Abstract
Thiocyanate hydrolase is a newly found enzyme from Thiobacillus thioparus THI 115 that converts thiocyanate to carbonyl sulfide and ammonia (Y. Katayama, Y. Narahara, Y. Inoue, F. Amano, T. Kanagawa, and H. Kuraishi, J. Biol. Chem. 267:9170–9175, 1992). We have cloned and sequenced the scn genes that encode the three subunits of the enzyme. The scnB, scnA, and scnC genes, arrayed in this order, contained open reading frames encoding sequences of 157, 126, and 243 amino acid residues, respectively, for the β, α, and γ subunits, respectively. Each open reading frame was preceded by a typical Shine-Dalgarno sequence. The deduced amino-terminal peptide sequences for the three subunits were in fair agreement with the chemically determined sequences. The protein molecular mass calculated for each subunit was compatible with that determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. From a computer analysis, thiocyanate hydrolase showed significant homologies to bacterial nitrile hydratases known to convert nitrile to the corresponding amide, which is further hydrolyzed by amidase to form acid and ammonia. The two enzymes were homologous over regions corresponding to almost the entire coding regions of the genes: the β and α subunits of thiocyanate hydrolase were homologous to the amino- and carboxyl-terminal halves of the β subunit of nitrile hydratase, and the γ subunit of thiocyanate hydrolase was homologous to the α subunit of nitrile hydratase. Comparisons of the catalytic properties of the two homologous enzymes support the model for the reaction steps of thiocyanate hydrolase that was previously presented on the basis of biochemical analyses.
Thiocyanate is a common compound found in natural environments. In crushed plant tissues, cellular glucosinolates (thioglucosides) are hydrolyzed by glucosidase to produce thiocyanate and other compounds (25). Glucosinolates are widely distributed in botanical families such as Cruciferae (12). Mammalian body fluids such as saliva and blood contain free thiocyanate that is mainly derived from plant glucosinolates in foodstuffs (25). Thiocyanate is also formed by the ubiquitous enzyme rhodanese (thiosulfate sulfurtransferase; EC 2.8.1.1) (24). The degradation of thiocyanate in mammalian tissues involves peroxidases such as lactoperoxidase and myeloperoxidase; methemoglobin and oxyhemoglobin have been reported to be involved in the degradation as well (23, 25).
In the microbial world, some chemoorganotrophic bacteria such as a Pseudomonas stutzeri-like bacterium and an Arthrobacter sp. have been reported to degrade thiocyanate (4, 22). These bacteria, however, do not utilize thiocyanate as an energy source. In contrast, Thiobacillus thioparus, which is a chemolithoautotrophic sulfur bacterium, has been shown to obtain energy by the degradation of thiocyanate (8, 10). T. thioparus THI 115 was isolated from an activated sludge that was used in the microbial processes of thiocyanate generated by factories during the gasification of coal (11).
Katayama et al. (11) isolated a unique enzyme for thiocyanate conversion from T. thioparus THI 115. This enzyme, named thiocyanate hydrolase, is composed of three different subunits, α (19 kDa), β (23 kDa), and γ (32 kDa). From the molecular mass (142 kDa) estimated for the native form, the subunit structure of the enzyme was deduced to be a heterohexamer (α2β2γ2). The enzyme catalyzes the degradation of thiocyanate (SCN−) to carbonyl sulfide (COS) and ammonia. From biochemical analysis, the stoichiometry of sulfur conversion from thiocyanate to carbonyl sulfide was found to be almost 1:1, and that of nitrogen conversion from thiocyanate to ammonia was also found to be 1:1. Several aspects of chemical thermodynamics argued against a single reaction mechanism for the degradation. Neither cyanide nor cyanate was detected as a reaction product; this excluded the possible involvement of degradation pathways similar to those reported for some mammalian systems (6) or for Thiobacillus thiocyanoxidans (26). Assuming the occurrence of unstable reaction intermediates, Katayama et al. (11) proposed a model for reaction steps in which the thiocyanate molecule is subjected to the sequential addition of H2O and hydrolysis, leading to the formation of carbonyl sulfide and ammonia.
To confirm the above model and further characterize the catalytic properties of each subunit of thiocyanate hydrolase, we have tried to clone the genes for this novel enzyme, which has been identified only in T. thioparus THI 115. A sequence analysis of cloned genes revealed a remarkable homology to bacterial nitrile hydratases, which have been shown to be adapted for the biodegradation of several nitrile compounds (15, 16, 18).
MATERIALS AND METHODS
Bacterial strains and culture conditions.
T. thioparus THI 115 (11) was grown in 3-liter culture flasks containing 800 ml of TC10 medium [0.5 g of K2HPO4, 0.05 g of MgSO4 · 7H2O, 0.01 g of FeCl3 · 6H2O, 0.01 g of CaCl2 · 2H2O, 1.0 g of (NH4)2SO4, 1.0 g of potassium thiocyanate, and 10 ml of trace metal solution per liter (10), pH 7.0]. Cells were grown aerobically with reciprocal shaking at 30°C. Escherichia coli DH5 and XL-1 Blue were grown in Luria-Bertani (LB) or terrific broth (Difco), which was supplemented with ampicillin (100 μg/ml) when required.
Enzyme purification and N-terminal protein sequencing.
Thiocyanate hydrolase was purified from T. thioparus THI 115 according to the method described previously (11). The purified enzyme was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and electroblotted onto a polyvinylidene difluoride membrane. α, β, and γ subunits were located by protein staining and subjected to microsequencing on an ABI 477A protein sequencer.
Antibody preparation.
The purified enzyme was separated by SDS-PAGE, and the β-subunit protein was electroeluted with 25 mM Tris-HCl (pH 8.3) containing 0.1% (wt/vol) SDS, precipitated in 80% (vol/vol) isopropanol, and then dialyzed against ethanol to remove SDS. The β-subunit protein thus prepared was dissolved in phosphate-buffered saline and mixed with Freund’s complete adjuvant for injection into rabbits. The polyclonal antibodies against α and γ subunits used in this work were from the same preparation as that described previously (11).
Genomic library.
A frozen bacterial pellet (3 g wet weight) was used to extract genomic DNA (about 0.5 mg) with extraction kit ISOGEN (Nippon Gene, Tokyo, Japan) containing benzyl chloride. Partial digestion of the DNA (23.5 μg) was performed with 10 U of EcoRI for 15 min at 37°C in 111 μl followed by phenol-chloroform extraction and ethanol precipitation. The pellet was resuspended in TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) and separated by electrophoresis through a 0.8% (wt/vol) agarose gel. A fraction of DNA ranging from 10 to 24 kbp was electrophoretically eluted from the gel. Four micrograms of the EcoRI partial digest was treated with calf intestine alkaline phosphatase. To construct a cosmid library, an aliquot of the dephosphorylated DNA (0.3 μg) was ligated with 1.5 μg of EcoRI-cut Charomid 9-28 vector (21) purchased from Nippon Gene. After ligation, aliquots of the mixture were subjected to in vitro packaging with Gigapack II Gold (Stratagene, La Jolla, Calif.) and the resultant phage suspension was used for infection of E. coli DH5.
Immunoscreening of a genomic library.
Bacterial colonies were transferred on nylon membranes (NY13N; Schleicher & Schuell, Dassel, Germany) and lysed with chloroform vapor for 20 min and further treated overnight at 30°C with a lysis buffer containing 50 mM Tris-HCl (pH 7.5), 150 mM NaCl, 5 mM MgCl2, 1 μg of DNase I per ml, 40 μg of lysozyme per ml, and 1% (wt/vol) gelatin. Membranes were then rinsed with the same buffer without DNase or lysozyme and treated for 30 min with a blocking solution containing 2% (wt/vol) gelatin in TBST (10 mM Tris-HCl [pH 8.0], 150 mM NaCl, 0.05% [wt/vol] Tween 20). They were further incubated for 2 h at 30°C in TBST containing the antibodies against the α, β, and γ subunits of thiocyanate hydrolase described above. Alkaline phosphatase-conjugated anti-rabbit immunoglobulin G antibody was used as a secondary antibody to detect positive colonies, which were visualized by the reaction with nitroblue tetrazolium–5-bromo-4-chloro-3-indolylphosphate.
Western blotting analysis.
Bacterial proteins were heat denatured in Laemmli’s buffer and separated by SDS-PAGE. Proteins were blotted on a nitrocellulose membrane and treated with antibodies to detect thiocyanate hydrolase subunits. The immunological detection of proteins was carried out by the same system used for screening colonies.
PCR analysis.
PCR was performed in a reaction mixture containing template DNA (10 ng), primers, dNTPs (400 μM each), Takara LA Taq (2.5 U), and the reaction buffer provided by the supplier (TAKARA, Tokyo, Japan). For each reaction, 100 pmol of degenerate primer mixture was used in combination with 10 pmol of reverse primer P3 (5′-GGAAACAGCTATGACCATG-3′) or universal primer P1 (5′-CCCAGTCACGACGTTGT-3′), which anneals upstream or downstream of the multicloning site of pUC118, respectively. The reaction mixture was first heated for 1 min at 94°C, and the amplification process (98°C for 20 s, 55°C for 1 min, 68°C for 5 min) was repeated for 30 cycles, followed by heating for 10 min at 75°C.
The PCR products were ligated with linearized TA cloning vector pCR2.1 (Invitrogen, Leek, The Netherlands) and used for transformation.
DNA preparation and sequencing.
Cosmid and plasmid DNA was prepared by using disposable columns with anion-exchange resin supplied from Qiagen (Hilden, Germany). The sequencing reaction was performed by a dye terminator method and was analyzed on an ABI Prism 377 sequencer. To sequence the coding strand of the cosmid clone, the following 10 primers were used: 5′-CGATCGGCATAGACCAGT-3′, 5′-CATCGTGGTCGTGTTCGA-3′, 5′-GCCTTATGTCAAACGTTC-3′, 5′-GCATAGTGAGCTTCATCA-3′, 5′-CGGCGGCGAGGCGGATGT-3′, 5′-GAGATGCGCGAGCGCGTT-3′, 5′-CCCGGCGGCGGAAGACGA-3′, 5′-CATCGAAAAAGGCCTGTT-3′, 5′-GGGCCAGTCGCCCGAGTG-3′, and 5′-CTGCCTGATCGGCGTCGC-3′. To sequence the opposite strand, the following 10 primers were used: 5′-GCGCGACGCTCGAGCGCA-3′, 5′-CCCTTCAAGCACGGGGAT-3′, 5′-GCGACGCCGATCAGGCAG-3′, 5′-TTCGGCGAGCACCTGACG-3′, 5′-CTTCTTGTACTCGGGGTC-3′, 5′-CGAACGTGTCGCTGTACT-3′, 5′-ATCTTGGCGTGGTGGGTT-3′, 5′-CGCGCCAGGCGAGGCATT-3′, 5′-GCGGCCTCTTTCACACAT-3′, and 5′-AACGTTTGACATAAGGCA-3′. The distance from one primer to the nearest neighbor was about 200 to 300 bases, so all determined sequences were overlapped on both strands. Sequence data were assembled and analyzed for coding capacity and homology by using computer program GENETYX (Software Development, Tokyo, Japan).
Enzyme assay.
Thiocyanate hydrolase activity was determined by the amount of ammonia or COS as described previously (11). Unless otherwise specified, the reaction was started by adding purified enzyme into the reaction mixture containing 30 mM potassium thiocyanate and incubating the mixture at 30°C for 15 min. Other assay conditions were as described previously (11).
Nucleotide sequence accession number.
The nucleotide sequence data for the scnB, scnA, and scnC genes in addition to the flanking regions have been submitted to the DDBJ database and will appear in the DDBJ, EMBL, and GenBank nucleotide sequence databases with the accession no. AB007989 (the sequence having this number includes 600 bp of the upstream sequence together with the 2,103-bp sequence reported in this paper).
RESULTS
N-terminal sequence analysis of subunit proteins.
Microsequencing of α, β, and γ subunits of the purified thiocyanate hydrolase revealed the N-terminal sequence of each subunit as shown in Fig. 1. None of the N termini were blocked, and 10 to 20 amino acid residues of each N terminus were unambiguously determined except for three residues of the γ subunit (Fig. 1). We reexamined the analytical data and confirmed the occurrence of significant peaks for His12 and Asp13 in addition to the prominent peaks for Lys12 and Pro13. There was also a minor peak for His14 in addition to a significant peak for Ala14. These peaks for Lys12-Pro13-Ala14 may be false ones that were followed by the much-higher second peaks (Lys15-Pro16-Ala17) of the same sequence. Disagreement with the DNA data is mentioned in the figure legend.
FIG. 1.
Amino-terminal sequences of thiocyanate hydrolase subunits and degenerate primers for PCR. The peptide sequences were chemically determined from the purified enzyme of T. thioparus THI 115. For the γ subunit, the italicized sequence (Lys12-Pro13-Ala14) was not concordant with the His-Asp-His sequence that was deduced from the nucleotide sequence. The regions used for α, β, and γ primer sequences were chosen to minimize the degeneracy.
Construction of a genomic library and immunoscreening.
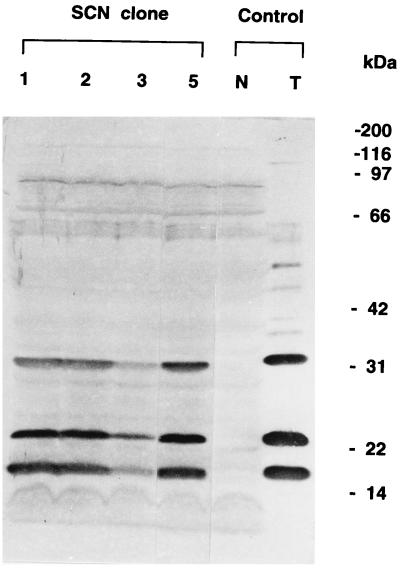
Partial EcoRI digestion of the genomic DNA of T. thioparus THI 115 produced DNA fragments of 3 to 25 kb. DNA fragments of 10 to 24 kb were purified and used for ligation to cosmid vector Charomid 9-28. After in vitro packaging, E. coli DH5 was infected with the resultant phage. A part of the infected bacterial suspension was plated on LB media containing ampicillin to obtain 6,810 independent colonies. Immunoscreening of this library with a mixture of two different rabbit antisera against α, β, and γ subunits yielded 25 candidate clones. Bacterial lysates derived from these clones were further screened by Western blotting analysis with the same antisera. Four of the isolated clones were found to produce all three subunits of thiocyanate hydrolase and were designated SCN-1, SCN-2, SCN-3, and SCN-5 (Fig. 2). The sizes of the subunits produced in E. coli were identical to those of T. thioparus THI 115.
FIG. 2.
Western blotting of the thiocyanate hydrolase subunits produced in E. coli clones carrying thiocyanate hydrolase genes. Positive E. coli clones SCN -1, -2, -3, and -5 in addition to a negative clone (N) that did not carry thiocyanate hydrolase genes were grown in LB medium with ampicillin. The bacterial cells were collected and denatured in Laemmli’s buffer. A partially purified enzyme sample prepared from T. thioparus THI 115 grown in TC10 medium was used as a positive control (T). Reported sizes for the three subunits (11) were 19 (α), 23 (β), and 32 kDa (γ).
Restriction analysis of DNA insert from positive clones.
Cosmid DNA was prepared from positive clones SCN-1, -2, -3, and -5 and digested with EcoRI. Each clone showed the linearized Charomid vector of 27.9 kb in addition to extra fragments derived from the DNA inserts: SCN-1 and SCN-5 contained 5-kbp fragments; SCN-2 contained 5-, 3.8-, and 2.3-kb fragments; and SCN-3 contained 5- and 3.8-kb fragments (data not shown). To find the coding region for the enzyme, EcoRI fragments of different sizes were prepared from SCN-2 and subcloned into pUC118. Western blotting analysis of the E. coli transformants revealed that some bacterial clones carrying 5-kb EcoRI inserts produced the α and β subunits but not the γ subunit, while other clones carrying a 5-, 3.8-, or 2.3-kb insert produced none of the three subunits (data not shown). Further analysis with other restriction enzymes suggested that the cosmid from SCN-2 had two different 5-kb EcoRI fragments that did not separate during agarose gel electrophoresis. The result indicated that one of the 5-kb EcoRI fragments should contain a promoter and complete coding sequences for at least the α and β subunits. This fragment may also contain a part of the coding sequence for the γ subunit; it has been suggested that this sequence is interrupted by the EcoRI site.
PCR analysis of subcloned DNA.
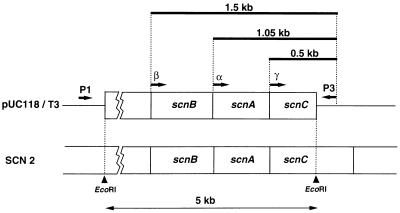
To confirm the sequence and organization of each gene, one of the positive subclones (pUC118/T3) with a 5-kb EcoRI fragment derived from SCN-2 was analyzed by PCR using degenerate primers with all possible combinations of codons for chemically determined N-terminal sequences of the α, β, and γ subunits of thiocyanate hydrolase (Fig. 1). In an optimized PCR condition, distinct PCR products were detected when each degenerate primer mixture was paired with reverse primer P3, while no significant PCR product was observed when the mixtures were paired with universal primer P1 (Fig. 3). This result indicated that all the coding sequences for the N termini of the three subunits were located within the 5-kb EcoRI fragment and were in the same orientation. The sizes of the PCR products amplified with primer sets α/P3, β1/P3, β2/P3, and γ/P3 were 1.05, 1.5, 1.5, and 0.5 kb, respectively. From these sizes, corresponding to the distances between the P3 primer and the N-terminal coding sequences, the gene organization in the EcoRI fragment was determined (Fig. 3); in Fig. 3, the genes coding for the α, β, and γ subunits of thiocyanate hydrolase are designated scnA, scnB, and scnC, respectively.
FIG. 3.
Schematic representation of PCR products and gene organization of scnA, scnB, and scnC. pUC118/T3 is a plasmid with a 5-kb EcoRI fragment derived from cosmid clone SCN-2. The annealing sites for universal (P1) and reverse (P3) primers of the pUC118 vector were located 16 bp downstream and 10 bp upstream, respectively, of the multicloning site. The α, β, and γ primers are shown in Fig. 1. Thick bars represent the sizes of the PCR products.
The PCR products were cloned into vector pCR2.1, and at least two plasmid clones with different orientations were isolated for each PCR product.
DNA sequences of the scnA, scnB, and scnC genes.
To determine the DNA sequences of the cloned PCR products, P1 and P3 primers were used for annealing with the pCR2.1 vector on both sides of the multicloning site. Thus, the entire sequences of scnA and scnB, and a part of the scnC sequence that is upstream of the EcoRI site were tentatively determined from the PCR clones. Based on this sequence information, a series of primers was synthesized to determine the original genomic sequence. The primer sequences were described in Materials and Methods. Cosmid clone SCN-2 was chosen as a representative and used for the template DNA.
Figure 4 shows the DNA sequence of the total 2,103 bp, which includes the extended sequence of the scnC gene downstream of the EcoRI site. As expected from the PCR experiment, the order of the gene array was scnB-scnA-scnC. The corresponding open reading frames (ORFs) for the β, α, and γ subunits encoded 157, 126, and 243 amino acid residues, respectively, giving the calculated molecular masses of 18, 15, and 28 kDa, respectively (Table 1). These values correlated with the apparent molecular masses of the three subunits (19, 23, and 32 kDa) estimated from SDS-PAGE (11). The deduced N-terminal sequences almost completely agreed with those obtained by chemical sequencing except for the γ subunit: amino acids 12 to 14 of the mature protein were chemically determined to be Lys-Pro-Ala (Fig. 1) instead of His-Asp-His, as indicated by the DNA analysis. The codon usage of the three genes showed a pattern similar to the one documented on the basis of 75 ORFs of Thiobacillus ferrooxidans (GenBank release 102), which is almost compatible with that of E. coli.
FIG. 4.
Nucleotide sequence of scnB-scnA-scnC series of genes. The deduced amino acid sequences for β (scnB), α (scnA), and γ (scnC) subunits are shown under each coding sequence. Asterisks indicate termination codons. The consensus Shine-Dalgarno sequences are underlined. Possible −35 and −10 regions are indicated by broken lines above the sequences. An EcoRI site was present at nucleotide 1606.
TABLE 1.
Numbers of amino acids and molecular masses of the three subunits of thiocyanate hydrolase as deduced from the structure of their genes
| Gene | Subunit | No. of amino acid residues | Molecular mass (kDa)
|
|
|---|---|---|---|---|
| Calculated | By SDS-PAGEa | |||
| scnA | α | 126 | 15 | 19 |
| scnB | β | 157 | 18 | 23 |
| scnC | γ | 243 | 28 | 32 |
Data from reference 11.
The three ORFs were located close to each other in different phases. As indicated in Fig. 4, all initiation codons were preceded by typical Shine-Dalgarno sequences (scnB: TAAGGAG; scnA: AxxGAGGT; scnC: TAAGGA). There were possible promoter-like sequences upstream of the scnB gene: the −35 sequence (TGGAA) spanned nucleotides 41 to 45, and the −10 sequence (CATAGT) spanned nucleotides 58 to 63. These features may suggest an operon structure. There was an unidentified ORF encoding at least 184 amino acids that terminated 246 bp upstream of the scnB gene; the DNA sequence constituting this ORF was not included in Fig. 4 but was included in the sequence submitted to DDBJ (accession no., AB007989).
Homology search for related protein sequences.
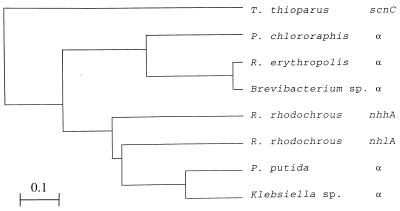
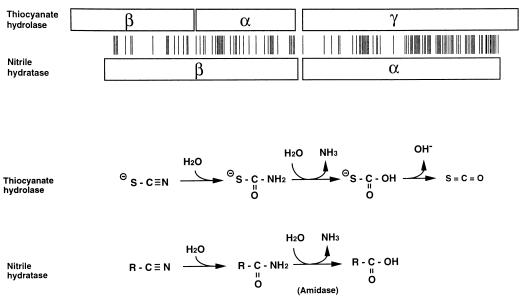
Amino acid sequences encoded by the three ORFs were subjected to a homology search by the tBLASTn program of the DDBJ database. A striking homology was found between the γ subunit of thiocyanate hydrolase and the α subunit of nitrile hydratase. More distant relationships were also found between the β and α subunits of thiocyanate hydrolase and the β subunit of nitrile hydratase: the β and α subunits of thiocyanate hydrolase were homologous to the N- and C-terminal halves, respectively, of the β subunit of nitrile hydratase (Fig. 5). A computer analysis was carried out to determine the phylogenetic relationships among the related genes of several bacterial species (Fig. 6). The scnC gene coding for the γ subunit of thiocyanate hydrolase showed a distinct position relative to a group of genes coding for the α subunits of nitrile hydratases derived from different bacteria. The closely related two species Pseudomonas chlororaphis and Rhodococcus erythropolis were chosen to illustrate the sequences homologous to the β and α subunits of thiocyanate hydrolase. Although the homology scores for these subunits were lower than that for the γ subunit, most of the identical amino acids matched with those conserved among β subunits of nitrile hydratases in the majority of the bacterial species shown in Fig. 6.
FIG. 5.
Alignment of amino acid sequences of each subunit of thiocyanate hydrolase and nitrile hydratases of different bacteria. Conserved amino acids among three (β- and α-subunits) or six (γ-subunit) sequences were boxed. Gaps (hyphens) were introduced to maximize the homology. The residue numbers of the carboxyl termini are indicated by -COOH. The accession numbers for each nitrile hydratase sequence are as follows: P. chlororaphis α and β subunits, D90216 (18); R. erythropolis α and β subunits, D14454 (7): Brevibacterium sp. α subunit, M60264 (17); R. rhodochrous α subunits of high- and low-molecular-mass enzymes (nhhA and nhlA), X64359 and X64360, respectively (14); P. putida α subunit, U89363 (20); Klebsiella sp. α subunit, E08305 (3).
FIG. 6.
Evolutionary relationships among the γ subunit of thiocyanate hydrolase and the α subunits of nitrile hydratases of different bacteria. The phylogenetic tree was made by the unweighted pair group method by arithmetic averaging with the computer program GENETYX. The bar indicates the genetic distance for 0.1 amino acid substitution/site.
Biochemical analysis of purified thiocyanate hydrolase.
To compare the substrate specificity of thiocyanate hydrolase with that of nitrile hydratase (1), 10 mM acetonitrile or propionitrile was used instead of thiocyanate. Ammonia formation was not observed, either in the presence or absence of 0.2 U of amidase (EC 3.5.1.4; Sigma; data not shown) per ml. To detect the possible accumulation of the amide intermediate (acetamide), the following experiments were performed. Thiocyanate hydrolase was preincubated with 5 mM acetonitrile at 0°C for 5 min prior to the enzyme reaction. The competitive inhibition of ammonia formation from thiocyanate by acetonitrile did not occur (data not shown). The conversion from acetamide (10 mM) to ammonia was not observed in our assay condition. Therefore, acetonitrile and its amide compound may not be utilized in the reaction of thiocyanate hydrolase. Furthermore, thiocyanate analogs such as methyl, ethyl, or benzyl derivatives did not serve as substrates, as determined on the basis of COS production (data not shown). The strict substrate specificity indicates that thiocyanate hydrolase is an enzyme distinct from nitrile hydratase and has a broad substrate specificity (1).
DISCUSSION
Thiocyanate hydrolase has a multimeric subunit structure with a molecular mass of 142 kDa. Polyclonal antibodies raised against the purified enzyme were shown to react with all three subunits, which have different sizes. By using these antibodies, we have screened a cosmid library constructed with EcoRI partial fragments of T. thioparus THI 115 genomic DNA. The four positive clones isolated by the immunoscreening showed expression of all three subunits strong enough for their detection by Western blotting analysis of E. coli lysate. Since the cosmid vector Charomid 9-28 does not have a promoter for the expression of inserted genes, the DNA inserts of the positive clones must have a promoter(s) for the downstream genes. It was previously observed that the enzyme was abundantly present in the cells of T. thioparus THI 115 grown in TC10 medium. The promoter should be strong and allow expression in a heterologous bacterial host.
A PCR analysis of the 5-kb EcoRI fragment subcloned in pUC118 revealed the presence of all the N-terminal coding sequences for β, α, and γ subunits, which were arrayed in this order in the same direction. Genes scnB, scnA, and scnC coding for these subunits were thus shown to be closely colocalized in the T. thioparus THI 115 genome. A Western blotting analysis of E. coli lysate of this subclone showed the production of the β and α subunits but not the γ subunit, whose coding sequence was indeed interrupted by an EcoRI site. The Western blot did not even show any smaller proteins related to the product of this distal ORF, indicating that the truncated γ subunit may degrade after translation; the α and β subunits may form a stable complex without the γ subunit. Alternatively, the truncated form of the γ subunit may not contain any epitopes for the polyclonal antibodies used in the experiments.
The nucleotide sequence of a part of the DNA insert of the positive cosmid clone revealed the total structures of the three ORFs estimated from the PCR analysis. Although there was one EcoRI site in the coding sequence of the γ subunit, the contiguous arrangement of the 5-kb sequence and its downstream EcoRI fragments was substantiated by the strong homology of the amino acid sequence corresponding to the codons in the vicinity of this site to the nitrile hydratase sequence described below. Normal codon usage and the occurrence of a typical Shine-Dalgarno sequence in each ORF are compatible properties for genes that show abundant expression. The thiocyanate hydrolase protein constitutes a few percent of the total proteins in T. thioparus THI 115 cells induced with the substrate thiocyanate, while enzyme production is inhibited by thiosulfate (11). There might be an operon structure comprising the subunit genes and their regulatory genes, as has been reported for genes of the nitrile-degrading enzymes described below (15, 16, 18).
The identities of the three ORFs were established by the almost perfect matching of the N-terminal sequences of the products to those chemically determined from the purified proteins. The calculated molecular masses for the three subunits were smaller than those determined from their migration on SDS-PAGE gel. We have confirmed the electrophoretic determination by using different size markers to obtain a consistent result. The difference may be due to possible protein modifications and/or some anomalous conformations of the denatured proteins.
The deduced amino acid sequences were strongly supported by the significant homologies to nitrile hydratase subunits that have been reported for a group of bacterial species. Comparisons with the matching regions of amino acid sequences of β, α, and γ subunits gave 21 (18 of 84), 31 (32 of 104), and 40% (72 of 180) identities, respectively, with P. chlororaphis sequences (18). The remarkable homology found in the γ subunit suggested that this subunit may share a common function with the α subunit of nitrile hydratase. Nitrile hydratase has been reported to contain cobalt or iron as a cofactor depending on the bacterial species (15). It was suggested that the conserved cysteine residues of the α subunit coordinated such metal ions (5, 20). The conserved cysteine residues were also present in the γ subunit of thiocyanate hydrolase (Cys128, Cys131, Cys133) in a region where the amino acid sequence showed the highest level of homology to that of the other enzyme. Although the occurrence of metal ions has not been evident for thiocyanate hydrolase, these domains in the two different enzymes should have similar functions. This might be another case of domain recruitment during evolution, which has been known to occur in a variety of enzyme families such as the amidotransferases (19).
In contrast to the strong homology found in the γ subunit, the β and α subunits showed only limited homologies to the β subunit of nitrile hydratase. It is obvious that the β subunit of nitrile hydratase was also less conserved among different bacteria than the α subunit. It should be noted, however, that 44% (8 of 18) of the identical amino acids found in even the least-homologous β subunit of thiocyanate hydrolase exactly matched those conserved among the different nitrile hydratases of P. chlororaphis (18), R. erythropolis (7), Rhodococcus rhodochrous (14), Pseudomonas putida (20), and a Klebsiella sp. (3) (data not shown). In the alignment of β subunits of different nitrile hydratases, the central regions were less conserved and required more gaps and insertions (data not shown) than the N- and C-terminal regions to which the β and α subunits, respectively, of thiocyanate hydrolase were homologous. This might be an indication of less important roles for the central regions as linker sequences after possible gene duplication and/or gene fusion events.
Katayama et al. (11) presented a model for the sequential reaction steps for thiocyanate hydrolase starting with the addition of H2O to the thiocyanate molecule, a step which is similar to the addition of H2O to the nitrile molecule (Fig. 7). This reaction step of the structurally related nitrile hydratase supports the model in which the conversion of a cyano group to an amide group was assumed to be the first step of thiocyanate hydrolase reactions. The model also contains the assumption of the production of ammonia and acid before the formation of carbonyl sulfide. For nitrile hydratase, the amide is hydrolyzed to ammonia and acid by an amidase that has been shown to be encoded by a gene within the same operon as that for nitrile hydratase (18). There is also another enzyme called nitrilase that catalyzes the direct hydrolysis of nitrile to ammonia and acid (13). We found a limited homology between amidase and nitrilase sequences, suggesting that they might be distantly related (data not shown). However, these enzymes did not show any significant sequence homology to thiocyanate hydrolase. Furthermore, we have also examined the reported sequence for cyanase (9); it converts cyanate (O−CN) to ammonia and CO2, which is structurally similar to carbonyl sulfide (COS). However, there were no significant homologies between cyanase and thiocyanate hydrolase or nitrile-degrading enzymes. These observations suggest that the subunit constitution of thiocyanate hydrolase is equivalent to that of nitrile hydratase but that thiocyanate hydrolase has unique extra functions to convert amide to produce ammonia and the corresponding acid or carbonyl. These functions may be new enzymic activities that have been acquired by thiocyanate hydrolase or they may be old ones that have been lost by nitrile hydratase during molecular evolution.
FIG. 7.
Structural homology and functional similarity between thiocyanate hydrolase and nitrile hydratase. Homologous subunits (boxes) are aligned with bars indicating positions of identical amino acids. The P. chlororaphis enzyme was chosen as the representative for nitrile hydratase. The lengths of the boxes and the spaces between bars are proportional to the actual numbers of amino acid residues. The array of the thiocyanate hydrolase subunits was drawn to fit the order of scn genes, but the gene organization of nitrile hydratase subunits varies among different bacteria. In the case of P. chlororaphis, the gene coding for the β subunit is downstream of that coding for the α subunit. Thiocyanate hydrolase converts thiocyanate (SCN−) to ammonia (NH3) and COS (SCO) through several intermediates; two intermediates assumed by Katayama et al. (11) are omitted in the figure for simpler comparison. Nitrile hydratase converts nitrile (RCN) to amide (RCONH2), which is hydrolyzed by a different enzyme, amidase, to produce carboxylic acid (RCOOH) and ammonia (2).
From a biochemical point of view, it may be possible to argue that those unidentified intermediates involved in the thiocyanate hydrolase reaction are extremely unstable, unstable enough to be converted nonenzymatically to the final product, COS. It is unlikely, however, that our enzyme fraction contained amidase contamination inherent to our purification procedure, because even a highly purified enzyme showed a stoichiometric conversion from thiocyanate to ammonia and COS. The hydrolysis of thiocyanate and ammonia formation occurred simultaneously for up to 60 min (11). Further biochemical and genetic analyses using cloned genes would help us to understand the reaction mechanisms and the properties of this novel enzyme.
ACKNOWLEDGMENTS
We thank Fumio Amano of National Institute of Health, Japan, for the preparation of antibodies.
This work was supported in part by a Grant-in Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan.
REFERENCES
- 1.Asano Y, Fujishiro K, Tani Y, Yamada H. Aliphatic nitrile hydratase from Arthrobacter sp. J-1. Purification and characterization. Agric Biol Chem. 1982;46:1165–1174. [Google Scholar]
- 2.Asano Y, Tani Y, Yamada H. A new enzyme, nitrile hydratase, which degrades acetonitrile in combination with amidase. Agric Biol Chem. 1980;44:2251–2252. [Google Scholar]
- 3.Baaku, P., K. Yamada, and H. Morimoto. November 1994. JP patent 1994303971-A.
- 4.Bett P M, Rinder D F, Fleeker J R. Thiocyanate utilization by an Arthrobacter. Can J Microbiol. 1979;25:1277–1282. doi: 10.1139/m79-201. [DOI] [PubMed] [Google Scholar]
- 5.Brennan B A, Cummings J G, Chase D B, Turner I M, Jr, Nelson M J. Resonance Raman spectroscopy of nitrile hydratase, a novel iron-sulfur enzyme. Biochemistry. 1996;35:10068–10077. doi: 10.1021/bi960163t. [DOI] [PubMed] [Google Scholar]
- 6.Chung J, Wood J L. Oxidation of thiocyanate to cyanide and sulfate by the lactoperoxidase-hydrogen peroxide system. Arch Biochem Biophys. 1970;141:73–78. doi: 10.1016/0003-9861(70)90108-6. [DOI] [PubMed] [Google Scholar]
- 7.Duran R, Nishiyama M, Horinouchi S, Beppu T. Characterization of nitrile hydratase genes cloned by DNA screening from Rhodococcus erythropolis. Biosci Biotechnol Biochem. 1993;57:1323–1328. doi: 10.1271/bbb.57.1323. [DOI] [PubMed] [Google Scholar]
- 8.Happold F C, Jones G L, Pratt D B. Utilization of thiocyanate by Thiobacillus thioparus and T. thiocyanoxidans. Nature (London) 1958;182:266–267. doi: 10.1038/182266a0. [DOI] [PubMed] [Google Scholar]
- 9.Harano Y, Suzuki I, Maeda S, Kaneko T, Tabata S, Omata T. Identification and nitrogen regulation of the cyanase gene from the cyanobacteria Synechocystis sp. strain PCC 6803 and Synechococcus sp. strain PCC 7942. J Bacteriol. 1997;179:5744–5750. doi: 10.1128/jb.179.18.5744-5750.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katayama Y, Kuraishi H. Characteristics of Thiobacillus thioparus and its thiocyanate assimilation. Can J Microbiol. 1978;24:804–810. doi: 10.1139/m78-135. [DOI] [PubMed] [Google Scholar]
- 11.Katayama Y, Narahara Y, Inoue Y, Amano F, Kanagawa T, Kuraishi H. A thiocyanate hydrolase of Thiobacillus thioparus: a novel enzyme catalyzing the formation of carbonyl sulfide from thiocyanate. J Biol Chem. 1992;267:9170–9175. [PubMed] [Google Scholar]
- 12.Kelly D P, Malin G, Wood A P. Microbial transformations and biogeochemical cycling of one-carbon substrates containing sulphur, nitrogen or halogens. In: Murrell J C, Kelly D P, editors. Microbial growth on C1 compounds. Andover, England: Intercept Ltd.; 1993. pp. 47–63. [Google Scholar]
- 13.Kobayashi M, Komeda H, Yanaka N, Nagasawa T, Tamada H. Nitrilase from Rhodococcus rhodochrous J1. Sequencing and overexpression of the gene and identification of an essential cysteine residue. J Biol Chem. 1992;267:20746–20751. [PubMed] [Google Scholar]
- 14.Kobayashi M, Nishiyama M, Nagasawa T, Horinouchi S, Beppu T, Yamada H. Cloning, nucleotide sequence and expression in Escherichia coli of two cobalt-containing nitrile hydratase genes from Rhodococcus rhodochrous J1. Biochim Biophys Acta. 1991;1129:23–33. doi: 10.1016/0167-4781(91)90208-4. [DOI] [PubMed] [Google Scholar]
- 15.Komeda H, Kobayashi M, Shimizu S. A novel gene cluster including the Rhodococcus rhodochrous J1 nhlBA genes encoding a low molecular mass nitrile hydratase (L-NHase) induced by its reaction product. J Biol Chem. 1996;271:15796–15802. doi: 10.1074/jbc.271.26.15796. [DOI] [PubMed] [Google Scholar]
- 16.Komeda H, Kobayashi M, Shimizu S. Characterization of the gene cluster of high-molecular-mass nitrile hydratase (H-NHase) induced by its reaction product in Rhodococcus rhodochrous J1. Proc Natl Acad Sci USA. 1996;93:4267–4272. doi: 10.1073/pnas.93.9.4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mayaux J F, Cerebelaud E, Soubrier F, Faucher D, Petre D. Purification, cloning, and primary structure of an enantiomer-selective amidase from Brevibacterium sp. strain R312: structural evidence for genetic coupling with nitrile hydratase. J Bacteriol. 1990;172:6764–6773. doi: 10.1128/jb.172.12.6764-6773.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nishiyama M, Horinouchi S, Kobayashi M, Nagasawa T, Yamada H, Beppu T. Cloning and characterization of genes responsible for metabolism of nitrile compounds from Pseudomonas chlororaphis B23. J Bacteriol. 1991;173:2465–2472. doi: 10.1128/jb.173.8.2465-2472.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nyunoya H, Lusty C J. Sequence of the small subunit of yeast carbamyl phosphate synthetase and identification of its catalytic domain. J Biol Chem. 1984;259:9790–9798. [PubMed] [Google Scholar]
- 20.Payne M S, Wu S, Fallon R D, Tudor G, Stieglitz B, Turner I M, Jr, Nelson M J. A stereoselective cobalt-containing nitrile hydratase. Biochemistry. 1997;36:5447–5454. doi: 10.1021/bi962794t. [DOI] [PubMed] [Google Scholar]
- 21.Saito I, Stark G R. Charomids: cosmid vectors for efficient cloning and mapping of larger or small restriction fragments. Proc Natl Acad Sci USA. 1986;83:8664–8668. doi: 10.1073/pnas.83.22.8664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stafford D A, Callely A G. The utilization of thiocyanate by a heterotrophic bacterium. J Gen Microbiol. 1969;55:285–289. doi: 10.1099/00221287-55-2-285. [DOI] [PubMed] [Google Scholar]
- 23.Vesey C J, Wilson J. Red cell cyanide. J Pharm Pharmacol. 1978;30:20–26. doi: 10.1111/j.2042-7158.1978.tb13147.x. [DOI] [PubMed] [Google Scholar]
- 24.Westley J. Cyanide and sulfane sulfur. In: Vennesland B, Conn E E, Knowles C J, Westley J, editors. Cyanide in biology. New York, N.Y: Academic Press; 1973. pp. 61–76. [Google Scholar]
- 25.Wood J L. Biochemistry. In: Newman A A, editor. Chemistry and biochemistry of thiocyanic acid and its derivatives. New York, N.Y: Academic Press; 1975. pp. 156–221. [Google Scholar]
- 26.Youatt J B. Studies on the metabolism of Thiobacillus thiocyanoxidans. J Gen Microbiol. 1954;11:139–149. doi: 10.1099/00221287-11-2-139. [DOI] [PubMed] [Google Scholar]