. Lane A, total cellular proteins; lane B, postribosomal fraction; lane C, SP Sepharose peak fractions; lane D, chromatofocusing peak fractions; lane E, phosphocellulose peak fractions. Positions of molecular mass standards (in kilodaltons) are indicated at the left. The gel (16.5% T and 6% C) was prepared and run as described by Schägger and von Jagow (26).
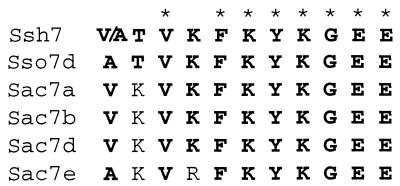
The N-terminal amino acid sequences of the 6.5-kDa proteins were determined by microsequencing. The first 11 residues of the S. shibatae proteins are similar to those of Sac7 and Sso7, 7-kDa DNA binding proteins isolated from Sulfolobus acidocaldarius and Sulfolobus solfataricus, respectively (Fig. 2). As found in Sac7 and Sso7, the S. shibatae proteins lack the initiator methionine and may contain methylated lysines at positions 4 and 6 since an unidentified compound, in addition to the lysine, was observed at these positions. Two different amino acid residues were found reproducibly in similar amounts at position 1 of the 6.5-kDa proteins, suggesting the presence of isoforms. Consistent with this result is the finding that, when genomic blots of EcoRI- and HindIII-digested S. shibatae DNA were probed with a mixture of degenerate oligonucleotides derived from the N-terminal amino acid sequences of the 6.5-kDa proteins, hybridization to two bands was observed in each case (data not shown). These data are not surprising in view of the observation that the native Sac7 proteins are a mixture of polypeptides of similar sizes and properties (11, 18, 19). Taken together, the N-terminal amino acid sequences, apparent molecular mass, and DNA binding ability of the 6.5 kDa proteins are consistent with the notion that the S. shibatae proteins are homologous to the Sac and Sso proteins. Therefore, we designate the S. shibatae proteins Ssh7.
FIG. 2.
Comparison of N-terminal amino acid sequences of the Ssh7 proteins, the Sso7d protein (7), and the Sac7a, -b, -d, and -e proteins (8, 14). Amino acids common to all six sequences are indicated by asterisks. Amino acids identical to the corresponding amino acids in Ssh7 are shown in boldface.
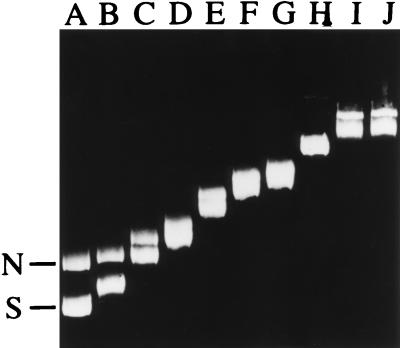
The Ssh7 proteins bind to both relaxed and negatively supercoiled DNAs (Fig. 3). The DNAs were maximally retarded by Ssh7 at protein/DNA mass ratios ≥2.6. A similar gel retardation pattern was obtained when negatively supercoiled DNA alone was used for Ssh7 binding in the assay. Presumably, the mass ratio of 2.6 represents an upper limit of the binding capacity of DNA for Ssh7. In other words, an Ssh7 molecule may occupy a binding site of at least 4 bp. This estimate is in good agreement with the binding site sizes determined in fluorescence studies for Sac7d (4 bp) and Sso7 (3 to 6 bp) (3, 19).
FIG. 3.
Binding of Ssh7 to negatively supercoiled and relaxed plasmid DNAs. Negatively supercoiled and nicked circular pBluescript KS(−) molecules (0.5 μg each) were mixed with the Ssh7 proteins at various Ssh7/DNA mass ratios. Following incubation for 10 min at 40°C, the protein-DNA complexes were subjected to electrophoresis in 0.8% agarose. Lanes A to J, DNAs bound by Ssh7 at the following Ssh7/DNA mass ratios: 0, 0.11, 0.22, 0.44, 0.66, 0.88, 1.32, 1.76, 2.64, and 3.52, respectively. Positions of free negatively supercoiled plasmid (S) and free nicked plasmid (N) are marked.
To estimate the cellular content of the Ssh7 proteins, S. shibatae cells were lysed as described above and the protein concentration of the lysate was determined by the trichloroacetic acid-Lowry protein assay using BSA as the standard (22). Various amounts of the lysate and pure Ssh7 were electrophoresed on a tricine-SDS-polyacrylamide gel. Following electrophoresis, the gel was stained with Coomassie brilliant blue R-250 (data not shown). A band in the lysate sample having the mobility of Ssh7 was well resolved in this gel system (Fig. 1, lane A). The putative Ssh7 band in each lysate lane and the Ssh7 band in each pure-protein lane were scanned with a densitometer (Hoefer), and the staining intensities of the bands were analyzed with the SigmaGel software (Jandel). We estimate that the Ssh7 proteins constitute approximately 4.8% of the total cellular protein assuming that no other proteins in the lysate comigrate with Ssh7. Clearly, the Ssh7 proteins are among the most abundant proteins in S. shibatae. We also measured the DNA concentration in the lysate using the fluorochrome bisbenzimidazole (Hoescht 33258) with a TKO 100-Mini Fluorometer (Hoefer). We found that the mass ratio of Ssh7 to chromosomal DNA is about 4 in S. shibatae. DNA binding proteins are known to exist in abundance in thermophilic archaea, including S. acidocaldarius and S. solfataricus (10). For instance, nucleoid fractions isolated from S. acidocaldarius contain protein and DNA at a weight ratio of 7:1 (24). Given the estimated binding capacity of DNA for Ssh7, the Ssh7 proteins are sufficiently abundant to coat the entire chromosomal DNA in S. shibatae. However, the Ssh7 proteins may be unevenly distributed in the nucleoid. Electron microscopic studies with Sso7 from S. solfataricus revealed that, at high protein-to-DNA ratios, large protein-DNA clusters were formed that were surrounded by protein-free DNA loops (7). Alternatively, binding of Ssh7 may be restricted to the periphery of the nucleoid as suggested for the distribution of HU in E. coli (27).
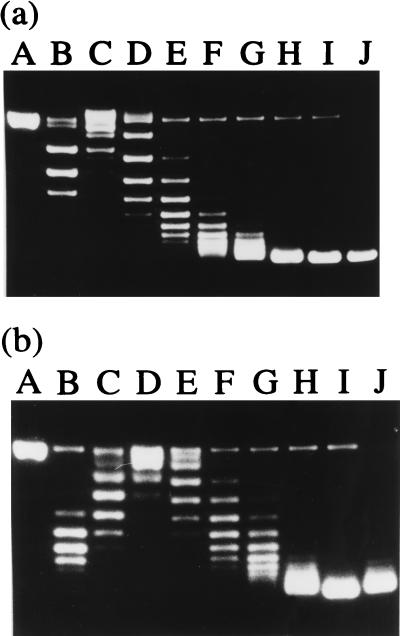
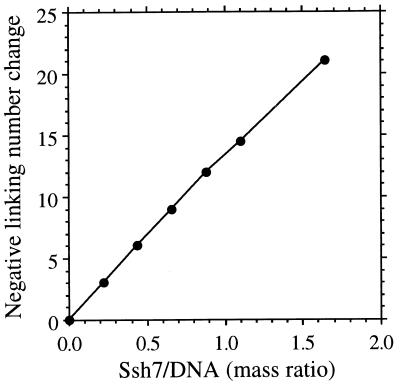
To study the effect of Ssh7 on superhelical tension in closed duplex DNA, we employed a nick closure assay. Plasmid pUC18 (16) containing a single nick per circular molecule was prepared using DNase I as described previously (9). The Ssh7 proteins were incubated for 15 min at 22°C with the nicked plasmid (4 μg) at various mass ratios in 20 mM Tris-HCl (pH 7.4)–50 mM KCl–10 mM MgCl2–1 mM DTT–100 μg of BSA per ml in a final volume of 40 μl. A ligation solution (20 μl) containing 20 mM Tris-HCl (pH 7.4)–50 mM KCl–10 mM MgCl2–1 mM DTT–3 mM ATP–200 U of T4 DNA ligase (New England Biolabs)–100 μg of BSA per ml was added to each mixture. The ligation reaction was for 15 min at 22°C and was stopped by the addition of SDS and EDTA to 0.6% and 30 mM, respectively. DNA-bound proteins were removed by digestion with proteinase K (3 mg/ml) for 30 min at 37°C. Aliquots (about 1.2 μg of DNA) of each sample were electrophoresed at a constant voltage (1.6 V/cm) at 22°C in 0.5× TPE (16) on 1.75% agarose gels containing different amounts of chloroquine (0 to 5 μg/ml). The linking number changes of the DNA were determined by band counting as described by Keller (13). As shown in Fig. 4a, when the nicked plasmid was ligated in the presence of Ssh7, the resulting closed circular DNA was supercoiled, suggesting that the Ssh7 proteins are capable of constraining DNA supercoils. Comparison of the mobilities of pUC18 topoisomers in the presence of 0.5 μg of chloroquine per ml and in the absence of chloroquine indicates that the DNA was constrained by Ssh7 in negative supercoils (Fig. 4b). The number of negative supercoils constrained by Ssh7 was proportional to the Ssh7/DNA mass ratio in the tested range (Fig. 5). Based on the estimated molecular weight of Ssh7, binding of about 20 Ssh7 molecules results in one negative supercoil being constrained. Since the above assay was conducted at 30°C, we asked if Ssh7 would have the same effect on the structure of DNA at the growth temperature of S. shibatae. To answer this question, we carried out the nick closure assay at 80°C using pfu DNA ligase (Stratagene), a thermostable enzyme from Pyrococcus furiosus. The capacity of Ssh7 to constrain negative supercoils at 80°C did not seem to differ measurably from that at 30°C (data not shown). Therefore, the Ssh7 proteins are different from HMf from M. fervidus in the direction of the supercoils that they constrain (20). It appears that DNA binding proteins from different hyperthermophilic archaea may affect DNA topology in different manners.
FIG. 4.
Nick closure analysis of the capacity of Ssh7 to constrain DNA supercoils. Plasmid pUC18 containing a single nick per molecule was mixed with Ssh7 at various Ssh7/DNA mass ratios and, after incubation, treated with T4 DNA ligase. The samples were deproteinized and subjected to agarose gel electrophoresis in the presence of 0.5 μg of chloroquine per ml (b) or in the absence of chloroquine (a). Lane A, single-nicked pUC18; lanes B to I, topoisomers of pUC18 ligated at the following Ssh7/DNA mass ratios: 0, 0.22, 0.44, 0.66, 0.88, 1.1, 1.65, and 2.2, respectively; lane J, native pUC18.
FIG. 5.
Linking number change of circular plasmid DNA covalently closed in the presence of Ssh7. Single-nicked plasmid pUC18 complexed with Ssh7 at various Ssh7/DNA ratios was ligated with T4 DNA ligase. The linking change of the plasmid was measured by resolving topoisomers on agarose gels in the presence of 0.5 to 6 μg of chloroquine per ml or in the absence of chloroquine and band counting.
Given their abundance, the Ssh7 proteins would probably take up a large number of DNA supercoils in vivo. If regions of free DNA exist in the S. shibatae nucleoid, they would presumably be positively supercoiled such that the chromosomal DNA is under no net superhelical tension. The presence of reverse gyrase in S. shibatae (4) seems to be consistent with the positive superhelicity in the protein-free regions of DNA. An equally plausible possibility would be that Ssh7, and perhaps other proteins, forms a protein core which is wrapped around by overtwisted DNA in a left-handed manner. In this model, a compensatory accumulation of positive supercoiling in the protein-free regions may not occur. If the chromosomal DNA is entirely bound by Ssh7 and other proteins, the model provides an explanation for the structural basis of the interaction of Ssh7 with DNA.
The in vivo function of the 7-kDa proteins from Sulfolobus remains to be elucidated. It has been speculated that, because of the low G+C content of genomic DNA and low cellular salt concentration, helix-stabilizing proteins may be necessary for these hyperthermophiles (24). The 7-kDa class of proteins has been considered a likely candidate (7, 8, 11). Previous studies have shown that the Sac7 and Sso7 proteins raise the melting temperature of duplex DNA by more than 30°C (3, 18). In this study, we found that the Ssh7 proteins are capable of constraining negative supercoils, a property that is presumably of importance to DNA packaging and duplex stabilization in Sulfolobus. Although covalently closed circular DNA is intrinsically resistant to irreversible thermodenaturation at temperatures up to 107°C in vitro (17), transient single-stranded DNA bubbles probably form more readily and extensively at temperatures suitable for the growth of hyperthermophiles (1, 2, 15). These single-stranded regions in DNA are likely sites of the initiation of abnormal or detrimental cellular processes. The 7-kDa proteins may thus serve to protect the chromosomal DNA of Sulfolobus from undergoing local denaturation at high growth temperatures through direct protein-DNA interaction as well as by its effect on the superhelicity of protein-free DNA regions.
Acknowledgments
We are grateful to David Becker, Peijin Zhou, and Youxin Song for valuable assistance and to Jindong Zhao of the State Key Laboratory of Protein Engineering at Peking University and Larry Eckler of Quality Controlled Biochemicals, Inc., for performing the N-terminal amino acid sequence analysis of the Ssh7 proteins.
This work was supported by a grant from the Irvine foundation and grant 39740009 from the National Science Foundation of China to L.H.
REFERENCES
- 1.Bauer W R, Benham C J. The free energy, enthalpy and entropy of native and of partially denatured closed circular DNA. J Mol Biol. 1993;234:1184–1196. doi: 10.1006/jmbi.1993.1669. [DOI] [PubMed] [Google Scholar]
- 2.Bauer W R, Ohtsubo H, Ohtsubo E, Benham C J. Energetics of coupled twist and writhe changes in closed circular pSM1 DNA. J Mol Biol. 1995;253:438–452. doi: 10.1006/jmbi.1995.0565. [DOI] [PubMed] [Google Scholar]
- 3.Baumann H, Knapp S, Lundback T, Ladenstein R, Hard T. Solution structure and DNA-binding properties of a thermostable protein from the archaeon Sulfolobus solfataricus. Struct Biol. 1994;1:808–819. doi: 10.1038/nsb1194-808. [DOI] [PubMed] [Google Scholar]
- 4.Bouthier de la Tour C, Potemer C, Huber R, Forterre P, Duguet M. Reverse gyrase, a hallmark of the hyperthermophilic archaebacteria. J Bacteriol. 1990;172:6803–6808. doi: 10.1128/jb.172.12.6803-6808.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brock T D, Brock K M, Belly R T, Weiss R L. Sulfolobus: a new genus of sulfur-oxidizing bacteria living at low pH and high temperature. Arch Mikrobiol. 1972;84:54–68. doi: 10.1007/BF00408082. [DOI] [PubMed] [Google Scholar]
- 6.Charbonnier F, Forterre P. Comparison of plasmid DNA topology among mesophilic and thermophilic eubacteria and archaebacteria. J Bacteriol. 1994;176:1251–1259. doi: 10.1128/jb.176.5.1251-1259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Choli T, Henning P, Wittmann-Liebold B, Reinhardt R. Isolation, characterization, and microsequence analysis of a small basic methylated DNA binding protein from the archaebacterium Sulfolobus solfataricus. Biochim Biophys Acta. 1988;950:193–203. doi: 10.1016/0167-4781(88)90011-5. [DOI] [PubMed] [Google Scholar]
- 8.Choli T, Wittmann-Liebold B, Reinhardt R. Microsequence analysis of DNA-binding proteins 7a, 7b, and 7e from the archaebacterium Sulfolobus acidocaldarius. J Biol Chem. 1988;263:7087–7093. [PubMed] [Google Scholar]
- 9.Clark D J, Felsenfeld G. Formation of nucleosomes on positively supercoiled DNA. EMBO J. 1991;10:387–395. doi: 10.1002/j.1460-2075.1991.tb07960.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grayling R A, Sandman K, Reeve J N. DNA stability and DNA binding proteins. In: Adams M W W, editor. Enzymes and proteins from hyperthermophilic microorganisms. San Diego, Calif: Academic Press; 1996. pp. 437–467. [Google Scholar]
- 11.Grote M, Dijk J, Reinhardt R. Ribosomal and DNA binding proteins of the thermoacidophilic archaebacterium Sulfolobus acidocaldarius. Biochim Biophys Acta. 1986;873:405–413. [Google Scholar]
- 12.Hüdepohl U, Reiter W-D, Zillig W. In vitro transcription of two rRNA genes of the archaebacterium Sulfolobus sp. B12 indicates a factor requirement for specific initiation. Proc Natl Acad Sci USA. 1990;87:5851–5855. doi: 10.1073/pnas.87.15.5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keller W. Determination of the number of superhelical turns in simian virus 40 DNA by gel electrophoresis. Proc Natl Acad Sci USA. 1975;72:4876–4880. doi: 10.1073/pnas.72.12.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura M, Kimura J, Davie P, Reinhardt R, Dijk J. The amino acid sequence of a small DNA binding protein from the archaebacterium Sulfolobus solfataricus. FEBS Lett. 1984;176:176–178. doi: 10.1016/0014-5793(84)80935-7. [DOI] [PubMed] [Google Scholar]
- 15.Kowalski D, Natale D, Eddy M. Stable DNA unwinding, not “breathing,” accounts for single-strand-specific nuclease hypersensitivity of specific A+T-rich sequences. Proc Natl Acad Sci USA. 1988;85:9464–9468. doi: 10.1073/pnas.85.24.9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 17.Marguet E, Forterre P. DNA stability at temperatures typical for hyperthermophiles. Nucleic Acids Res. 1994;22:1681–1686. doi: 10.1093/nar/22.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McAfee J G, Edmondson S P, Dalta P K, Shriver J W, Gupla R. Gene cloning, expression, and characterization of the Sac7 proteins from the hyperthermophile Sulfolobus acidocaldarius. Biochemistry. 1995;34:10063–10077. doi: 10.1021/bi00031a031. [DOI] [PubMed] [Google Scholar]
- 19.McAfee J G, Edmondson S P, Zegar I, Shriver J W. Equilibrium DNA binding of Sac7d protein from the hyperthermophile Sulfolobus acidocaldarius: fluorescence and circular dichroism studies. Biochemistry. 1996;35:4034–4045. doi: 10.1021/bi952555q. [DOI] [PubMed] [Google Scholar]
- 20.Musgrave D M, Sandman K M, Reeve J N. DNA binding by the archaeal histone HMf results in positive supercoiling. Proc Natl Acad Sci USA. 1991;88:10397–10401. doi: 10.1073/pnas.88.23.10397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nadal M, Mirambeau G, Forterre P, Reiter W-D, Duguet M. Positively supercoiled DNA in a virus-like particle of an archaebacterium. Nature. 1986;321:256–258. [Google Scholar]
- 22.Peterson G L. A simplification of the protein assay method of Lowry et al. which is more generally applicable. Anal Biochem. 1977;83:346–356. doi: 10.1016/0003-2697(77)90043-4. [DOI] [PubMed] [Google Scholar]
- 23.Pruss G J, Drlica K. DNA supercoiling and prokaryotic transcription. Cell. 1989;56:521–523. doi: 10.1016/0092-8674(89)90574-6. [DOI] [PubMed] [Google Scholar]
- 24.Reddy T R, Suryanarayana T. Archaebacterial histone-like proteins: purification and characterization of helix stabilizing DNA binding proteins from the acidothermophile Sulfolobus acidocaldarius. J Biol Chem. 1989;264:17298–17308. [PubMed] [Google Scholar]
- 25.Rouviere-Yaniv J, Germond J, Yaniv M. E. coli DNA binding protein HU forms nucleosome-like structure with circular double-stranded DNA. Cell. 1979;17:265–274. doi: 10.1016/0092-8674(79)90152-1. [DOI] [PubMed] [Google Scholar]
- 26.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 27.Shellman V L, Pettijohn D E. Introduction of proteins into living bacterial cells: distribution of labeled HU protein in Escherichia coli. J Bacteriol. 1991;173:3047–3059. doi: 10.1128/jb.173.10.3047-3059.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sinden R R, Carlson J O, Pettijohn D E. Torsional tension in the DNA double helix measured with trimethylpsoralen in living E. coli cells. Cell. 1980;21:773–783. doi: 10.1016/0092-8674(80)90440-7. [DOI] [PubMed] [Google Scholar]
- 29.Wang J C, Peck L J, Becherer K. DNA supercoiling and its effects on DNA structure and function. Cold Spring Harbor Symp Quant Biol. 1983;47:85–91. doi: 10.1101/sqb.1983.047.01.011. [DOI] [PubMed] [Google Scholar]
- 30.Wang J C. DNA topoisomerases. Annu Rev Biochem. 1985;54:665–697. doi: 10.1146/annurev.bi.54.070185.003313. [DOI] [PubMed] [Google Scholar]