ABSTRACT
Lysinibacillus fusiformis PwPw_T2 isolated from deteriorating Ananas comosus sample collected from Lagos State, Nigeria putatively possesses genomic features like potential enzymes catalyzing acetic acid production and xenobiotic compounds degradation via various pathways as indicated by its genome sequences. These could make the organism relevant in food waste valorization and micro-biotechnology.
KEYWORDS: Lysinibacillus, xenobiotics, acetic acid, Ananas comosus, genome
ANNOUNCEMENT
Lysinibacillus fusiformis PwPw_T2 was isolated from deteriorating Ananas comosus collected from Mushin Market, Lagos State, Nigeria (N 6o 31’ 59.9988”, E 3o 21’ 0”) as a potential vinegar producer while scavenging for acetic acid (vinegar) producers during food waste valorization. Before isolation, deteriorating Ananas comosus pulp was enriched on glucose, yeast extract, peptone, ethanol (GYPE) medium for 3 days followed by centrifugation at 250 rpm/2 min; pellets obtained were serially diluted. Dilutions 104, 106, and 108 were inoculated on YCEA medium (yeast extract, calcium carbonate, and ethanol) for 3 days at 30°C. After observing visible zone of clearing, colonies were re-inoculated on YBE medium (yeast extract, bromocresol, and ethanol) to ascertain acetic acid production (1). Interest in organism’s genomic features has expanded (2–4). Hence, L. fusiformis PwPw_T2 genome was sequenced to further explore its genomic features and confirm presence of genes involved in acetic acid production.
Genomic DNA was extracted from fresh pure cultures on yeast extract peptone and glucose broth at 28°C–30°C for 48 hours with Quick-DNA fungal/bacterial miniprep kit (Zymo Research, Irvine, CA, USA) (5, 6). Library preparation and 2 × 150 bp paired-end sequencing were done with Nextera XT Index kit v2 (FC-131-2001 to -2004) and a NextSeq 500 system (Illumina, San Diego, CA, USA) at Quadram Institute Bioscience (Norwich, UK). Preassembly trimming was done by Trim Galore v0.6.5 (7). QUAST v5.0.2 was used to ascertain the quality of reads (8). The sequence reads were assembled into contigs with SPAdes v3.15.3 on KBase platform (9). Contigs from KBase platform were uploaded on online servers of NCBI prokaryotic Genome Automatic Annotation Pipeline (PGAAP v6.5), Bacterial and Viral Bioinformatics Resource Center (BV-BRC) (v3.30.19) (10), Rapid Annotations Subsystems Technology v2.0 (RAST), and SEED Viewer v2.0 (11, 12) for automated annotation and comparison. For all bioinformatics analyses, default settings were employed.
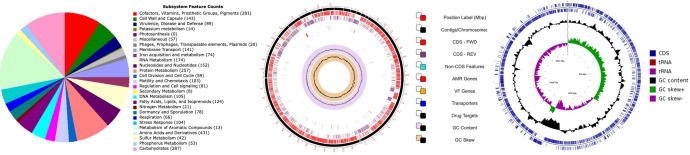
Details of L. fusiformis PwPw_T2 genome sequence are summarized in Table 1. The overview of subsystems unique to this bacteria’s genome is highlighted in Fig. 1. Based on putative genome exploration on BV-BRC/Pathosystems Resource Integration Center (PATRIC) online server, L. fusiformis PwPw_T2 genome possesses metabolic enzymes capable of degrading different types of xenobiotics via specific degradation pathway. Putatively, the isolate has potential for geraniol degradation by acetyl-CoA acetyltransferase [EC 2.3.1.9; pathway ID (PID) 00281] and fluorobenzoate degradation by N-acetylglucosamine deacetylase (EC 3.5.1; PID 00364). L. fusiformis PwPw_T2 also has the potential to biosynthesize alcohol dehydrogenase via glycolytic/glyconeogenetic pathway (EC 1.1.1.1; PID 00010) and aldehyde dehydrogenase via pyruvate metabolism (EC.1.2.1.3; ID 00620). Furthermore, in L. fusiformis PwPw_T2 genome, AntiSmash 7.0 run at default predicted biosynthetic clusters for the production of kijanimicin, beta-lactones, fengycin, petrobactin, terpenes, bacillibactin, and thiopeptides (13). The biosynthetic clusters of L. fusiformis PwPw_T2 make it a good candidate for further biotechnological manipulation, notably in the food manufacturing industry.
TABLE 1.
| Parameter | Value |
|---|---|
| Genome size | 4,820,104 bp |
| Genes (total) | 4,920 |
| Number of contigs | 21 |
| Number of scaffolds | 18 |
| tRNA, rRNA | 36, 5 |
| GC percent | 37 |
| Hypothetical proteins | 1,547 |
| CDSs (total)b | 4,874 |
| CDSs (with protein) | 4,824 |
| Genes (RNA) | 46 |
| Contig N50 | 1.5 Mb |
| Contig L50 | 2 |
NCBI Prokaryotic Genome Automatic Annotation Pipeline (PGAAP v6.5) (https://www.ncbi.nlm.nih.gov/genome/annotation_prok/).
CDSs, coding sequences.
Fig 1.
Overview of genome subsystems and circular views of Lysinibacillus fusiformis PwPw_T2 were created on RAST, Kbase, and BV-BRC online servers (9, 10, 12).
Contributor Information
Oyetayo Olaoluwa Adefiranye, Email: oyetayooo@gmail.com.
Olubukola Oluranti Babalola, Email: olubukola.babalola@nwu.ac.za.
John J. Dennehy, Department of Biology, Queens College, Queens, New York, USA
DATA AVAILABILITY
The draft whole genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number JAUIZN000000000.1. The version described in this paper is version JAUIZN010000000. The project data are available under BioProject accession number PRJNA991757 and BioSample accession number SAMN36317136 as well as Sequence Read Archive Accession number SRR25938945.
REFERENCES
- 1. Diba F, Alam F, Talukder AA. 2015. Screening of acetic acid producing microorganisms from decomposed fruits for vinegar production. Adv Microbiol 5:291–297. doi: 10.4236/aim.2015.55028 [DOI] [Google Scholar]
- 2. Passera A, Rossato M, Oliver JS, Battelli G, Shahzad G-I-R, Cosentino E, Sage JM, Toffolatti SL, Lopatriello G, Davis JR, Kaiser MD, Delledonne M, Casati P. 2021. Characterization of Lysinibacillus fusiformis strain S4C11:in vitro, in planta, and in silico analyses reveal a plant-beneficial microbe. Microbiol Res 244:126665. doi: 10.1016/j.micres.2020.126665 [DOI] [PubMed] [Google Scholar]
- 3. Pudova DS, Lutfullin MT, Shagimardanova EI, Hadieva GF, Shigapova L, Toymentseva AA, Kabanov DA, Mardanova AM, Vologin SG, Sharipova MR. 2018. Draft genome sequence data of Lysinibacillus fusiformis strain GM, isolated from potato phyllosphere as a potential probiotic. Data Brief 21:2504–2509. doi: 10.1016/j.dib.2018.11.107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhao L, Bao G, Geng B, Song J, Li Y. 2015. Draft genome sequence of Lysinibacillus fusiformis strain SW-B9, a novel strain for biotransformation of isoeugenol to vanillin. Genome Announc 3:e00289-15. doi: 10.1128/genomeA.00289-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Diaz M, Kellingray L, Akinyemi N, Adefiranye OO, Olaonipekun AB, Bayili GR, Ibezim J, du Plessis AS, Houngbédji M, Kamya D, et al. 2019. Comparison of the microbial composition of African fermented foods using amplicon sequencing. Sci Rep 9:1386–1395. doi: 10.1038/s41598-019-50190-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Petros C, Walls D, Munger H, Wei C. 2019. An unbiased comparison of BigDye Terminator and BrilliantDye Terminator. J Biomol Tech 30:S4. [Google Scholar]
- 7. Krueger F. 2015. TrimGalore: a wrapper tool around Cutadapt and FastQC to consistently apply quality and adapter trimming to FastQ files. Babraham Bioinformatics; 516. https://www.bioinformatics.babraham.ac.uk/projects/trim_galore. [Google Scholar]
- 8. Mikheenko A, Prjibelski A, Saveliev V, Antipov D, Gurevich A. 2018. Versatile genome assembly evaluation with QUAST-LG. Bioinformatics 34:i142–i150. doi: 10.1093/bioinformatics/bty266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Arkin AP, Stevens RL, Cottingham RW, Maslov S, Henry CS, Dehal P, Ware D, Perez F, Harris NL, Canon S, et al. 2016. The DOE systems biology Knowledgebase (KBase). bioRxiv. doi: 10.1101/096354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Olson RD, Assaf R, Brettin T, Conrad N, Cucinell C, Davis JJ, Dempsey DM, Dickerman A, Dietrich EM, Kenyon RW, et al. 2023. Introducing the bacterial and viral bioinformatics resource center (BV-BRC): a resource combining PATRIC, IRD and ViPR. Nucleic Acids Res 51:D678–D689. doi: 10.1093/nar/gkac1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, et al. 2008. The RAST server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2013. The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Blin K, Shaw S, Augustijn HE, Reitz ZL, Biermann F, Alanjary M, Fetter A, Terlouw BR, Metcalf WW, Helfrich EJN, van Wezel GP, Medema MH, Weber T. 2023. antiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res 51:W46–W50. doi: 10.1093/nar/gkad344 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The draft whole genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number JAUIZN000000000.1. The version described in this paper is version JAUIZN010000000. The project data are available under BioProject accession number PRJNA991757 and BioSample accession number SAMN36317136 as well as Sequence Read Archive Accession number SRR25938945.