Abstract
Background
Acyl-CoA-binding domain-containing 3 (ACBD3) is a multifunctional protein, that plays essential roles in cellular signaling and membrane domain organization. Although the precise roles of ACBD3 in various cancers remain unclear. Thus, we aimed to determine the diverse roles of ACBD3 in pan-cancers.
Methods
Relevant clinical and RNA-sequencing data for normal tissues and 33 tumors from The Cancer Genome Atlas (TCGA) database, the Human Protein Atlas, and other databases were applied to investigate ACBD3 expression in various cancers. ACBD3-binding and ACBD3-related target genes were obtained from the STRING and GEPIA2 databases. The possible functions of ACBD3-binding genes were explored using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses. We also applied the diagnostic value and survival prognosis analysis of ACBD3 in pan-cancers using R language. The mutational features of ACBD3 in various TCGA cancers were obtained from the cBioPortal database.
Results
When compared with normal tissues, ACBD3 expression was statistically upregulated in eleven cancers and downregulated in three cancers. ACBD3 expression was remarkably different among various pathological stages of tumors, immune and molecular subtypes of cancers, cancer phosphorylation levels, and immune cell infiltration. The survival of four tumors was correlated with the expression level of ACBD3, including pancreatic adenocarcinoma, adrenocortical carcinoma, sarcoma, and glioma. The high accuracy in diagnosing multiple tumors and its correlation with prognosis indicated that ACBD3 may be a potential biomarker of pan-cancers.
Conclusion
According to our pan-cancer analysis, ACBD3 may serve as a remarkable prognostic and diagnostic biomarker of pan-cancers as well as contribute to tumor development. ACBD3 may also provide new directions for cancer treatment targets in the future.
Supplementary Information
The online version contains supplementary material available at 10.1186/s40001-023-01576-8.
Keywords: ACBD3, GCP60, GOCAP1, Pan-cancer, Bioinformatic analysis, Biomarker
Background
Most cancers are diagnosed at an advanced stage with a low chance of cure. The recent research on cancer biomarkers has provided a basis for cancer diagnosis and prognostic assessment, offering new opportunities for the survival of cancer patients [1–3]. Therefore, pan-cancer analyses of any possible genes are necessary to further investigate their molecular mechanisms and to determine their correlation with cancer prognosis.
Acyl-CoA binding domain-containing proteins (ACBDs) are made up of seven ACBD proteins that are essential for the transport and stabilization of acyl-CoA, cellular lipid metabolism, and organelle contact sites, and thus plays an important role in cell metabolism [4]. ACBDs have recently been considered key regulators in the development and progression of some cancers, including breast cancer, hepatocellular carcinoma, etc. [5–7] Acyl-CoA-binding domain-containing 3 (ACBD3) is a part of the ACBD family and a 528 amino acid residue protein [8]. Its vital biological feature is its interaction with different proteins [1]. ACBD3 is also known as Golgi complex-associated protein 1 (GOCAP1), Golgi phosphoprotein 1(GOLPH1), Golgi complex-associated protein of 60 kDa(GCP60), and cAMP-dependent protein kinase and peripheral-type benzodiazepine receptor-associated protein 7(PAP7) [6, 9]. The various designations for ACBD3 reflect its most prominent biological properties, such as transport and transfer of lipids, maintenance of Golgi integrity, regulation of steroidogenesis, and replication of the picornavirus family [4, 6, 10]. ACBD3 is also a crucial player in membrane domain organization and cellular signaling [3]. Existing studies have shown that ACBD3 mediates the malignant process of breast cancer by regulating the intracellular β-84 catenin signaling pathway [11]. ACBD3 also affects the replication of gastric cancer cells in an AKT-dependent manner [12]. In addition, ACBD3 may be involved in the progress of gefitinib on lung cancer cells [13].
Previous researches have demonstrated that ACBD3 played a significant role in the development and treatment of different cancers [11–13]. No studies have explored the association between ACBD3 and pan-cancer development until now. The aim of this study was to further explore whether ACBD3 could serve as a new pan-cancer biomarker and to further determine its molecular mechanism and prognostic relevance. This research was conducted to investigate ACBD3 expression among pan-cancers, investigate the prognostic and diagnostic value of ACBD3 among various tumors, and determine the correlation between ACBD3 mutation characteristics and pan-cancer prognosis.
Methods
Gene expression analysis
We used the “TISSUE” module of Human Protein Atlas (THPA) database (https://www.proteinatlas.org) in December 2022, which works to provide information on the tissue and cellular distribution of all 24,000 human proteins, to investigate ACBD3 expression from the normal tissue atlas and tumor cell lines. And then explored ACBD3 expression in cells using the “SUBCELL” module of THPA database. We downloaded the relevant clinical and RNA-sequencing (RNA-seq) data for normal tissues and 33 tumors from The Cancer Genome Atlas (TCGA) database on 23 February 2023 [14]. R software (version 4.2.1) and ggplot2 package (version 3.3.6) were performed to generate statistical analysis and visualizations, respectively. The Wilcoxon rank-sum test and T test were performed to compare the differences in ACBD3 expression levels between cancer and normal tissues, and between cancer and paracancerous tissues. The Cancer Cell Line Encyclopedia (CCLE) database was used to verify gene expression in pan-cancers on 14 May 2023 [15]. Immunohistochemistry images of different cancers and normal tissues were obtained from the “TISSUE” and “PATHOLOGY” module of THPA database [16]. The data on the relationship between ACBD3 expression and immune infiltrates among various tumors were obtained from the “Immune-Gene” module of TIMER2.0 database in December 2022, which used six state-of-the-art algorithms to provide more reliable estimates of immune infiltration levels for tumor profiles [17].
The ACBD3 expression among various tumor pathological stages was visualized using the “Stage Plot” module of the gene expression profiling interactive analysis (GEPIA2) database (Used on 28 February 2023). ACBD3 phosphoprotein levels in normal tissues and different cancers were obtained by searching for “PhosphoProtein” module of ACBD3 from the Clinical Proteomic Tumor Analysis Consortium (CPTAC) database in March 2023 [18]. Furthermore, we entered ACBD3 in the “Gene Symbol” module of the TISIDB (an integrated repository portal for tumor-immune system interactions) database [19], a website for analyzing interactions between tumors and the immune system, to observe the correlation between ACBD3 expression and various molecular and immune subtypes among different cancers.
ACBD3-related gene enrichment analyses
We obtained 50 available experimentally determined ACBD3-binding proteins from the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database in February 2023 by searching ACBD3 and Homo sapiens organism. We regulated the parameters as follows: “full STRING network”, “evidence”, “Experiments, Textmining, Databases”, “medium confidence (0.400)”, and “no more than 50 interactors” in the 1st shell. Subsequently, the protein − protein interaction (PPI) network was visualized using Cytoscape (version 3.9.1). Next, we retrieved the top 100 ACBD3-related target genes from the “Similar Gene Detection” module of the GEPIA2 database. A Venn diagram was constructed to compare ACBD3-binding and ACBD3-related target genes. Moreover, the possible functions of ACBD3-binding genes were explored using Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses by the R package “ClusterProfiler” (version 4.4.4).
Diagnostic value and survival prognosis analysis
We investigated the diagnostic and prognostic values of ACBD3 using relevant pan-cancer clinical data from TCGA database. The diagnostic performance of ACBD3 in pan-cancers was assessed using the receiver-operating characteristic (ROC) curve and the area under the ROC curve (AUC) applied by the “pROC” package (version 1.18.0). The included AUC values ranged from 0.7 to 1. As a measure of diagnostic accuracy, when the value of the AUC is closer to 1, the diagnostic value is higher. Furthermore, we constructed Kaplan–Meier (K-M) plots using the “survival” and “survminer” package (version 3.3.1) to demonstrate the relationship between ACBD3 expression level and the prognosis [OS (overall survival), DSS (disease-specific survival), and PFI (progress-free interval)] of various tumors [20], the Gene Expression Omnibus (GEO) database was used to verify this relationship [21].
Genetic alteration and DNA methylation analysis
The cBioPortal website was used to investigate genetic alterations in ACBD3 on February 2023. Next, we searched out the mutation type, the alteration frequency, and the copy number alteration (CNA) of all TCGA tumors from the “Cancer Types Summary” module. Furthermore, we obtained the three-dimensional (3D) diagram of ACBD3 structure by using the “Mutations” module. In addition, we also accessed the correlation between ACBD3 alternation and clinical outcomes in different tumors by performing the “Comparison/Survival” module. “Meth-Exp correlation” module of DNA Methylation Interactive Visualization Database (DNMIVD) (http://www.unimd.org/dnmivd/) was used to assess the relationship between promoter methylation and ACBD3 expression, and “MethSurv” (https://biit.cs.ut.ee/methsurv) was used to explore the effect of DNA methylation on tumor prognosis and the relationship between ACBD3 expression and methylation levels [22, 23].
Results
ACBD3 expression in pan-cancers
These results suggested that ACBD3 was moderately expressed in the majority of normal tissues. ACBD3 was highly expressed in the cerebral cortex, hippocampus, duodenum, small intestine, colon, gallbladder, pancreas, prostate, placenta, appendix, and bone marrow. ACBD3 was expressed at low levels in the oral mucosa, liver, ovary, soft tissue, and adipose tissue (Additional file 1: Fig. S1A). The expression of ACBD3 ranked high in breast cancer, kidney cancer, and myeloma tumor cell lines (Additional file 1: Fig. S1B). Intracellular ACBD3 was mainly distributed in the Golgi apparatus (Additional file 1: Fig. S1C, D).
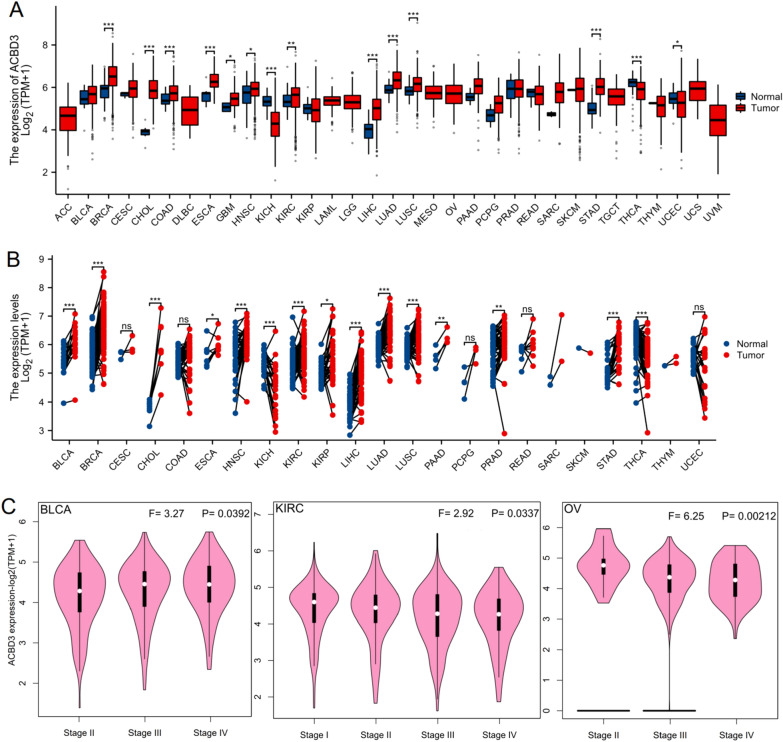
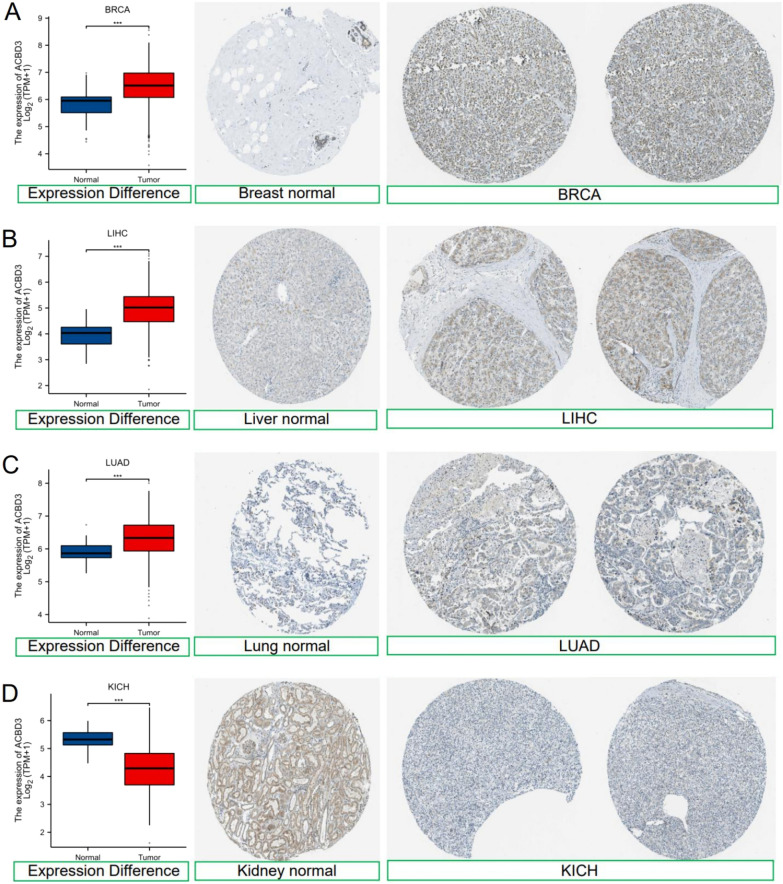
Furthermore, we found that ACBD3 expression was statistically upregulated in eleven cancer types as compared to normal tissues, including stomach adenocarcinoma (STAD), lung squamous cell carcinoma (LUSC), lung adenocarcinoma (LUAD), liver hepatocellular carcinoma (LIHC), kidney renal clear cell carcinoma (KIRC), head and neck squamous cell carcinoma (HNSC), glioblastoma multiforme (GBM), esophageal carcinoma (ESCA), colon adenocarcinoma (COAD), cholangiocarcinoma (CHOL), and breast invasive carcinoma (BRCA), and downregulated in uterine corpus endometrial carcinoma (UCEC), kidney chromophobe (KICH), and thyroid carcinoma (THCA) (Fig. 1A). The CCLE database revealed that ACBD3 was highly expressed in skin cutaneous melanoma (SKCM), BRCA, GBM, KIRC, brain lower grade glioma (LGG), and THCA (Additional file 1: Fig. S1E). In comparison with the adjacent normal tissues, ACBD3 expression was statistically upregulated in thirteen cancers, including STAD, prostate adenocarcinoma (PRAD), pancreatic adenocarcinoma (PAAD), LUSC, LUAD, LIHC, kidney renal papillary cell carcinoma (KIRP), KIRC, HNSC, ESCA, CHOL, BRCA, and bladder urothelial carcinoma (BLCA), and downregulated in KICH and THCA (Fig. 1B). We also constructed a violin diagram of the relationship between various pathological stages of tumors and ACBD3 expression (Fig. 1C). Immunohistochemistry images of normal breast tissue, liver tissue, lung tissue, kidney tissue, BRCA, LIHC, LUAD, and KICH were displayed (Fig. 2).
Fig. 1.
The expression level of ACBD3 in normal tissues and different TCGA tumors. (*p < 0.05, **p < 0.01, ***p < 0.001). adrenocortical cancer (ACC), bladder cancer (BLCA), breast cancer (BRCA), cervical cancer (CESC), bile duct cancer (CHOL), colon cancer (COAD), large B-cell lymphoma (DLBC), esophageal cancer (ESCA), Glioblastoma (GBM), head and neck cancer (HNSC), kidney chromophobe (KICH), kidney clear cell carcinoma (KIRC), kidney papillary cell carcinoma (KIRP), acute myeloid leukemia (LAML), lower grade glioma (LGG), liver hepatocellular carcinoma (LIHC), lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), mesothelioma (MESO), ovarian cancer (OV), pancreatic cancer (PAAD), pheochromocytoma & paraganglioma (PCPG), prostate cancer (PRAD), rectal cancer (READ), sarcoma (SARC), melanoma (SKCM), stomach cancer (STAD), testicular cancer (TGCT), thyroid cancer (THCA), thymoma (THYM), endometrioid cancer (UCEC), uterine carcinosarcoma (UCS), ocular melanomas (UVM). A ACBD3 expression was statistically upregulated in eleven cancer types as compared to normal tissues, including BRCA, CHOL, COAD, ESCA, GBM, HNSC, KIRC, LIHC, LUAD, LUSC, STAD, and downregulated in KICH, THCA, and UCEC; B In comparison with the adjacent normal tissues, ACBD3 expression was statistically upregulated in thirteen cancers, including STAD, PRAD, PAAD, LUSC, LUAD, LIHC, KIRP, KIRC, HNSC, ESCA, CHOL, BRCA, and BLCA, and downregulated in KICH and THCA; C The expression level of ACBD3 in various pathological stages of BLCA, KIRC, and OV. ACBD3 expressed the highest in stage III of BLCA, stage I of KIRC, and stage II of OV
Fig. 2.
The expression level of ACBD3 in different tumors and normal tissues (*p < 0.05, **p < 0.01, ***p < 0.001) and corresponding immunohistochemistry images. A ACBD3 expression was statistically upregulated in breast invasive carcinoma (BRCA), B ACBD3 expression was statistically upregulated in liver hepatocellular carcinoma (LIHC), C ACBD3 expression was statistically upregulated in lung adenocarcinoma (LUAD), D ACBD3 expression was statistically downregulated in kidney chromophobe (KICH)
We assumed that the different expression levels of ACBD3 might affect the immune infiltration of various tumors. Therefore, we explored the correlation between ACBD3 expression and immune infiltration in different cancers using the TIMER2.0 database. Remarkably, the expression of ACBD3 was actively correlated with cancer-associated fibroblast (CAF) infiltration in HNSC (Additional file 1: Fig. S2A) and positively related to neutrophil infiltration in BLCA, COAD, and THCA (Additional file 1: Fig. S2B). ACBD3 expression also positively related to endothelial cell infiltration in COAD, HNSC, and KIRC (Additional file 1: Fig. S2C).
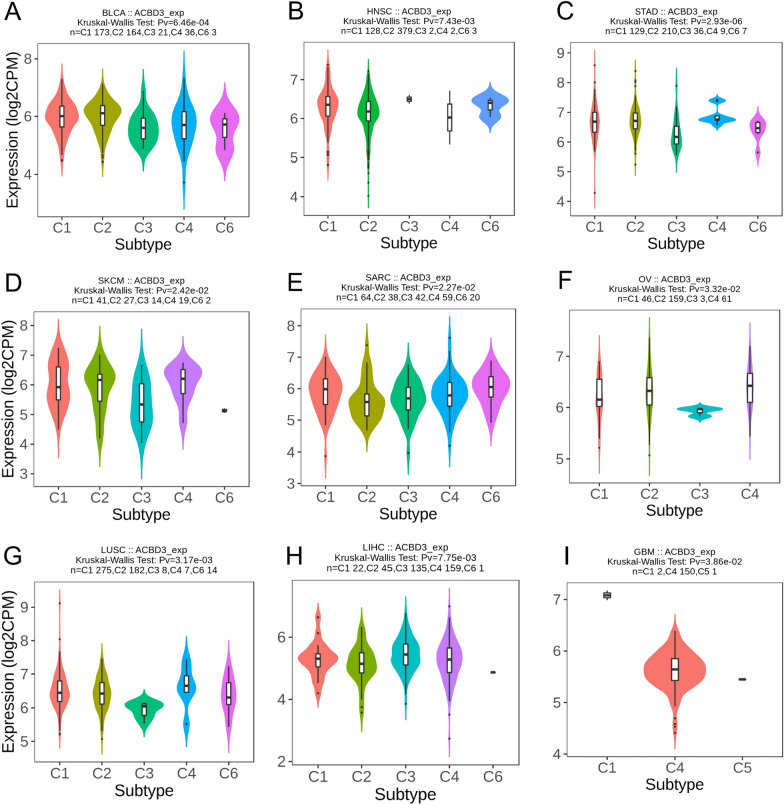
Figure 3 showed that ACBD3 expression was statistically different among the immune subtypes of nine tumors, including BLCA (Fig. 3A), HNSC (Fig. 3B), STAD (Fig. 3C), SKCM (Fig. 3D), sarcoma (SARC) (Fig. 3E), ovarian serous cystadenocarcinoma (OV) (Fig. 3F), LUSC (Fig. 3G), LIHC (Fig. 3H), and GBM (Fig. 3I). Moreover, for immune subtype C1 (wound healing), ACBD3 expressed high in BLCA, SKCM, SARC, and LUSC. For immune subtype C2 (IFN-gamma dominant), ACBD3 expressed high in HNSC, STAD, and OV. For immune subtype C4 (lymphocyte depleted), ACBD3 expressed high in LIHC, and GBM.
Fig. 3.
ACBD3 expression in different immune subtypes of various TCGA cancers. Wound healing (C1), IFN-gamma dominant (C2), inflammatory (C3), lymphocyte depleted (C4), immunologically quiet (C5), TGF-b dominant (C6). A bladder urothelial carcinoma (BLCA), B head and neck squamous cell carcinoma (HNSC), C stomach adenocarcinoma (STAD), D skin cutaneous melanoma (SKCM), E sarcoma (SARC), F ovarian serous cystadenocarcinoma (OV), G lung squamous cell carcinoma (LUSC), H liver hepatocellular carcinoma (LIHC), I glioblastoma multiforme (GBM). For the immune subtype of C1, ACBD3 expressed high in BLCA, SKCM, SARC, and LUSC. For the immune subtype of C2, ACBD3 expressed high in HNSC, STAD, and OV. For the immune subtype of C4, ACBD3 expressed high in LIHC, and GBM
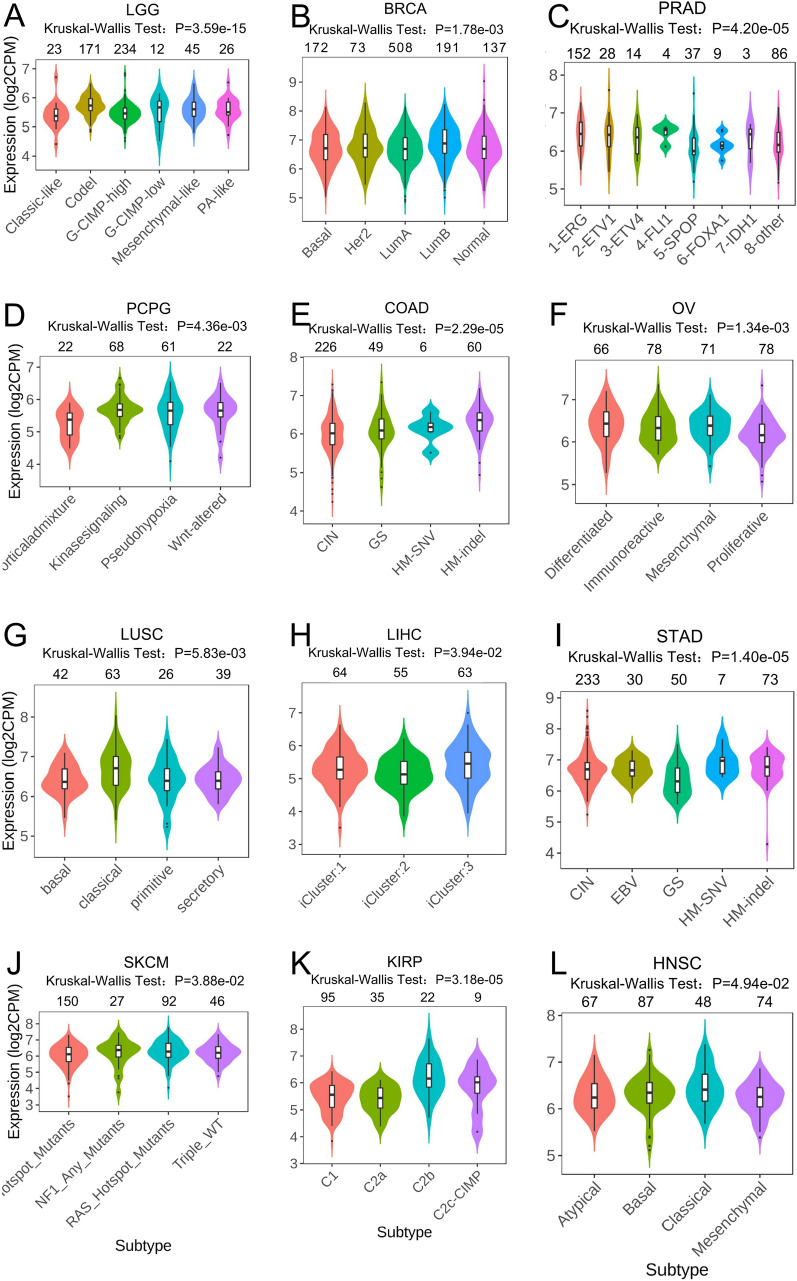
Furthermore, we identified twelve tumors with molecular subtypes associated with ACBD3 expression. Among the molecular subtypes of G-CIMP-high in LGG, ACBD3 showed the highest expression (Fig. 4A). Among the molecular subtypes of LunA in BRCA, ACBD3 showed the highest expression (Fig. 4B). Among the molecular subtypes of 1-ERG in PRAD, ACBD3 showed the highest expression (Fig. 4C). Among the molecular subtypes of Kinasesignaling in pheochromocytoma & paraganglioma (PCPG), ACBD3 showed the highest expression (Fig. 4D). Among the molecular subtype of CIN in COAD, ACBD3 showed the highest expression (Fig. 4E). Among the molecular subtypes of Immunoreactive and Proliferative in OV, ACBD3 showed the highest expression (Fig. 4F). Among the molecular subtypes of classical in LUSC, ACBD3 showed the highest expression (Fig. 4G). Among the molecular subtypes of iCluster:1 in LIHC, ACBD3 showed the highest expression (Fig. 4H). Among the molecular subtypes of CIN in STAD, ACBD3 showed the highest expression (Fig. 4I). Among the molecular subtypes of BRAF_Hotspot_Mutants in SKCM, ACBD3 showed the highest expression (Fig. 4J). Among the molecular subtypes of C1in KIRP, ACBD3 showed the highest expression (Fig. 4K). Among the molecular subtypes of Basal in HNSC, ACBD3 showed the highest expression (Fig. 4L).
Fig. 4.
ACBD3 expression in different molecular subtypes of various TCGA cancers. brain lower grade glioma (LGG), breast invasive carcinoma (BRCA), prostate adenocarcinoma (PRAD), pheochromocytoma and paraganglioma (PCPG), colon adenocarcinoma (COAD), ovarian serous cystadenocarcinoma (OV), lung squamous cell carcinoma (LUSC), liver hepatocellular carcinoma (LIHC), stomach adenocarcinoma (STAD), skin cutaneous melanoma (SKCM), kidney renal papillary cell carcinoma (KIRP), head and neck squamous cell carcinoma (HNSC). ACBD3 expressed the highest in the molecular subtype of A G-CIMP-high in LGG, B LunA in BRCA, C 1-ERG in PRAD, D Kinasesignaling in PCPG, E CIN in COAD, F Immunoreactive and Proliferative in OV, G classical in LUSC, H iCluster:1 in LIHC, I CIN in STAD, J BRAF_Hotspot_Mutants in SKCM, K C1in KIRP, and L Basal in HNSC
ACBD3-related gene enrichment analyses
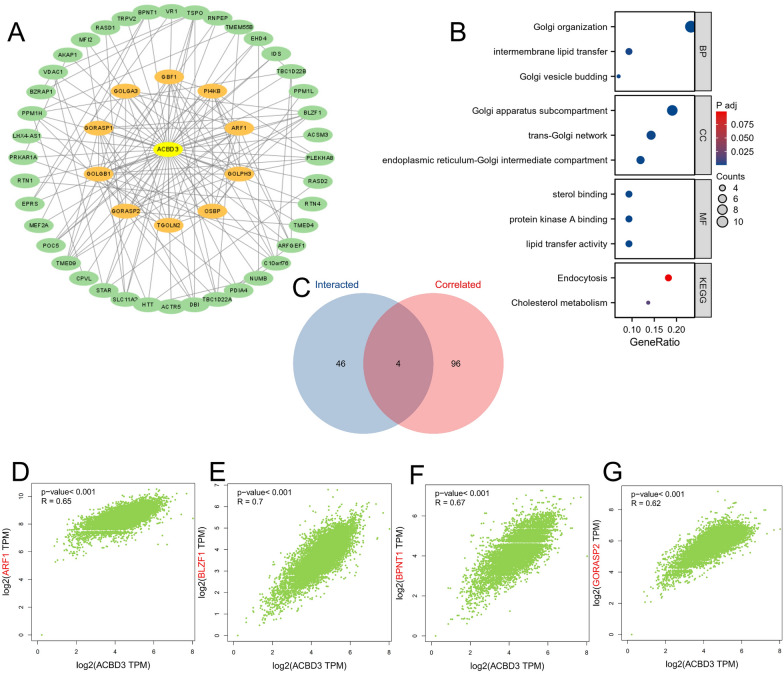
We identified 50 ACBD3-binding proteins from the STRING database (Fig. 5A). Next, GO|KEGG enrichment analysis was conducted on the 50 proteins, revealing that the biological processes (BP) of the 50 proteins included Golgi organization, intermembrane lipid transfer, and Golgi vesicle budding. The cellular components (CC) of the 50 proteins were mainly involved in the Golgi apparatus subcompartment, endoplasmic reticulum-Golgi intermediate compartment, and trans-Golgi network. The molecular function (MF) of 50 proteins were primarily enriched in sterol binding, protein kinase A binding, and lipid transfer activity. The KEGG pathway enrichment of the 50 proteins was primarily related to endocytosis and cholesterol metabolism (Fig. 5B).
Fig. 5.
The correlated and interacted genes of ACBD3. A 50 targeted binding proteins of ACBD3 involved in the protein–protein interaction network; B the Gene Ontology and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses of the 50 interacted genes of ACBD3, including biological process (BP), cellular component (CC), molecular function (MF), and KEGG; C The Venn diagram of the correlated and interacted genes of ACBD3; Correlation analysis between four intersection genes and ACBD3, including D ARF1 (R = 0.65), E BLZF1 (R = 0.70), F BPNT1 (R = 0.67), and G GORASP2 (R = 0.62)
In addition, the top 100 ACBD3-related target genes were retrieved, and a Venn diagram identified the four genes at the intersection of the ACBD3-binding and ACBD3-related target genes as: ARF1, BLZF1, BPNT1, and GORASP2 (Fig. 5C). The expression levels of these four genes were closely related to those of ACBD3: ARF1 (R = 0.65), BLZF1 (R = 0.70), BPNT1 (R = 0.67), and GORASP2 (R = 0.62) (Fig. 5D–G).
Diagnostic value of ACBD3 in pan-cancer
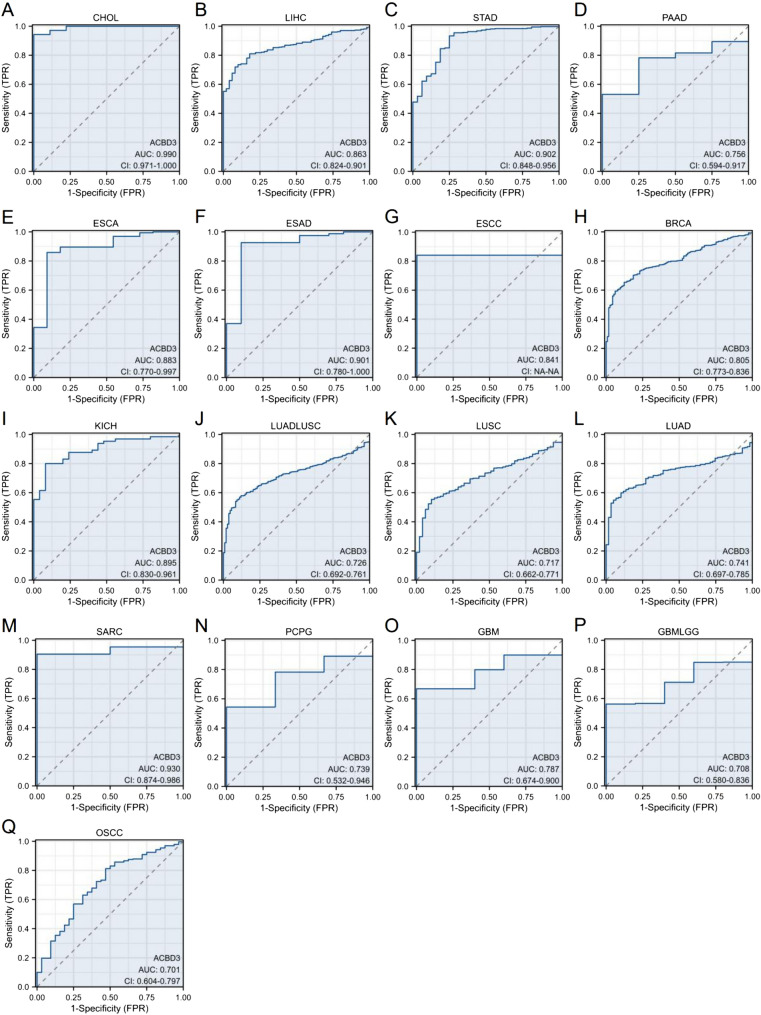
The ROC curve was drawn to explore the diagnostic value of ACBD3 in different cancers, and an AUC value > 0.7 was considered to have a diagnostic value. As shown, ACBD3 had an accurate diagnostic value for 17 kinds of cancers, including CHOL (AUC = 0.990), LIHC (AUC = 0.863), STAD (AUC = 0.902), PAAD (AUC = 0.756), ESCA (AUC = 0.883), esophagus adenocarcinoma (ESAD) (AUC = 0.901), esophagus squamous cell carcinoma (ESCC) (AUC = 0.841), BRCA (AUC = 0.805), KICH (AUC = 0.895), Lung cancer (LUADLUSC) (AUC = 0.726), LUSC (AUC = 0.717), LUAD (AUC = 0.741), SARC (AUC = 0.930), PCPG (AUC = 0.739), GBM (AUC = 0.787), Glioma (GBMLGG) (AUC = 0.708), and Oral squamous cell carcinoma (OSCC) (AUC = 0.701) (Fig. 6).
Fig. 6.
Correlations between ACBD3 expression and receiver operating characteristic (ROC) curve across TCGA tumors, area under the ROC curve (AUC) value > 0.7 was considered to have a diagnostic value. As a measure of diagnostic accuracy, the value of AUC is closer to 1, the diagnostic value is higher. A cholangiocarcinoma (CHOL) (AUC = 0.990), B liver hepatocellular carcinoma (LIHC) (AUC = 0.863), C stomach adenocarcinoma (STAD) (AUC = 0.902), D pancreatic adenocarcinoma (PAAD) (AUC = 0.756), E esophageal carcinoma (ESCA) (AUC = 0.883), F esophagus adenocarcinoma (ESAD) (AUC = 0.901), G esophagus squamous cell carcinoma (ESCC) (AUC = 0.841), H breast invasive carcinoma (BRCA) (AUC = 0.805), I kidney chromophobe (KICH) (AUC = 0.895), J Lung cancer (LUADLUSC) (AUC = 0.726), K lung squamous cell carcinoma (LUSC) (AUC = 0.717), L lung adenocarcinoma (LUAD) (AUC = 0.741), M sarcoma (SARC) (AUC = 0.930), N pheochromocytoma and paraganglioma (PCPG) (AUC = 0.739), O glioblastoma multiforme (GBM) (AUC = 0.787), P Glioma (GBMLGG) (AUC = 0.708), Q Oral squamous cell carcinoma (OSCC) (AUC = 0.701)
Prognostic value of ACBD3 in pan-cancer
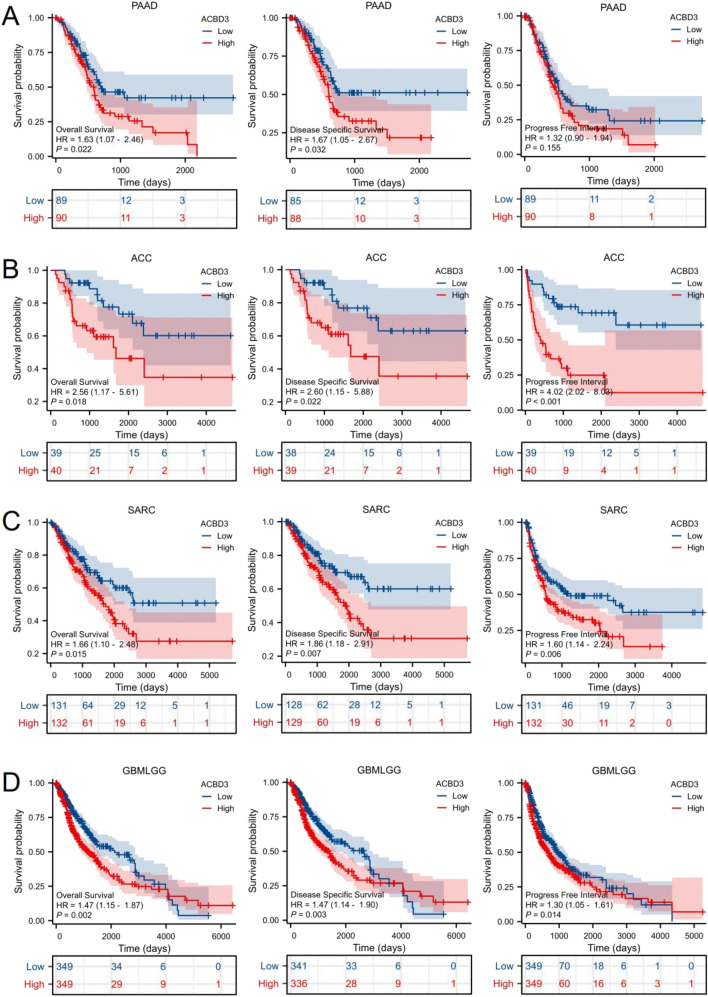
Figure 7 showed that the OS, DSS, and PFI of four tumors were closely correlated with the expression levels of ACBD3, including PAAD, adrenocortical carcinoma (ACC), SARC, and GBMLGG. As for PAAD, the prognosis was negatively related to ACBD3 expression, including OS [hazard ratio (HR) = 1.63, 95% confidence interval (CI): 1.07–2.46, p = 0.022], DSS (HR = 1.67, 95% CI: 1.05–2.67, p = 0.032), and PFI (HR = 1.32, 95% CI: 0.90–1.94, p = 0.155). As for ACC, the prognosis was negatively related to ACBD3 expression, including OS (HR = 2.56, 95% CI: 1.17–5.61, p = 0.018), DSS (HR = 2.60, 95% CI: 1.15–5.88, p = 0.022), and PFI (HR = 4.02, 95% CI: 2.02–8.03, p < 0.001). Also, for SARC, the prognosis was negatively related to ACBD3 expression, including OS (HR = 1.66, 95% CI: 1.10–2.48, p = 0.015), DSS (HR = 1.86, 95% CI: 1.18–2.91, p = 0.007), and PFI (HR = 1.60, 95% CI: 1.14–2.24, p = 0.006). And for GBMLGG, the prognosis was negatively related to ACBD3 expression, including OS (HR = 1.47, 95% CI: 1.15–1.87, p = 0.002), DSS (HR = 1.47, 95% CI: 1.14–1.90, p = 0.003), and PFI (HR = 1.30, 95% CI: 1.05–1.61, p = 0.014).
Fig. 7.
Kaplan–Meier plots [Overall Survival (OS), Disease Specific Survival (DSS), and Progress Free Interval (PFI)]for ACBD3 expression in pan-cancers. A For pancreatic adenocarcinoma (PAAD), the prognosis was negatively related to ACBD3 expression, including OS [hazard ratio (HR) = 1.63, 95% confidence interval (CI): 1.07–2.46, p = 0.022], DSS (HR = 1.67, 95% CI: 1.05–2.67, p = 0.032), and PFI (HR = 1.32, 95% CI: 0.90 -1.94, p = 0.155). B For adrenocortical carcinoma (ACC), the prognosis was negatively related to ACBD3 expression, including OS (HR = 2.56, 95% CI: 1.17–5.61, p = 0.018), DSS (HR = 2.60, 95% CI: 1.15–5.88, p = 0.022), and PFI (HR = 4.02, 95% CI: 2.02–8.03, p < 0.001). C For sarcoma (SARC), the prognosis was negatively related to ACBD3 expression, including OS (HR = 1.66, 95% CI: 1.10–2.48, p = 0.015), DSS (HR = 1.86, 95% CI: 1.18–2.91, p = 0.007), and PFI (HR = 1.60, 95% CI: 1.14–2.24, p = 0.006). D For glioma (GBMLGG), the prognosis was negatively related to ACBD3 expression, including OS (HR = 1.47, 95% CI: 1.15–1.87, p = 0.002), DSS (HR = 1.47, 95% CI: 1.14–1.90, p = 0.003), and PFI (HR = 1.30, 95% CI: 1.05–1.61, p = 0.014
We used the GSE57495, GSE83300, and GSE19750 in the GEO database for validation and found that the OS prognosis was negatively related to ACBD3 expression in GBMLGG (HR = 2.30, 95% CI: 1.19–4.43, p = 0.013). However, there was no statistically significant relationship between ACBD3 expression and prognosis in PAAD and ACC (Additional file 1: Fig. S3).
Genetic alteration and DNA methylation analysis of ACBD3 in pan-cancers
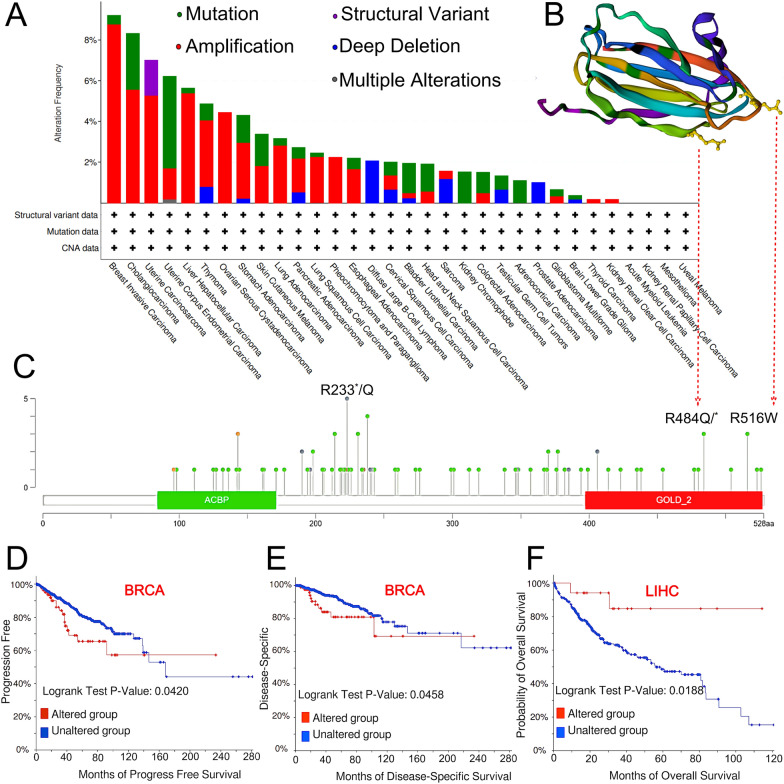
The gene alteration characteristics of ACBD3 in TCGA pan-cancer atlas were obtained. We found that among the many alteration types of ACBD3, “Amplification” was the most common type in patients with BRCA (9.22% alteration frequency) and LIHC (5.38% alteration frequency) (Fig. 8A). It is worth noting that, “Amplification” was the only type of genetic alteration in ovarian serous cystadenocarcinoma, pheochromocytoma, and paraganglioma (Fig. 8A). “Mutation” is the only type of genetic alteration in kidney chromophobe and adrenocortical carcinoma cases (Fig. 8A). “Deep Deletion” is the only type of genetic alteration in diffuse large B-cell lymphoma and prostate adenocarcinoma (Fig. 8A). Figure 8B displayed the 3D structure of ACBD3 protein. Figure 8C displayed the number, sites, and types of the ACBD3 genetic alteration. As shown in Fig. 8C, “Missense Mutation” was the most dominant type of mutation. Alteration in R484Q/ * and R516W in GOLD-2 could have led to the missense mutations in ACBD3.
Fig. 8.
ACBD3 mutation in various tumors of TCGA. A The alteration type and frequency. Among the many alteration types of ACBD3, “Amplification” was the most common type in patients with breast invasive carcinoma (BRCA) (9.22% alteration frequency) and liver hepatocellular carcinoma (LIHC) (5.38% alteration frequency). B The 3D structure of ACBD3 protein. C The number, sites, and types of the ACBD3 genetic alteration. “Missense Mutation” was the most dominant type of mutation. The alteration of R484Q/* and R516W in GOLD-2 is able to lead to the missense mutation of ACBD3. D–F The correlation between ACBD3 alternation and clinical outcomes in different tumors. For BRCA, cases with ACBD3 alternation had a worse prognosis in both progression-free survival and disease-specific survival. Patients with LIHC with ACBD3 alternation had a better prognosis in overall survival
In addition, we determined the relationship between ACBD3 alterations and clinical outcomes in BRCA and LIHC. For BRCA, patients with ACBD3 alterations had a worse prognosis in terms of progression-free survival (Fig. 8D) and disease-specific survival (Fig. 8E) than those patients without ACBD3 alterations. Patients with LIHC and ACBD3 alteration had a better prognosis in overall survival than those patients without ACBD3 alterations (Fig. 8F).
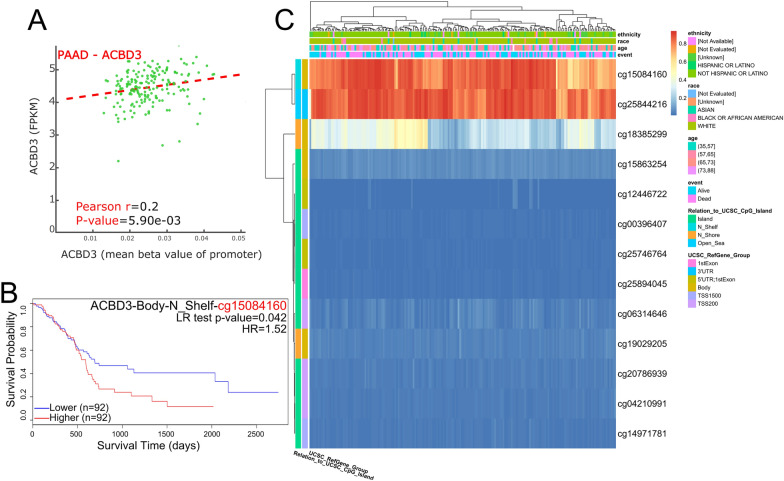
Furthermore, we accessed the relationship between DNA methylation and ACBD3 expression and found that promoter methylation was positively correlated with ACBD3 expression in PAAD (Pearson r = 0.2) (Fig. 9A). In addition, higher methylation β values of ACBD3-Body-N_Shelf-cg15084160 led to a worse OS prognosis in PAAD [HR = 1.52, CI(1.014;2.281), P < 0.05] (Fig. 9B and Table 1).
Fig. 9.
The correlation between the methylation level and ACBD3 expression in pancreatic adenocarcinoma (PAAD). A ACBD3 expression was positive related with ACBD3 promoter methylation in PAAD (Pearson r = 0.2). B Kaplan–Meier plots for ACBD3 promoter methylation in PAAD. the higher methylation β values of ACBD3-Body-N_Shelf-cg15084160 lead to a worse Overall Survival prognosis in PAAD. C The heat map of the relationship between ACBD3 expression and the methylation level in PAAD
Table 1.
Association of ACBD3 with methylation sites in pan-cancers
| CpG site | Cancer | Gene | Group | CpG Island | HR | CI | P |
|---|---|---|---|---|---|---|---|
| cg20786939 | KIRC | ACBD3 | TSS200 | Island | 0.362 | (0.208;0.632) | < 0.001 |
| cg18385299 | LGG | ACBD3 | Body | N_Shore | 0.382 | (0.264;0.551) | < 0.001 |
| cg25844216 | LGG | ACBD3 | 3'UTR | Open_Sea | 0.573 | (0.394;0.834) | 0.004 |
| cg15084160 | PAAD | ACBD3 | Body | N_Shelf | 1.52 | (1.014;2.281) | 0.043 |
HR hazard ratio, CI confidence interval, KIRC kidney renal clear cell carcinoma, LGG brain lower grade glioma, PAAD pancreatic adenocarcinoma
Protein phosphorylation analysis
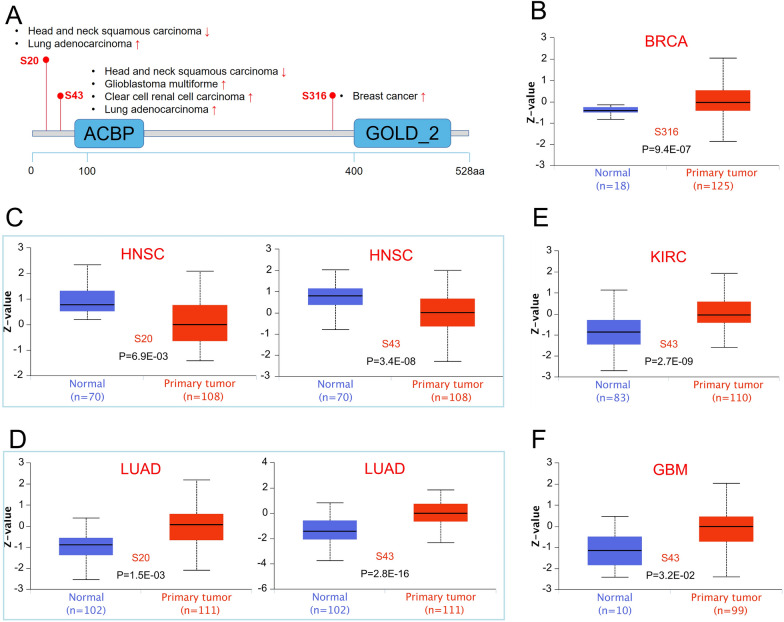
Figure 10A summarized the correlation between various cancers and ACBD3 phosphorylation sites. The S316 locus showed an increased phosphorylation level in BRCA when compared with normal tissues (Fig. 10B). Compared with normal tissues, the S20 locus exhibited a lower phosphorylation level in HNSC (Fig. 10C), with LUAD (Fig. 10D) exhibited the opposite trend. The phosphorylation levels at S43 were higher in KIRC (Fig. 10E), LUAD (Fig. 10D), and GBM (Fig. 10F) and decreased in HNSC (Fig. 10C).
Fig. 10.
Phosphorylation levels of ACBD3 protein in various tumors. A Summary of the correlation between various cancers and ACBD3 phosphorylation sites; B breast invasive carcinoma (BRCA); C head and neck squamous cell carcinoma (HNSC); D lung adenocarcinoma (LUAD); E kidney renal clear cell carcinoma (KIRC); F glioblastoma multiforme (GBM). The S316 locus expressed an upper phosphorylation level in BRCA when compared with normal tissues. The S20 locus exhibited a lower phosphorylation level in HNSC, and LUAD is the opposite. The S43 locus expressed an upper phosphorylation level in KIRC, LUAD, and GBM, and decreased in HNSC
Discussion
Known as GCP60, ACBD3 majored in maintaining the structure and function of the Golgi apparatus. Changes in Golgi structure and function are closely related to cancer development, and Golgi-associated proteins may help diagnose cancer and guide treatment [24–26]. Since the Golgi apparatus is mainly involved in the synthesis and redistribution of new proteins, we speculate that ACBD3 promotes protein binding and thus plays significant roles in the occurrence and development of many tumors with different characteristics. Previous studies demonstrated that ACBD3 was involved in the development and treatment of various types of cancers [11–13]. Nevertheless, no studies have explored the function of ACBD3 in pan-cancers systematically. In order to gain a more comprehensive understanding of ACBD3, we are the first to explore its function and expression of ACBD3 in pan-cancers from the perspective of bioinformatic analysis.
By exploring the TCGA database, we found that ACBD3 expression was remarkably upregulated in eleven cancers, and downregulated in three cancers. This finding suggests that ACBD3 regulates the formation and replication of tumors and facilitates the development of most cancers by acting as an oncogenic gene. We investigated the correlation between ACBD3 expression and the molecular and immune subtypes of TCGA tumors and found that the molecular and immune subtypes of HNSC, STAD, SKCM, OV, LUSC, and LIHC were related to ACBD3 expression. We have assumed that ACBD3 might play a potential role in the occurrence of tumor subtypes, and more experimental results are needed to support this theory. In addition, analysis of the molecular and immune subtypes of various malignant tumors provides a research direction for new tumor therapeutic targets.
We identified ten proteins that interacted most closely with ACBD3: GOLGA3, PI4KB, ARF1, GOLPH3, OSBP, TGOLN2, GORASP2, GOLGB1, GORASP1, and GBF1. The GO|KEGG pathway enrichment analysis suggested that “Golgi organization” and “protein kinase A binding” is the main function of ACBD3, which confirms our hypothesis about the function of the ACBD3-binding proteins. Previous studies had revealed that protein kinase A (PKA) is involved in cancer transformation [27]. The occurrence and development of LIHC, OV, GBM, and ESCC are closely related to PKA [28–31], which reflects the potential association between ACBD3 and various tumors.
What’s more, exploration of the relationship between ACBD3 expression and the diagnostic value of various malignant tumors with different biological characteristics showed that ACBD3 can be used to diagnose a variety of cancers, including CHOL, LIHC, STAD, PAAD, ESCA, ESAD, ESCC, BRCA, KICH, LUADLUSC, LUSC, LUAD, SARC, PCPG, GBM, GBMLGG, and OSCC. Notably, ACBD3 had high diagnostic value (AUC > 0.9) for CHOL, STAD, ESAD, and SARC. In addition, the K-M survival curve for various cancers revealed that ACBD3 was closely associated with the prognosis in PAAD, ACC, SARC, and GBMLGG. Because of the difficulty of integrating all sarcoma-related data, we verified the remaining three results using the GEO database and found that only the prognosis of GBMLGG was correlated with ACBD3 expression. We cannot rule out that this negative result is due to the small amount of data in the GEO database. Furthermore, we found that higher methylation β values of ACBD3-Body-N_Shelf-cg15084160 led to worse OS prognosis in PAAD. These discoveries indicate that ACBD3 has a very important diagnostic and prognostic significance in most cancers, and is expected to be a new biomarker for pan-carcinoma.
In addition, ACBD3 gene mutation analysis has shown that ACBD3 mutations exist in a variety of tumor cells, with missense mutations being the most common. The missense mutations of R484Q/ * and R516W in GOLD-2 can lead to missense mutations in ACBD3. In BRCA, the ACBD3 altered group had poor prognosis, whereas the reverse was true for LIHC. These results provide a new direction for evaluating the prognosis.
Phosphorylation is one of the most extensive post-translational modifications and plays an important role in regulating cell growth, differentiation, apoptosis, and cell signaling [32]. Kinase inhibitors are also considered valuable for the treatment of tumors [33]. Thus, we investigated the phosphorylation levels of ACBD3 in BRCA, HNSC, KIRC, LUAD, and GBM. We found that the phosphorylation levels of ACBD3 at various phosphorylation sites decreased in HNSC, but increased in BRCA, KIRC, LUAD, and GBM. This discovery could lead to further research on the molecular mechanisms and potential therapeutic targets for tumors. However, we cannot get rid of the possibility that the difference in phosphorylation levels is a by-product of meaningless signal dysregulation. Therefore, further experimental verification is required.
Previous research had revealed that CAF was involved in various cancers developing [34]. We discovered that ACBD3 expression positively related to CAF infiltration in HNSC. Besides, ACBD3 expression was also different in various immune subtypes of HNSC, which may indicate a correlation between the occurrence and development of HNSC and the infiltration of CAF.
The advantage of this study is that we reflected the expression and clinical value of ACBD3 in pan-cancers using a variety of databases in a comprehensive and systematic manner. Secondly, this was the first study to analyze the biological significance of ACBD3 in pan-cancers and obtain relatively comprehensive results.
However, our study has several limitations. First of all, we only used the existing RNA-seq and clinical data of cancers in online databases for analysis but lacked actual clinical data. Secondly, there is need to conduct further biological experiments to verify our conclusions. Currently, various bioinformatic analysis methods are available. In future studies, we plan to combine various learning methods, such as machine learning, to further understand the function of ACBD3 in various cancers.
In summary, we found a statistically significant relationship between ACBD3 expression and immune subtypes, molecular subtypes, diagnosis, prognosis, tumor mutation burden, protein phosphorylation levels, and immune cell infiltration in pan-cancers. This comprehensive and systematic pan-cancer analysis of ACBD3 supports further explorations into the critical role of ACBD3 during the development of tumors and offer a comprehensive analytical basis for further molecular, biological, and experimental verification in future clinical decisions.
Conclusion
According to our pan-cancer analysis of ACBD3, ACBD3 may serve as a novel prognostic and diagnostic biomarker for pan-cancers as it contributes to tumor development. As such, ACBD3 may also provide new directions for cancer treatment targets in the future.
Supplementary Information
Additional file 1: Figure S1 The expression level of ACBD3. A In normal tissues, ACBD3 expressed highly in cerebral cortex, hippocampus, duodenum, small intestine, colon, gallbladder, pancreas, prostate, placenta, appendix, and bone marrow. Moreover, ACBD3 expressed lowly in oral mucosa, liver, ovary, soft tissue, and adipose tissue; B In tumor cell lines, The expression of ACBD3 ranks high in breast cancer, kidney cancer, and myeloma in tumor cell lines; C, D Intracellular ACBD3 is mainly distributed in Golgi; E ACBD3 expressed high in SKCM, BRCA, GBM, KIRC, LGG, and THCA in Cancer Cell Line Encyclopedia (CCLE) database. Small cell lung cancer (SCLC), colon and rectal cancer (COADREAD), large b-cell lymphoma (DLBC), sarcoma (SARC), multiple myeloma (MM), acute myeloid leukemia (LAML), skin cutaneous melanoma (SKCM), breast cancer (BRCA), liver hepatocellular carcinoma (LIHC), mesothelioma (MESO), ovarian cancer (OV), esophageal cancer (ESCA), endometrioid cancer (UCEC), glioblastoma multiforme (GBM), pancreatic adenocarcinoma (PAAD), neuroblastoma (NB), lung adenocarcinoma (LUAD), stomach adenocarcinoma (STAD), kidney clear cell carcinoma (KIRC), non-small cell lung carcinoma (NSC), acute lymphoeytic leukemia (ALL), lung squamous cell carcinoma (LUSC), brain lower grade glioma (LGG), head and neck cancer (HNSC), thyroid cancer (THCA), chronic myelocytic leukemia (LCML), Human medulloblastoma cells (MB), prostate cancer (PRAD), cervical cancer (CESC), chronic lymphocytic leukemia (CLL). Figure S2 Correlation between immune infiltrates and ACBD3 expression in different tumors. head and neck cancer (HNSC), bladder cancer (BLCA), colon cancer (COAD), thyroid cancer (THCA), kidney clear cell carcinoma (KIRC). A The expression of ACBD3 was actively correlated with the cancer-associated fibroblast (CAF) infiltration for HNSC. B The expression of ACBD3 was positively related to neutrophil infiltration for BLCA, COAD, and THCA. C ACBD3 expression was positively related to endothelial cell infiltration for COAD, HNSC, and KIRC. Figure S3 Kaplan–Meier plots ([Overall Survival (OS)]) for ACBD3 expression in pan-cancers. A Pancreatic For pancreatic adenocarcinoma (PAAD), there was no statistically significant relationship between ACBD3 expression and prognosis. B For Glioma (GBMLGG), the OS prognosis was negatively related to ACBD3 expression, HR = 2.30, 95% CI: 1.19–4.43, p = 0.013. C For adrenocortical carcinoma (ACC), there was no statistically significant relationship between ACBD3 expression and prognosis.
Acknowledgements
This work was supported by the following grants: Natural Science Foundation of Sichuan Province (No. 2022NSFSC1378). Xiaowei Tang has received the research support.
Author contributions
All authors contributed to the study conception and design. Material preparation, data collection and analysis were performed by XM, SH and HS. The first draft of the manuscript was written by XM and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the following grants: Natural Science Foundation of Sichuan Province (No. 2022NSFSC1378). Xiao-Wei Tang has received the research support.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Xinyue Ma, Shu Huang and Huiqin Shi share the first authorship.
Contributor Information
Xia Chen, Email: 970217858@qq.com.
Xiaowei Tang, Email: solitude5834@hotmail.com.
References
- 1.Mino-Kenudson M, Schalper K, Cooper W, Dacic S, Hirsch FR, Jain D, Lopez-Rios F, Tsao MS, Yatabe Y, Beasley MB, Yu H, Sholl LM, Brambilla E, Chou TY, Connolly C, Wistuba I, Kerr KM, Lantuejoul S, IASLC Pathology Committee Predictive biomarkers for immunotherapy in lung cancer: perspective from the international association for the study of lung cancer pathology committee. J Thorac Oncol. 2022;17(12):1335–1354. doi: 10.1016/j.jtho.2022.09.109. [DOI] [PubMed] [Google Scholar]
- 2.Zhong H, Shi Q, Wen Q, Chen J, Li X, Ruan R, Zeng S, Dai X, Xiong J, Li L, Lei W, Deng J. Pan-cancer analysis reveals potential of FAM110A as a prognostic and immunological biomarker in human cancer. Front Immunol. 2023;27(14):1058627. doi: 10.3389/fimmu.2023.1058627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pradat Y, Viot J, Yurchenko AA, Gunbin K, Cerbone L, Deloger M, Grisay G, Verlingue L, Scott V, Padioleau I, Panunzi L, Michiels S, Hollebecque A, Jules-Clément G, Mezquita L, Lainé A, Loriot Y, Besse B, Friboulet L, André F, Cournède PH, Gautheret D, Nikolaev SI. Integrative pan-cancer genomic and transcriptomic analyses of refractory metastatic cancer. Cancer Discov. 2023;13(5):1116–1143. doi: 10.1158/2159-8290.CD-22-0966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Islinger M, Costello JL, Kors S, Soupene E, Levine TP, Kuypers FA, et al. The diversity of ACBD proteins - From lipid binding to protein modulators and organelle tethers. Biochim Biophys Acta Mol Cell Res. 2020;1867(5):118675. doi: 10.1016/j.bbamcr.2020.118675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Houghton-Gisby J, Kerslake R, Karteris E, Mokbel K, Harvey AJ. ACBD3 bioinformatic analysis and protein expression in breast cancer cells. Int J Mol Sci. 2022;23(16):8881. doi: 10.3390/ijms23168881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yue X, Qian Y, Gim B, Lee I. Acyl-CoA-binding domain-containing 3 (ACBD3; PAP7; GCP60): a multi-functional membrane domain organizer. Int J Mol Sci. 2019;20(8):2028. doi: 10.3390/ijms20082028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang H, Liao X, Zhu G, Han C, Wang X, Yang C, et al. Acyl-CoA binding domain containing 4 polymorphism rs4986172 and expression can serve as overall survival biomarkers for hepatitis B virus-related hepatocellular carcinoma patients after hepatectomy. Pharmgenomics Pers Med. 2022;29(15):277–300. doi: 10.2147/PGPM.S349350.PMID:35378899;PMCID:PMC8976523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Neess D, Bek S, Engelsby H, Gallego SF, Færgeman NJ. Long-chain acyl-CoA esters in metabolism and signaling: role of acyl-CoA binding proteins. Prog Lipid Res. 2015;59:1–25. doi: 10.1016/j.plipres.2015.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Yue X, Qian Y, Zhu L, Gim B, Bao M, Jia J, Jing S, Wang Y, Tan C, Bottanelli F, Ziltener P, Choi S, Hao P, Lee I. ACBD3 modulates KDEL receptor interaction with PKA for its trafficking via tubulovesicular carrier. BMC Biol. 2021;19(1):194. doi: 10.1186/s12915-021-01137-7.PMID:34493279;PMCID:PMC8424950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liao J, Guan Y, Chen W, Shi C, Yao D, Wang F, et al. ACBD3 is required for FAPP2 transferring glucosylceramide through maintaining the Golgi integrity. J Mol Cell Biol. 2019;11(2):107–117. doi: 10.1093/jmcb/mjy030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang Y, Yang L, Pei YY, Wang J, Wu H, Yuan J, et al. Overexpressed ACBD3 has prognostic value in human breast cancer and promotes the self-renewal potential of breast cancer cells by activating the Wnt/beta-catenin signaling pathway. Exp Cell Res. 2018;363(1):39–47. doi: 10.1016/j.yexcr.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 12.Zheng Y, Pei Y, Tang R, Zhou X, Feng Z, Li D, et al. ACBD3 is up-regulated in gastric cancer and promotes cell cycle G1-to-S transition in an AKT-dependent manner. Exp Cell Res. 2021;406(2):112752. doi: 10.1016/j.yexcr.2021.112752. [DOI] [PubMed] [Google Scholar]
- 13.Kakiuchi S, Daigo Y, Ishikawa N, Furukawa C, Tsunoda T, Yano S, et al. Prediction of sensitivity of advanced non-small cell lung cancers to gefitinib (Iressa, ZD1839) Hum Mol Genet. 2004;13(24):3029–3043. doi: 10.1093/hmg/ddh331. [DOI] [PubMed] [Google Scholar]
- 14.Ganini C, Amelio I, Bertolo R, Bove P, Buonomo OC, Candi E, Cipriani C, Di Daniele N, Juhl H, Mauriello A, Marani C, Marshall J, Melino S, Marchetti P, Montanaro M, Natale ME, Novelli F, Palmieri G, Piacentini M, Rendina EA, Roselli M, Sica G, Tesauro M, Rovella V, Tisone G, Shi Y, Wang Y, Melino G. Global mapping of cancers: the Cancer Genome Atlas and beyond. Mol Oncol. 2021;15(11):2823–2840. doi: 10.1002/1878-0261.13056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nusinow DP, Szpyt J, Ghandi M, Rose CM, McDonald ER, 3rd, Kalocsay M, Jané-Valbuena J, Gelfand E, Schweppe DK, Jedrychowski M, Golji J, Porter DA, Rejtar T, Wang YK, Kryukov GV, Stegmeier F, Erickson BK, Garraway LA, Sellers WR, Gygi SP. Quantitative proteomics of the cancer cell line encyclopedia. Cell. 2020;180(2):387–402.e16. doi: 10.1016/j.cell.2019.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gong K, Zhou H, Liu H, Xie T, Luo Y, Guo H, et al. Identification and integrate analysis of key biomarkers for diagnosis and prognosis of non-small cell lung cancer based on bioinformatics analysis. Technol Cancer Res Treat. 2021;20:15330338211060202. doi: 10.1177/15330338211060202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li T, Fu J, Zeng Z, Cohen D, Li J, Chen Q, Li B, Liu XS. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–W514. doi: 10.1093/nar/gkaa407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu P, Heins ZJ, Muller JT, Katsnelson L, de Bruijn I, Abeshouse AA, et al. Integration and analysis of cptac proteomics data in the context of cancer genomics in the cBioportal. Mol Cell Proteomics. 2019;18(9):1893–1898. doi: 10.1074/mcp.TIR119.001673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, et al. TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics. 2019;35(20):4200–4202. doi: 10.1093/bioinformatics/btz210. [DOI] [PubMed] [Google Scholar]
- 20.Zhang C, Guo C, Li Y, Liu K, Zhao Q, Ouyang L. Identification of claudin-6 as a molecular biomarker in pan-cancer through multiple omics integrative analysis. Front Cell Dev Biol. 2021;2(9):726656. doi: 10.3389/fcell.2021.726656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, et al. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41:D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Modhukur V, Iljasenko T, Metsalu T, Lokk K, Laisk-Podar T, Vilo J. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics. 2018;10(3):277–288. doi: 10.2217/epi-2017-0118. [DOI] [PubMed] [Google Scholar]
- 23.Xing C, Wang Z, Zhu Y, Zhang C, Liu M, Hu X, et al. Integrate analysis of the promote function of Cell division cycle-associated protein family to pancreatic adenocarcinoma. Int J Med Sci. 2021;18(3):672–684. doi: 10.7150/ijms.53243.PMID:33437202;PMCID:PMC7797531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X. Alterations of Golgi structural proteins and glycosylation defects in cancer. Front Cell Dev Biol. 2021;12(9):665289. doi: 10.3389/fcell.2021.665289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ali OM, El Amin HA, Sharkawy YL, Mohamed Ali AA, Kholef EFM, Elsewify WAE. Golgi protein 73 versus alpha-fetoprotein as a new biomarker in early diagnosis of hepatocellular carcinoma. Int J Gen Med. 2020;18(13):193–200. doi: 10.2147/IJGM.S253622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bajaj R, Warner AN, Fradette JF, Gibbons DL. Dance of the Golgi: understanding Golgi dynamics in cancer metastasis. Cells. 2022;11(9):1484. doi: 10.3390/cells11091484.PMID:35563790;PMCID:PMC9102947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ahmed MB, Alghamdi AAA, Islam SU, Lee JS, Lee YS. cAMP signaling in cancer: a PKA-CREB and EPAC-centric approach. Cells. 2022;11(13):2020. doi: 10.3390/cells11132020.PMID:35805104;PMCID:PMC9266045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang F, Ma G, Zhou X, Zhu X, Yu X, Ding F, et al. Depletion of LAMP3 enhances PKA-mediated VASP phosphorylation to suppress invasion and metastasis in esophageal squamous cell carcinoma. Cancer Lett. 2020;1(479):100–111. doi: 10.1016/j.canlet.2020.03.014. [DOI] [PubMed] [Google Scholar]
- 29.Jiang K, Yao G, Hu L, Yan Y, Liu J, Shi J, et al. MOB2 suppresses GBM cell migration and invasion via regulation of FAK/Akt and cAMP/PKA signaling. Cell Death Dis. 2020;11(4):230. doi: 10.1038/s41419-020-2381-8.PMID:32286266;PMCID:PMC7156523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang H, Yang S, Wang J, Jiang Y. Blockade of AMPK-mediated cAMP-PKA-CREB/ATF1 signaling synergizes with aspirin to inhibit hepatocellular carcinoma. Cancers (Basel) 2021;13(7):1738. doi: 10.3390/cancers13071738.PMID:33917483;PMCID:PMC8038809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kilanowska A, Ziółkowska A, Stasiak P, Gibas-Dorna M. cAMP-dependent signaling and ovarian cancer. Cells. 2022;11(23):3835. doi: 10.3390/cells11233835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh V, Ram M, Kumar R, Prasad R, Roy BK, Singh KK. Phosphorylation: implications in cancer. Protein J. 2017;36(1):1–6. doi: 10.1007/s10930-017-9696-z. [DOI] [PubMed] [Google Scholar]
- 33.Ardito F, Giuliani M, Perrone D, Troiano G, Lo ML. The crucial role of protein phosphorylation in cell signaling and its use as targeted therapy (Review) Int J Mol Med. 2017;40(2):271–280. doi: 10.3892/ijmm.2017.3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Biffi G, Tuveson DA. Diversity and biology of cancer-associated fibroblasts. Physiol Rev. 2021;101(1):147–176. doi: 10.1152/physrev.00048.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1 The expression level of ACBD3. A In normal tissues, ACBD3 expressed highly in cerebral cortex, hippocampus, duodenum, small intestine, colon, gallbladder, pancreas, prostate, placenta, appendix, and bone marrow. Moreover, ACBD3 expressed lowly in oral mucosa, liver, ovary, soft tissue, and adipose tissue; B In tumor cell lines, The expression of ACBD3 ranks high in breast cancer, kidney cancer, and myeloma in tumor cell lines; C, D Intracellular ACBD3 is mainly distributed in Golgi; E ACBD3 expressed high in SKCM, BRCA, GBM, KIRC, LGG, and THCA in Cancer Cell Line Encyclopedia (CCLE) database. Small cell lung cancer (SCLC), colon and rectal cancer (COADREAD), large b-cell lymphoma (DLBC), sarcoma (SARC), multiple myeloma (MM), acute myeloid leukemia (LAML), skin cutaneous melanoma (SKCM), breast cancer (BRCA), liver hepatocellular carcinoma (LIHC), mesothelioma (MESO), ovarian cancer (OV), esophageal cancer (ESCA), endometrioid cancer (UCEC), glioblastoma multiforme (GBM), pancreatic adenocarcinoma (PAAD), neuroblastoma (NB), lung adenocarcinoma (LUAD), stomach adenocarcinoma (STAD), kidney clear cell carcinoma (KIRC), non-small cell lung carcinoma (NSC), acute lymphoeytic leukemia (ALL), lung squamous cell carcinoma (LUSC), brain lower grade glioma (LGG), head and neck cancer (HNSC), thyroid cancer (THCA), chronic myelocytic leukemia (LCML), Human medulloblastoma cells (MB), prostate cancer (PRAD), cervical cancer (CESC), chronic lymphocytic leukemia (CLL). Figure S2 Correlation between immune infiltrates and ACBD3 expression in different tumors. head and neck cancer (HNSC), bladder cancer (BLCA), colon cancer (COAD), thyroid cancer (THCA), kidney clear cell carcinoma (KIRC). A The expression of ACBD3 was actively correlated with the cancer-associated fibroblast (CAF) infiltration for HNSC. B The expression of ACBD3 was positively related to neutrophil infiltration for BLCA, COAD, and THCA. C ACBD3 expression was positively related to endothelial cell infiltration for COAD, HNSC, and KIRC. Figure S3 Kaplan–Meier plots ([Overall Survival (OS)]) for ACBD3 expression in pan-cancers. A Pancreatic For pancreatic adenocarcinoma (PAAD), there was no statistically significant relationship between ACBD3 expression and prognosis. B For Glioma (GBMLGG), the OS prognosis was negatively related to ACBD3 expression, HR = 2.30, 95% CI: 1.19–4.43, p = 0.013. C For adrenocortical carcinoma (ACC), there was no statistically significant relationship between ACBD3 expression and prognosis.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.