Abstract
Earlier studies from our laboratory on randomly isolated transcriptional signals of mycobacteria had revealed that the −10 region of mycobacterial promoters and the corresponding binding domain in the major sigma factor are highly similar to their Escherichia coli counterparts. In contrast, the sequences in −35 regions of mycobacterial promoters and the corresponding binding domain in the major sigma factor are vastly different from their E. coli counterparts (M. D. Bashyam, D. Kaushal, S. K. Dasgupta, and A. K. Tyagi, J. Bacteriol. 178:4847–4853, 1996). We have now analyzed the role of the TGN motif present immediately upstream of the −10 region of mycobacterial promoters. Sequence analysis and site-specific mutagenesis of a Mycobacterium tuberculosis promoter and a Mycobacterium smegmatis promoter reveal that the TGN motif is an important determinant of transcriptional strength in mycobacteria. We show that mutation in the TGN motif can drastically reduce the transcriptional strength of a mycobacterial promoter. The influence of the TGN motif on transcriptional strength is also modulated by the sequences in the −35 region. Comparative assessment of these extended −10 promoters in mycobacteria and E. coli suggests that functioning of the TGN motif in promoters of these two species is similar.
Transcription is the first and often the principal step in regulation of gene expression in bacteria. Comparisons of promoter sequences recognized by the major forms of RNA polymerases of several bacterial species have identified two conserved 6-bp canonical sequences located approximately 10 and 35 bp upstream from the transcription start point. Genetic analysis and protein-DNA interaction studies have confirmed that these two hexameric sequences (called the −10 and −35 regions) are necessary for initiation of transcription. Another region, an AT-rich up element (a target for the C-terminal domain of the α subunit of RNA polymerase), has been reported to enhance the promoter activity 2- to 20-fold (25). An interesting exception to this classical structure of promoters was elucidated by studies on the λ PRE promoter and the Escherichia coli galP1 promoter. It was shown that RNA polymerase could initiate transcription (albeit suboptimally) from promoters lacking a functional −35 region sequence, provided an “extended −10” motif was present in these promoters (6, 14, 16, 23). The extended −10 motif comprises the sequence TGN present immediately upstream of the −10 region. It was also shown that the thermal-energy requirement for open complex formation in an extended −10 promoter was less than that for a conventional −10/−35 promoter (5). Other promoters that have been shown to function as extended −10 promoters include the E. coli cysG promoter (3), promoter of the DpnII operon of Streptococcus pneumoniae (26), the ferredoxin gene promoter of Clostridium pasteurianum (11), and the amyP gene promoter of Bacillus subtilis (29). Kenney and Churchward have reported that the TGN motif present upstream of the −10 hexamer can play a role in the activity of the rpsL promoter of Mycobacterium smegmatis (15).
We had earlier initiated studies on randomly isolated transcriptional signals of mycobacteria (7) and shown that the −10 region of these promoters and the corresponding binding domain in the principal sigma factor of mycobacteria (ςA) are almost identical to those of E. coli (2). However, the −35 region of mycobacterial promoters and the corresponding binding domain in the principal sigma factor were found to be vastly different from those of E. coli (2). We also reported that the −35 region of mycobacterial promoters can tolerate a greater variety of sequences compared to other bacterial promoters, owing to the presence of multiple constitutive sigma factors having different or overlapping binding specificities for the −35 region of promoters (2). We have carried out analysis of mycobacterial promoters that belong to the class of extended −10 promoters. We show that the TGN motif located upstream of the −10 region in mycobacterial promoters is an important determinant of transcriptional activity. Our studies suggest that the TGN motif may play similar roles in the initiation of transcription in mycobacteria and E. coli.
Mycobacterial promoters containing the TGN motif immediately upstream of the Pribnow box.
We have analyzed the sequences of 59 mycobacterial promoters (for which the transcriptional start points have been experimentally determined in our laboratory or elsewhere) for the presence of the TGN motif immediately upstream of the −10 hexamer. These included 16 M. smegmatis promoters and 12 M. tuberculosis promoters from our mycobacterial promoter library (2). The other promoters included in the analysis belonged to the mpb70 gene of M. bovis BCG (20), the rpsL gene of M. smegmatis (15), the recA genes of M. tuberculosis and M. smegmatis (21, 22), the DNA gyrase genes of M. smegmatis and M. tuberculosis (18, 19), the purC and purL genes of M. tuberculosis (13), the 18-kDa gene of M. leprae (8), the oxyR and ahpC genes of M. leprae (9), and the repA gene of plasmid pAL5000 from M. fortuitum (27). The remaining 16 promoters have been listed earlier (2). Thirteen of the 59 promoters analyzed in this study contained the TGN motif (Fig. 1). These included six promoters from our M. smegmatis promoter library and two from our M. tuberculosis promoter library in addition to the hsp60 P2 promoter of M. bovis BCG (28), the rpsL promoter of M. smegmatis (15), the repA promoter from the M. fortuitum plasmid pAL5000 (27), the rRNA gene promoter of M. leprae (30), and the 18-kDa gene promoter of M. leprae (8). Thus, based on a small sample size of 59, 22% of mycobacterial promoters contain the TGN motif. Analysis of 183 promoters from various species of gram-positive bacteria (11, 12, 26) reveals that frequency of occurrence of the TGN motif in these promoters is around 60%. In E. coli promoters, the TGN motif occurs with a frequency of about 16% (16).
FIG. 1.
Alignment of putative extended −10 promoters of mycobacteria. Fifty-nine mycobacterial promoters for which the TSP had been experimentally determined were analyzed. The sequences of the 13 promoters which contained the TGN motif are shown. (A) Promoters from our mycobacterial promoter library. S5, S6, S16, S19, S21, and S119 are from M. smegmatis, and T101 and T129 are from M. tuberculosis. (B) rpsL gene promoter from M. smegmatis, hsp60 P2 promoter from M. bovis, repA promoter from the plasmid pAL5000 of M. fortuitum, and promoters of the 18-kDa gene and the rRNA operon of M. leprae. The extended −10 sequences are underlined.
Functional analysis of the S16 promoter of M. smegmatis.
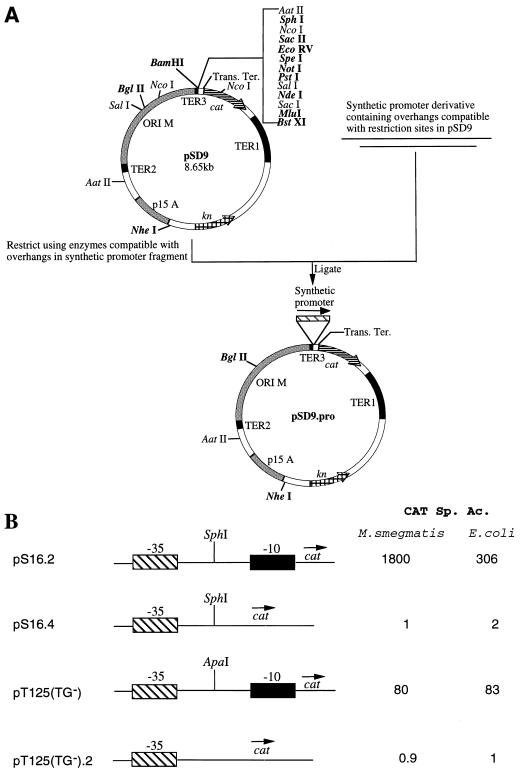
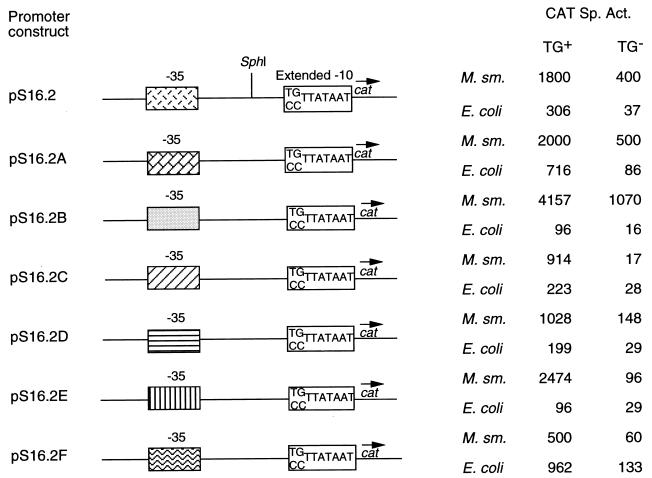
We chose the S16 promoter for further analysis because (i) it contained the perfect extended −10 motif (TGTTATAAT), (ii) it represented one of the strongest promoters in our library, and (iii) it exhibited suboptimal but significant activity in E. coli, an organism in which the importance of the TGN motif has been well demonstrated. We have earlier shown that the −10 region in mycobacterial promoters is essential for initiation of transcription (2). In order to determine its importance in the putative extended −10 promoters of mycobacteria, we synthesized two truncated derivatives of S16—S16.2 and S16.4—and cloned them separately into pSD9, generating pS16.2 and pS16.4, respectively (Table 1). The cloning strategy is depicted in Fig. 2A. The S16.2 derivative harbored both the −10 and the −35 regions, whereas the S16.4 derivative did not harbor the −10 region. In addition, three bases were changed to incorporate an SphI restriction site at the −18 position in both derivatives. Since (i) for mycobacterial promoters including S16, the same transcription start point (TSP) is used in mycobacteria and E. coli (2), (ii) the conserved hexameric sequence TATAAT is located 10 bp upstream of the TSP in both E. coli and mycobacterial promoters (2), and (iii) the −10 binding domain in the sigma factor of E. coli is identical to the corresponding domain in the mycobacterial sigma factor (2), the effects of deletion of the −10 region should be similar in the two hosts. M. smegmatis and E. coli were transformed with pS16.2 and pS16.4 separately as described earlier (7). Deletion of the −10 region completely abolished promoter activity in both mycobacteria and E. coli (pS16.4 in Fig. 2B). This result confirmed our earlier observation that the −10 region is an important functional determinant of promoter activity in mycobacteria as in other eubacteria. In order to determine the role of the TGN motif in mycobacterial promoters, we decided to study the effect of mutating this motif. For this purpose, pS16.2(TG−) was generated by using synthetic oligonucleotides in which TG of the extended −10 motif of pS16.2 was replaced by CC. M. smegmatis and E. coli were separately transformed with the constructs pS16.2 and pS16.2(TG−), and the chloramphenicol acetyltransferase (CAT)-specific activities supported by each of these constructs were determined as described earlier (7). As shown in Fig. 3, mutation of the TGN motif resulted in a fourfold reduction in the CAT activity supported by the promoter in mycobacteria. In E. coli, there was an eightfold decrease in the activity when the TGN motif was mutated (Fig. 3). These results suggested that the TGN motif may be an important determinant of transcriptional strength in mycobacteria as is the case in E. coli. To determine whether the effect of mutation of the TGN motif would be different for different sequences in the −35 region, six modified pS16.2 constructs (pS16.2A to pS16.2F) and six modified pS16.2(TG−) constructs [pS16.2(TG−)A to pS16.2(TG−)F] were generated as described in Table 1. These six pS16.2 constructs as well as the six pS16.2(TG−) constructs contained identical −10 regions but different −35 regions. M. smegmatis and E. coli were transformed by using each of these 12 constructs separately. The CAT specific activities supported by the pS16.2 constructs were compared to those of the corresponding pS16.2(TG−) constructs to determine the contribution of the TGN motif in the context of different sequences in the −35 region. The results are given in Fig. 3. All the pS16.2(TG−) constructs supported lower levels of CAT activity in mycobacteria compared to their corresponding pS16.2 derivatives. There was a 13-fold reduction in the case of pS16.2C, whereas in the case of pS16.2E the decrease was twofold. Similar results were obtained when the constructs were tested in E. coli (Fig. 3). In each case, there was a decrease in the activity when the TGN motif was mutated. The level of reduction varied, ranging from about eightfold for pS16.2, pS16.2A, and pS16.2C to about twofold for pS16.2E. These results further substantiate the importance of the TGN motif in initiation of transcription in mycobacteria. Additionally, in both mycobacteria and E. coli, the levels of reduction following the mutation in the TGN motif were different for different constructs depending on the sequences generated in the −35 region due to the various insertions.
TABLE 1.
Promoter constructs used in this study
| Plasmid(s) | Description |
|---|---|
| pS16.2 | Synthetic S16 promoter, representing promoter sequences from −42 to +5, cloned into pSD9 |
| pS16.4 | Synthetic S16 promoter, representing sequences from −42 to −17, cloned into pSD9 |
| pS16.2(TG−) | pS16.2 with CC instead of TG 1 bp upstream of the −10 region of the promoter |
| pS16.2A to -F and pS16.2(TG−)A to −F | Contain various fragments (44 to 276 bp) derived from the multiple cloning region of plasmid pGEM5Zf(+) cloned into the SphI restriction site of pS16.2 [or pS16.2(TG−)], giving rise to six mosaic pS16.2 [or pS16.2(TG−)] constructs containing identical −10 but different −35 regions |
| pT125(TG−) | Original T125 promoter from pSD7.T125, which does not contain the TGN motif, cloned into pSD9 |
| pT125(TG−).2 | Construct containing the T125 promoter sequence till the −29 position and lacking the −10 region |
| pT125 | Synthetic T125 promoter fragment, containing the sequence TG instead of GA 1 bp upstream of the −10 region sequence, cloned into pT125(TG−) as described in the text |
| pT125A to -F and pT125(TG−)A to −F | Contain various fragments (44 to 276 bp) derived from the multiple cloning region of plasmid pGEM5Zf(+) cloned into the ApaI restriction site of pT125 [or pT125(TG−)], giving rise to six mosaic pT125 [or pT125(TG−)] constructs containing identical −10 but different −35 regions |
FIG. 2.
(A) Strategy for cloning of various modified promoter derivatives in pSD9. pSD9 was generated by cloning the end-repaired ApaI-NsiI fragment derived from the multiple cloning region of pGEM5Zf(+) into the PstI restriction site present upstream of the reporter gene encoding CAT in pSD7; the other PstI restriction site present in the inessential region downstream to TER1 in pSD7 was inactivated prior to this cloning. The synthetic promoter fragments were annealed in 1× NEB 2 buffer (10 mM Tris-HCl [pH 7.9], 10 mM MgCl2, 50 mM NaCl, 1 mM dithiothreitol) and cloned into pSD9 restricted with appropriate enzymes. Trans. Ter., transcription terminus. (B) Functional dissection of the S16 promoter of M. smegmatis and the T125 promoter of M. tuberculosis. The construct pS16.2 represents the synthetic S16.2 promoter, containing sequences from −42 to +5, cloned into pSD9, pS16.4 represents the synthetic S16.4 promoter, containing sequences from −42 to −17 cloned into pSD9. pT125(TG−) represents the original T125 promoter from pSD7.T125 cloned into pSD9, and pT125(TG−).2 represents the T125 promoter harboring only the −35 region cloned into pSD9. The CAT specific activity (Sp. Ac.) supported by each construct in M. smegmatis and E. coli is expressed as nanomoles of chloramphenicol converted into its acetylated derivatives per minute per milligram of protein.
FIG. 3.
A comparative assessment of CAT activities supported by various modified pS16.2 constructs with and without the TGN motif in mycobacteria and E. coli. The unique SphI restriction site in pS16.2 and pS16.2(TG−) was used to generate the various modified promoter derivatives as described in Table 1. The extended −10 sequence (identical for all constructs) and the −35 region (different for each construct) are indicated. The CAT specific activity (Sp. Act.) was determined as described elsewhere (7) and is expressed as nanomoles of chloramphenicol converted into its acetylated derivatives per minute per milligram of protein. M. sm., M. smegmatis.
Functional analysis of the M. tuberculosis promoter T125.
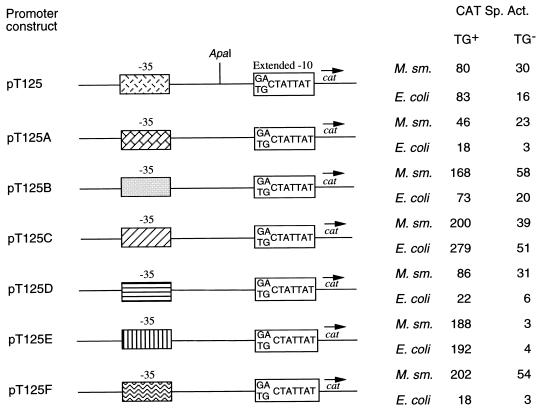
In the case of the S16 promoter of M. smegmatis, we had mutated the TGN motif to evaluate its importance in transcription initiation. Following this, we proceeded to determine the effect of introducing the TGN motif in a conventional −10/−35 promoter that does not harbor the TGN motif. This was done to ensure that the results obtained with the M. smegmatis S16 promoter were applicable to other promoters of mycobacteria. For this purpose, we chose the M. tuberculosis promoter T125 because it contained a near-perfect −10 region sequence (TATTAT) and provided a unique ApaI restriction site between the −10 and −35 regions of the promoter for carrying out manipulations (2). In order to facilitate modifications in the promoter, the T125 promoter fragment from pSD7.T125 was cloned into pSD9. This promoter construct lacking the TGN motif was designated pT125(TG−) (Table 1). We first determined the effect of deletion of the −10 region. The ApaI restriction site was used to delete promoter sequences downstream of the −29 position, and the resulting construct was designated pT125(TG−).2 (Table 1). As shown in Fig. 2B, deletion of the −10 region completely abolished promoter activity in both mycobacteria and E. coli. For introducing the TGN motif, synthetic oligonucleotides containing the sequence TG instead of GA 1 bp upstream of the −10 region were used, resulting in the generation of pT125 (Table 1). M. smegmatis and E. coli were transformed with pT125 and pT125(TG−) separately, and the CAT activity supported by each construct was determined. Introduction of the TGN motif resulted in threefold and fivefold enhancements in CAT activity in M. smegmatis and E. coli, respectively (Fig. 4). These results suggest that the TGN motif may be an important determinant of promoter activity in mycobacteria. In order to determine the role of the TGN motif vis-à-vis different −35 regions in the promoter T125, six modified pT125 constructs (pT125A to pT125F) and six modified pT125(TG−) constructs [pT125(TG−)A to pT125(TG−)F] containing identical −10 regions but different −35 regions were generated (Table 1). M. smegmatis and E. coli were separately transformed with all these mosaic promoter constructs. The CAT activities supported by each construct in M. smegmatis and E. coli are listed in Fig. 4. Introduction of the TGN motif into various modified pT125(TG−) constructs significantly raised the levels of CAT activity in M. smegmatis (Fig. 4). The level of enhancement varied from 75-fold in the case of pT125E to about 2-fold in the case of pT125A. Similar results were obtained when the CAT activities supported by the various constructs were determined for the respective E. coli transformants. The level of enhancement for different constructs in E. coli varied from 45-fold in the case of pT125E(TG−) to about 4-fold in the case of pT125B(TG−) and pT125D(TG−) (Fig. 4). In fact, the CAT activity supported by pT125E(TG−) in mycobacteria was extremely low, comparable to the basal activity supported by pSD9 (1 nmol/min/mg of protein). Introduction of the TGN motif upstream of the −10 region raised the activity to 188 nmol/min/mg of protein. Therefore, as observed with the S16 promoter, the effect of the TGN motif on transcriptional activity of a promoter varies with different sequences in the −35 region, in both M. smegmatis and E. coli.
FIG. 4.
A comparative assessment of CAT activities supported by various modified pT125 constructs with and without the TGN motif in mycobacteria and E. coli. The unique ApaI restriction site in pT125 and pT125(TG−) was used to generate the various modified promoter derivatives as described in Table 1. The extended −10 sequence (identical for all constructs) and the −35 region (different for each construct) are indicated. The CAT specific activity (Sp. Act.) was determined as described elsewhere (7) and is expressed as nanomoles of chloramphenicol converted into its acetylated derivatives per minute per milligram of protein. M. sm., M. smegmatis.
Concluding remarks.
We have screened 59 mycobacterial promoters for which the TSPs have been experimentally determined and carried out a functional analysis of the S16 promoter of M. smegmatis and the T125 promoter of M. tuberculosis. Our results suggest that the TGN motif may be an important determinant of transcriptional strength in mycobacteria as in other bacterial species. Therefore, the mechanism of basal transcription in mycobacteria appears similar to that of conventional bacterial promoters. Presently, it seems unlikely that the mycobacterial extended −10 promoters are responsible for transcription of a specific set of genes. Genes like rpsL (which encodes the S12 ribosomal protein) of M. smegmatis, hsp60 of M. bovis, repA from pAL5000, the rRNA gene of M. leprae, and the 18-kDa gene of M. leprae do not belong to any particular category. However, presence of the extended −10 promoter may be required in particular regions of the bacterial chromosome having sequence constraints, where it may be difficult to maintain two specific hexameric sequences (e.g., within an open reading frame), as has been already suggested for the E. coli cysG promoter (3). Another role of extended −10 promoters could be to maintain a basal level of transcription in the case of promoters that contain a weak −35 region and are regulated by protein-DNA interactions in the −35 region (as in the case of the galP1 promoter of E. coli) (6). One possible advantage of the TGN motif could be to facilitate transcription initiation at cold temperatures or when the sigma factor is proteolytically cleaved (under stress conditions). As has been suggested earlier, the present transcription machinery may have evolved from a primitive apparatus containing just the extended −10 motif in the promoters and the −10 binding domain of the ς factor along with a catalytic subunit constituting the RNA polymerase (4). One may need to look at the −10 region not as a hexamer but as a nonamer, and the presence of perfect nucleotides in all the nine positions (as opposed to six) enables RNA polymerase to undertake transcription initiation due to the extended contact it makes with DNA in the −10 region (nine bases equal almost one full turn of the DNA helix), even though it may make poor contacts with the −35 region. Kumar and coworkers have actually shown that RNA polymerase can initiate transcription from an extended −10 promoter in the absence of the −35 binding domain of the sigma factor (16). Transcription initiation based on such an extended −10 region alone, however, may not represent the optimal strength of the promoter. As shown in the present study, the sequences present in the −35 region modulate initiation of transcription even in the presence of a TGN motif. Chan and Busby as well as Keilty and Rosenberg have shown that −35 region sequences can apparently modulate overall promoter strength in the λ PRE extended −10 promoter (6, 14). With suppression genetics, it has been shown that the region of ς70 immediately downstream of the region 2.4 (termed region 2.5) is actually responsible for contacting the TGN motif (1). We have compared the amino acid sequence of region 2.5 of ς70 with that of the corresponding region of the M. smegmatis sigma factors ςA and ςB (Fig. 5) (24). The two amino acids, glutamic acid and histidine, implicated in interaction with the TGN motif in E. coli (1) are also conserved in ςA and ςB. The region of mycobacterial ς factors corresponding to the 2.5 region of E. coli ς70 (spanning 22 amino acids) contains only four nonconserved substitutions. Also, the amino acids that exhibited a high frequency of occurrence, as deduced from a comparison of 30 ς factor sequences (4), are conserved in both the ς factors of M. smegmatis. The amino acid sequences in the 2.5 regions of ςA and ςB of M. tuberculosis and M. smegmatis are also identical (10). These observations further substantiate the similarities between the transcriptional machineries of E. coli and mycobacteria, as far as the extended −10 region is concerned. Footprinting analysis and studies on methylation and ethylation interference in combination with site-specific mutagenesis would further enhance our understanding of the role of the TGN motif in initiation of transcription in mycobacteria and help in the development of tools for high-level expression of genes in mycobacteria.
FIG. 5.
Comparison of the amino acid sequences in the region 2.5 of the sigma factors of M. smegmatis (ςA and ςB) and the principal sigma factor of E. coli (RpoD). The two amino acids previously implicated in making specific contacts with the TGN motif (filled circles), identical amino acids (dashes), conserved substitutions (plain letters), and nonconserved substitutions (boldfaced letters) are indicated. The amino acid sequences are from reference 17 for RpoD of E. coli and from reference 24 for ςA and ςB of M. smegmatis. The consensus sequence is from reference 4, wherein the amino acids that exhibit a high-frequency occurrence (based on a compilation of 30 sigma factors) are indicated. Conserved substitutions are defined as the following groups: I, L, M, and V; A and G; S and T; K, H, and R; D, E, N, and Q; F, Y, and W; C; and P. The numbers do not represent actual amino acid positions.
Acknowledgments
We thank Manisha Sharma for excellent technical assistance and J. S. Tyagi and Shruti Jain for critical reading of the manuscript.
This work was supported by a financial grant from the Department of Biotechnology of India. M.D.B. is thankful to the Council of Scientific and Industrial Research and the Department of Biotechnology for financial assistance.
REFERENCES
- 1.Barne K A, Brown J A, Busby S J E, Minchin S D. Region 2.5 of the Escherichia coli RNA polymerase ς70 subunit is responsible for the recognition of the ‘extended −10’ motif at promoters. EMBO J. 1997;16:4034–4040. doi: 10.1093/emboj/16.13.4034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bashyam M D, Kaushal D, DasGupta S K, Tyagi A K. A study of the mycobacterial transcriptional apparatus: identification of novel features in promoter elements. J Bacteriol. 1996;178:4847–4853. doi: 10.1128/jb.178.16.4847-4853.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Belyaeva T, Griffiths L, Minchin S, Cole J, Busby S. The Escherichia coli cysG promoter belongs to the ‘extended −10’ class of bacterial promoters. Biochem J. 1993;296:851–857. doi: 10.1042/bj2960851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bown J A, Barne K A, Minchin S D, Busby S J W. Extended −10 promoters. Nucleic Acids Mol Biol. 1997;11:41–52. [Google Scholar]
- 5.Burns H, Minchin S. Thermal energy requirement for strand separation during transcription initiation: the effect of supercoiling and extended protein DNA contacts. Nucleic Acids Res. 1994;22:3840–3845. doi: 10.1093/nar/22.19.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chan B, Busby S. Recognition of nucleotide sequences at the Escherichia coli galactose operon P1 promoter by RNA polymerase. Gene. 1989;84:227–236. doi: 10.1016/0378-1119(89)90496-4. [DOI] [PubMed] [Google Scholar]
- 7.DasGupta S K, Bashyam M D, Tyagi A K. Cloning and assessment of mycobacterial promoters by using a plasmid shuttle vector. J Bacteriol. 1993;175:5186–5192. doi: 10.1128/jb.175.16.5186-5192.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dellagostin O A, Esposito G, Eales L-J, Dale J W, McFadden J. Activity of mycobacterial promoters during intracellular and extracellular growth. Microbiology. 1995;141:1785–1792. doi: 10.1099/13500872-141-8-1785. [DOI] [PubMed] [Google Scholar]
- 9.Dhandayuthapani S, Mudd M, Deretic V. Interactions of OxyR with the promoter region of the oxyR and ahpC genes from Mycobacterium leprae and Mycobacterium tuberculosis. J Bacteriol. 1997;179:2401–2409. doi: 10.1128/jb.179.7.2401-2409.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doukhan L, Predich M, Nair G, Dussurget O, Mandic-Mulec I, Cole S T, Smith D R, Smith I. Genomic organization of the mycobacterial sigma gene cluster. Gene. 1995;165:67–70. doi: 10.1016/0378-1119(95)00427-8. [DOI] [PubMed] [Google Scholar]
- 11.Graves M C, Rabinowitz J C. In vivo and in vitro transcription of the Clostridium pasteurianum ferredoxin gene. J Biol Chem. 1986;261:11409–11415. [PubMed] [Google Scholar]
- 12.Helman J. Compilation and analysis of Bacillus subtilis ς A-dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA. Nucleic Acids Res. 1995;23:2351–2360. doi: 10.1093/nar/23.13.2351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jackson M, Berthet F-X, Otal I, Rauzier J, Martin C, Gicquel B, Guilhot C. The Mycobacterium tuberculosis purine biosynthetic pathway: isolation and characterization of the purC and purL genes. Microbiology. 1996;142:2439–2447. doi: 10.1099/00221287-142-9-2439. [DOI] [PubMed] [Google Scholar]
- 14.Keilty S, Rosenberg M. Constitutive function of a positively regulated promoter reveals new sequences essential for activity. J Biol Chem. 1987;262:6389–6395. [PubMed] [Google Scholar]
- 15.Kenney T J, Churchward G. Genetic analysis of the Mycobacterium smegmatis rpsL promoter. J Bacteriol. 1996;178:3564–3571. doi: 10.1128/jb.178.12.3564-3571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kumar A, Malloch R A, Fujita N, Smillie D A, Ishihama A, Hayward R S. The minus 35-recognition region of Escherichia coli sigma 70 is inessential for initiation of transcription at an “extended minus 10” promoter. J Mol Biol. 1993;232:406–418. doi: 10.1006/jmbi.1993.1400. [DOI] [PubMed] [Google Scholar]
- 17.Lonetto M, Gribskov M, Gross C A. The ς70 family: sequence conservation and evolutionary relationships. J Bacteriol. 1992;174:3843–3849. doi: 10.1128/jb.174.12.3843-3849.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Madhusudan K, Nagaraja V. Mycobacterium smegmatis DNA gyrase: cloning and overexpression in Escherichia coli. Microbiology. 1995;141:3029–3037. doi: 10.1099/13500872-141-12-3029. [DOI] [PubMed] [Google Scholar]
- 19.Madhusudan K, Ramesh V, Nagaraja V. Molecular cloning of gyrA and gyrB genes of Mycobacterium tuberculosis: analysis of nucleotide sequence. Biochem Mol Biol Int. 1994;33:651–660. [PubMed] [Google Scholar]
- 20.Matsuo T, Matsumoto S, Ohara N, Kitaura H, Mizuno A, Yamada T. Differential transcription of the MPB70 genes in two major groups of Mycobacterium bovis BCG substrains. Microbiology. 1995;141:1601–1607. doi: 10.1099/13500872-141-7-1601. [DOI] [PubMed] [Google Scholar]
- 21.Movahedzadeh F, Colston M J, Davis E O. Determination of DNA sequences required for regulated Mycobacterium tuberculosis RecA expression in response to DNA-damaging agents suggests that two modes of regulation exist. J Bacteriol. 1997;179:3509–3518. doi: 10.1128/jb.179.11.3509-3518.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papavinasasundaram K G, Movahedzadeh F, Keer J T, Stoker N G, Colston M J, Davis E O. Mycobacterial recA is cotranscribed with a potential regulatory gene called recX. Mol Microbiol. 1997;24:141–153. doi: 10.1046/j.1365-2958.1997.3441697.x. [DOI] [PubMed] [Google Scholar]
- 23.Ponnambalam S, Webster C, Bingham A, Busby S. Transcription initiation at the Escherichia coli galactose operon promoters in the absence of the normal −35 region sequences. J Biol Chem. 1986;261:16043–16048. [PubMed] [Google Scholar]
- 24.Predich M, Doukhan L, Nair G, Smith I. Characterization of RNA polymerase and two sigma-factor genes from Mycobacterium smegmatis. Mol Microbiol. 1995;15:355–366. doi: 10.1111/j.1365-2958.1995.tb02249.x. [DOI] [PubMed] [Google Scholar]
- 25.Ross W, Gosink K, Solomon J, Igarashi K, Zou C, Ishihama A, Severinov K, Gourse R. A third recognition element in bacterial promoters: DNA binding by the α subunit of RNA polymerase. Science. 1993;262:1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- 26.Sabelnikov A G, Greenberg B, Lacks S A. An extended −10 promoter alone directs transcription of the DpnII operon of Streptococcus pneumoniae. J Mol Biol. 1995;250:144–155. doi: 10.1006/jmbi.1995.0366. [DOI] [PubMed] [Google Scholar]
- 27.Stolt P, Stoker N G. Protein-DNA interactions in the ori region of the Mycobacterium fortuitum plasmid pAL5000. J Bacteriol. 1996;178:6693–6700. doi: 10.1128/jb.178.23.6693-6700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stover C K, de la Cruz V F, Fuerst T R, Burlein J E, Benson L A, Bennett L T, Bansal G P, Young J F, Lee M H, Hatfull G F, Snapper S B, Barletta R G, Jacobs W R, Jr, Bloom B R. New use of BCG for recombinant vaccines. Nature (London) 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 29.Voskuil M I, Voepel K, Chambliss G H. The −16 region, a vital sequence for the utilization of a promoter in Bacillus subtilis and Escherichia coli. Mol Microbiol. 1995;17:271–279. doi: 10.1111/j.1365-2958.1995.mmi_17020271.x. [DOI] [PubMed] [Google Scholar]
- 30.Yuan-en J, Colston M J, Cox R A. Nucleotide sequence and secondary structures of precursor 16S rRNA of slow-growing mycobacteria. Microbiology. 1994;140:123–132. doi: 10.1099/13500872-140-1-123. [DOI] [PubMed] [Google Scholar]