Abstract
Objective:
To build a novel classifier using an optimized 3D-convolutional neural network for predicting high-grade small bowel obstruction (HGSBO).
Summary background data:
Acute SBO is one of the most common acute abdominal diseases requiring urgent surgery. While artificial intelligence and abdominal computed tomography (CT) have been used to determine surgical treatment, differentiating normal cases, HGSBO requiring emergency surgery, and low-grade SBO (LGSBO) or paralytic ileus is difficult.
Methods:
A deep learning classifier was used to predict high-risk acute SBO patients using CT images at a tertiary hospital. Images from three groups of subjects (normal, nonsurgical, and surgical) were extracted; the dataset used in the study included 578 cases from 250 normal subjects, with 209 HGSBO and 119 LGSBO patients; over 38 000 CT images were used. Data were analyzed from 1 June 2022 to 5 February 2023. The classification performance was assessed based on accuracy, sensitivity, specificity, and area under the receiver operating characteristic curve.
Results:
After fivefold cross-validation, the WideResNet classifier using dual-branch architecture with depth retention pooling achieved an accuracy of 72.6%, an area under receiver operating characteristic of 0.90, a sensitivity of 72.6%, a specificity of 86.3%, a positive predictive value of 74.1%, and a negative predictive value of 86.6% on all the test sets.
Conclusions:
These results show the satisfactory performance of the deep learning classifier in predicting HGSBO compared to the previous machine learning model. The novel 3D classifier with dual-branch architecture and depth retention pooling based on artificial intelligence algorithms could be a reliable screening and decision-support tool for high-risk patients with SBO.
Keywords: 3D CNN, deep learning, small bowel obstruction
Introduction
Highlights
We built an artificial intelligence-based model with abdominal computed tomography images to help detect high-risk patients with acute small bowel obstruction (ASBO).
A 3D-convolutional neural network model detected high-risk ASBO patients with high accuracy and efficiency.
Artificial intelligence models can accurately detect high-risk ASBO patients, making them reliable screening and decision-support tools for high-risk patients with ASBO.
Acute small bowel obstruction (ASBO) is one of the most common acute abdominal diseases that may require urgent surgery. In emergent surgical cases, any delay in operation is known to be related to significant morbidity and mortality1. According to reports from the United States, ASBO accounts for 12–16% of surgical hospitalizations, amounting to 300 000 surgeries and $2.3 billion in medical costs annually2. The most common cause of acute intestinal obstruction is postoperative adhesion (70%), followed by cancer, inflammatory bowel disease, and hernias3,4. The pathophysiology of ASBO progresses from the onset of intestinal obstruction due to various causes, followed by the proximal dilatation of the bowel to the occluded area being deteriorated by the accumulation of fermented gas and shifted fluids5. Prolonged intestinal obstruction and distension can lead to hypovolemia and increased intramural pressure, leading to intestinal ischemia or necrosis. Ischemia and bowel wall dilatation weaken the intestinal barrier, increasing the risk of bowel perforation. This risk increases with clinical deterioration and the duration of unsuccessful medical treatment6.
Treatment options for ASBO vary considerably depending on pathophysiological progression. Emergency surgery is essential when an intestinal infarction or peritonitis is suspected. In addition, early surgery is required if intestinal ischemia worsens. However, in ~70% of cases, nonoperative management (NOM) is successful, and ~20% of patients who have undergone NOM will eventually undergo surgery owing to clinical deterioration1. Recently, abdominal computed tomography (CT) has been playing a critical role in predicting the failure of NOM treatment. Several papers have reported using abdominal CT for imaging findings and modeling7,8. However, despite abdominal CT’s crucial role in deciding ASBO surgical treatment, the radiologic findings that require surgery are only discovered by experienced radiologists, and the interobservation variation is relatively high9,10. Recently, artificial intelligence (AI) has been widely applied to various tasks in medical imaging, and AI can make predictions as accurately as professional human interpreters11,12. Most studies on ASBO and AI have been conducted using simple radiography. Furthermore, AI studies using abdominal CT are scarce, and recent ones depend on radiologists’ interpretations rather than AI-based approaches13. Although papers are reporting the use of AI in abdominal CT, several problems have been encountered. For example, a recent study conducted by Vanderbecq et al.14 successfully detected the transition zone of ASBO by CT, but it used only ASBO CT and not the normal CT. The study diagnosed several instances of ASBO from CT; however, the proposed method cannot classify between normal and abnormal instances because the study only used abnormal data. Previously, a pilot study was performed to determine whether an AI can distinguish between normal and ASBO X-ray images11. However, the imbalance between normal and abnormal data was significant, and the study used non-CT images. It is often clinically challenging to diagnose patients that are at risk of requiring emergency surgery. In addition, distinguishing high-risk cases, such as closed-loop obstructions or SBO by band adhesion, is a laborious and challenging task for radiologists and clinicians.
In recognition of these problems, this study aimed to establish a novel AI model that can effectively diagnose ASBO in normal subjects using only CT images and assist in the early diagnosis and identification of patients at risk of surgery.
Methods
Dataset
This study has been reported in accordance with the strengthening the reporting of cohort, cross-sectional, and case–control studies in surgery (STROCSS) standards15 (Supplemental Digital Content 1, http://links.lww.com/JS9/B23). This study was registered at cris.nih.go.kr. A single-center retrospective medical record study at a tertiary institution was designed for this diagnostic investigation. The ethical review board authorized this study. In accordance with the university’s requirements for retrospective analyses, informed consent was waived. The research was conducted in accordance with the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD)16 , the Checklist for Artificial Intelligence in Medical Imaging (CLAIM)17 and the Standards for Reporting of Diagnostic Accuracy Studies (STARD)18 (Supplemental Digital Content 2, http://links.lww.com/JS9/B24).
The following dataset refinement process was conducted to identify a high-risk group that may require emergency surgery for intestinal obstruction. The participants consisted of three groups, including the nonsurgical group: patients who visited the emergency department or were admitted to the hospital for ileus between 1 January 2000 and 31 December 2021; surgical group: patients who underwent lysis of adhesion for ASBO during the same period; and the normal subjects: healthy individuals without any abnormal abdominal CT findings during the health screenings in 2019. Their images were extracted after the anonymization process. The inclusion criteria were: 18 years and older, CT image findings with SBO, and mechanism of obstruction caused by the adhesion. Before this process, we identified normal subjects (n=1000) without any abnormal abdominal CT findings during the health screenings conducted in 2019. Since the cause and clinical aspects of intestinal obstruction are very heterogeneous, a detailed review of the case was required. Appendices 1 (Supplemental Digital Content 3, http://links.lww.com/JS9/B25) and 2 (Supplemental Digital Content 4, http://links.lww.com/JS9/B26) describe the exclusion cases during the refinement process. Exclusions were made because of nonadhesion mechanisms; other anatomical locations; or pathological conditions such as malignancy, peritonitis, and inflammatory bowel disease. In addition, cases with gastrografin use and postoperative ileus within one month were excluded. Overt ischemia and necrosis of the small bowel were also excluded. The patient’s demographics are described in Appendix 3 (Supplemental Digital Content 5, http://links.lww.com/JS9/B27). The surgical and nonsurgical groups were investigated to identify patients with high-risk intestinal obstruction. First, in the nonsurgical group, the ileus-related disease classification code (similar to the ICD code) was used. Cases for which abdominal CT images were unavailable and cases of nonadhesive ASBO were excluded. Cases of high-grade SBO (HGSBO) were accompanied by one of the closed-loop findings, adhesive bands, or complete or incomplete high-grade obstruction with abrupt luminal narrowing. In the case of low-grade SBO (LGSBO), it was determined that there was fluid-filled distension of the small bowel, accompanied by one of the following findings: a low possibility of obstruction or a low-grade or partial obstruction. In the surgical group, all patients who underwent abdominal surgery were investigated at the hospital. Cases were refined according to the criteria of adhesiolysis due to adhesive SBO. Finally, high-risk surgical findings, such as adhesive bands or closed loops, consistent with preoperative radiologic findings, such as high-grade obstruction or strangulation, were included in the final cohort. Figure 1 shows a flowchart of the above process. Thus, 209 HGSBO, 119 LGSBO, and 250 normal cases were obtained. The dataset was divided into 462 training sets and 116 test sets to build the model, and fivefold cross-validation was performed.
Figure 1.
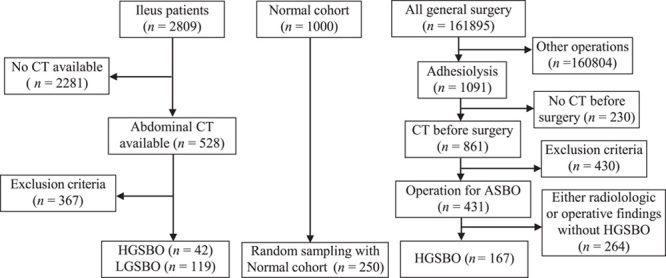
Patient selection process. CT, computed tomography; HGSBO, high-grade small bowel obstruction; LGSBO, low-grade small bowel obstruction.
Development and training of AI system
3D image classification is widely used in the medical field. Some works19–23 have used 3D-convolutional neural networks (CNN) for classifying Alzheimer’s disease. Other studies24–26 have used 3D image classification for brain diseases. However, little research has been done on using 3D image classification to distinguish ASBO cases.
The proposed network aims to distinguish between CT images of normal cases, cases of HGSBO that require emergency surgery, and cases of LGSBO or paralytic ileus. HGSBO and LGSBO are clinically distinct because a delay in surgical intervention in the case of HGSBO is known to increase the risk of morbidity and mortality, particularly in older patients1,27; however, the CT images for both appear similar. Therefore, it is challenging to classify HGSBO and LGSBO on CT images using a simple network structure. Thus, we propose two approaches for the classification method: dual-branch architecture (DBA) and depth retention pooling. For DBA, to enrich the class information of HGSBO and LGSBO, we trained the features by learning the base classifier and finer classifier simultaneously; this was to enrich the class information of HGSBO and LGSBO. In the second approach, we preserved the depth information in the last feature map to intensify the subtle information in CT images. Our networks based on these approaches effectively distinguish all three class labels: normal, HGSBO, and LGSBO. A detailed description of the proposed network is presented in the following subsections.
DBA
Suppose the architecture simply uses a normal three-label classifier as the last fully connected (FC) layer. In that case, it cannot distinguish HGSBO from LGSBO, and it cannot properly learn the conflicting characteristics. Some networks are split into branches to improve performance28–31. Zhang et al.31 used multiple branches and outputs resembling ensembles, and Xie et al.30 converted one 3×3 convolution into multiple 3×3 convolution branches using cardinality. Zhang et al.31 also split feature maps using cardinality and by sub-grouping one cardinality into several subsamples. Wu et al.29 split a branch using different kernel sizes to extract different features. All of these studies split branches in various ways to extract more diverse and sophisticated features and improve performance. Inspired by this, we introduce a DBA. Although a simple FC layer cannot distinguish and handle similar features, our DBA can learn the difference between two cases of SBO by synchronously training the classifier into two branches.
In the DBA, each branch is not trained separately through fine-tuning but by training from scratch at a time. We trained a network as a generic multiclass classification32 to discriminate between normal and two SBOs, as shown in Figure 2B. Figure 2C shows a finer classifier trained to separate HGSBO and LGSBO, which are indistinguishable from each other, as in the case of binary classification. The parameters of the finer classifier were also updated and used to make more accurate predictions. During inference, the base classifier distinguishes the normal and two SBO images, and the anticipated abnormal images are sent into the finer classifier as input. The finer classifier then categorizes the abnormal images into HGSBO and LGSBO. We use probabilities from both classifiers if the base classifier predicts that the input image is abnormal using Equation 1.
Figure 2.
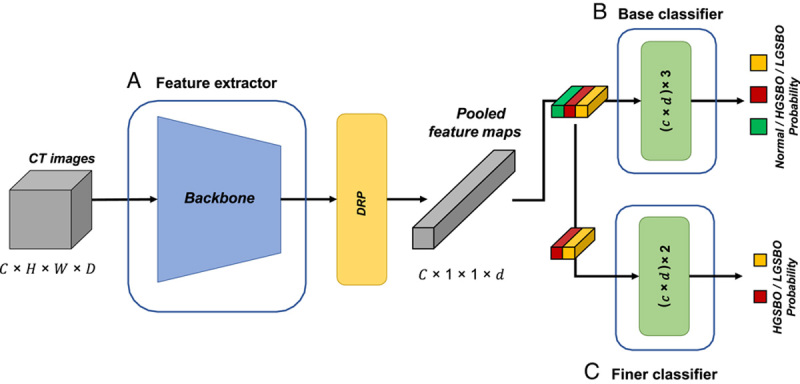
Workflow of the proposed Acute SBO diagnosis network. (A) refers to a feature extractor such as WideResNet; (B) is the base classifier to distinguish normal, HGSBO, and LGSBO; and (C) denotes a finer classifier that differentiates HGSBO and LGSBO. (C) trains only HGSBO and LGSBO. Green, red, and yellow pooled feature maps represent normal, HGSBO, and LGSBO, respectively. CT, computed tomography; HGSBO, high-grade small bowel obstruction; LGSBO, low-grade small bowel obstruction.
| (1) |
DRP
Most diseases are concentrated in extremely small areas of CT images. Therefore, to capture the information in a small area, it is essential to retain as much information as possible in the last pooling layer. However, the conventional method does not consider this point and even ignores the depth information of CT images in the pooling layer33. Therefore, traditional pooling squashes the depth information while pooling the C×D×7×7 feature map into C×1×1×1, where C denotes the channel size and D represents the depth size.
Instead of losing depth information, we contend that 3D images should retain depth information to learn 3D image features appropriately. In other words, lesions such as HGSBO and LGSBO may present only at a specific depth in 3D pictures, and it is critical to be able to learn features by depth in such circumstances. Therefore, we propose a method of retaining such depth information to collect the information present in such small regions. It performs pooling only in the vertical and horizontal spatial spaces of the deep features, except for the channel of the feature map, which represents the depth information of the CT image. We refer to the proposed method as DRP.
Network architecture with DBA and DRP
The proposed two-stage training strategy, DRP, is a generic method that can be applied to most deep neural network architectures. Therefore, we exploited representative CNN models30,34–36 for the feature extractor, as shown in Figure 2A. Once the features of the CT images were extracted, all features were fed to two FC layers that classified the images as normal, abnormal, HGSBO, and LGSBO. Notably, the second FC layer receives features from the output of the DRP in addition to the features generated by the first FC layer. Therefore, the second layer contains rich feature information, including CT depth information.
To clarify DBA and DRP in network architecture, we can say that DBA is the architectural approach of the multibranch classifier layer, whereas DRP is the feature pooling approach used to increase the feature representation power.
Training setup
In the training process, we used a stochastic gradient descent optimizer with a learning rate of 0.1, a 5e-4 weight decay, and 0.01 learning rate decay using a cosine annealing scheduler for 50 epochs. We also applied gradient clipping and efficient data augmentation methods, such as flip, gamma, noise, motion, bias field, random affine, and elastic deformation. The input image was cropped and padded to 224×224×112 pixels after resampling the voxel space to two. The area under receiver operating characteristic (AUROC) curve was used as the evaluation metric in both training and inference. In addition, performance measures, such as specificity and sensitivity, were also evaluated via macro reduction, which independently calculates the metric for each class and then averages the metrics across classes. The code and models are publicly released at https://github.com/SoongE/DBADRP_Classifier.
Results
We compared the performance achieved by applying DBA and DRP using DenseNet12135, which is frequently used in the medical domain. We also used EfficientNet-B036 and the ResNet family30,34,37, which are frequently used as a backbone to achieve good performance in CNNs.
Performance of ASBO diagnosis
We evaluated the classification performance of the proposed network for normal, HGSBO, and LGSBO images. Table 1 shows that the proposed network performed well in terms of accuracy, specificity, sensitivity, and AUROC. The AUC was 0.896 (95% CI: 0.895–0.897) for WideResNet, 0.873 (95% CI: 0.866–0.880) for DenseNet, and 0.873 (95% CI: 0.865–0.871) for EfficientNet. The proposed networks use DBA and DRP methods together to perform better than naive networks with a performance increase of 0.25 AUROC.
Table 1.
Performance comparison of the naive method and our method with various backbones.
| Backbone | Methods | Accuracy (%) | Specificity (%) | Sensitivity (%) | AUROC |
|---|---|---|---|---|---|
| ResNet | Naive | 66.15 | 83.08 | 66.15 | 0.848±0.05 |
| DBA+DRP | 70.26 | 85.13 | 70.26 | 0.876±0.02 | |
| ResNext | Naive | 68.46 | 84.23 | 68.46 | 0.874±0.02 |
| DBA+DRP | 66.41 | 83.21 | 66.41 | 0.883±0.01 | |
| WideResNet | Naive | 65.13 | 82.56 | 65.13 | 0.861±0.02 |
| DBA+DRP | 72.56 | 86.28 | 72.56 | 0.896±0.01 | |
| DenseNet | Naive | 75.12 | 83.83 | 62.81 | 0.868±0.05 |
| DBA+DRP | 72.68 | 83.28 | 63.41 | 0.873±0.07 | |
| EfficientNet | Naive | 71.22 | 82.64 | 60.41 | 0.841±0.04 |
| DBA+DRP | 71.71 | 83.16 | 63.36 | 0.868±0.03 |
Regardless of the backbone, DBA+DRP always outperformed the naive approach.
The values were all obtained through fivefold cross-validation.
DBA, dual-branch architecture; DRP, depth retention pooling.
Precise comparison of class-specific performance
Although the AUROC decreased when each proposed method was used alone, examining this in more detail is necessary. The measurement of AUROC for each class in the networks indicates that DRP helps better distinguish each class; however, the overall performance may appear degraded. Figure 3 shows the ROC curves of naive and DBA+DRP in WideResNet. Figure 3A shows that the performance of HGSBO was poor when the threshold was high, whereas the ROC curve of the DRP+DRP network Figure 3B shows that HGSBO exhibited high performance even at a high threshold. Even though the average AUROC of all classes can be lowered, DRP+DRP better distinguishes features between classes, as shown in Figure 3B.
Figure 3.
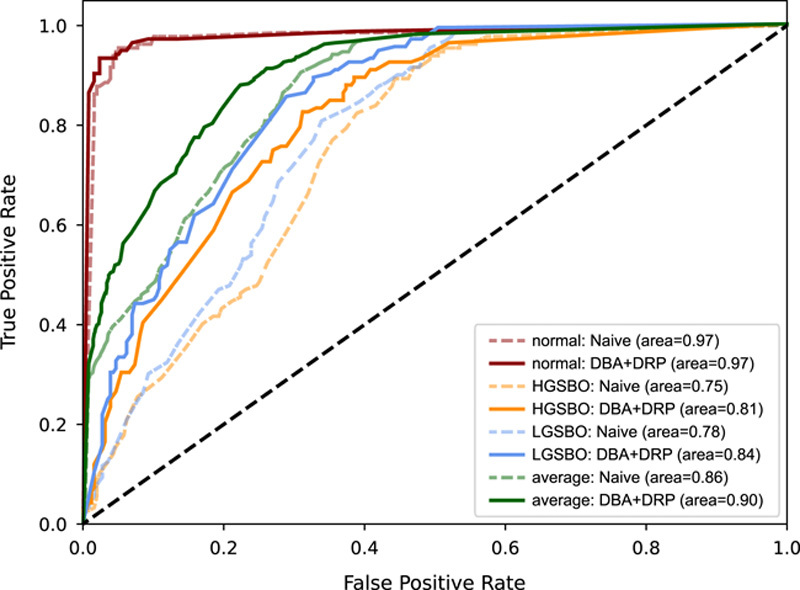
Receiver operating characteristic curve when applying naive and proposed methods with WideResNet. Solid line and dotted lines are the receiver operating characteristics of DBA+DRP and Naïve, respectively. Regardless of each class, the solid line is always above the dotted line. DBA, dual-branch architecture; DRP, depth retention pooling; HGSBO, high-grade small bowel obstruction; LGSBO, low-grade small bowel obstruction.
We also compared the performance by class in terms of various metrics. The proposed model predicts normal, HGSBO, LGSBO, and ASBO classes as a set of HGSBOs and LGSBOs. Table 2 presents the detection performance of three classes using our model. The detection performance of ASBO was significantly better than that of the HGSBO and LGSBO classes in terms of all the metrics.
Table 2.
Precision matrix of our proposed methods, WideResNet with DBA+DRP, for each class from the entire cohort.
| Class | Accuracy | Specifity | Sensitivity | PPV | NPV | F1 |
|---|---|---|---|---|---|---|
| HGSBO | 0.731 | 0.719 | 0.754 | 0.573 | 0.854 | 0.651 |
| LGSBO | 0.759 | 0.892 | 0.492 | 0.696 | 0.779 | 0.577 |
| ASBO | 0.962 | 0.977 | 0.931 | 0.953 | 0.966 | 0.942 |
We measured the metrics for each class from the entire cohort. HGSBO and LGSBO in the Class column indicate the performances of predicting each class on the entire cohort, while ASBO indicates the performance of predicting HGSBO or LGSBO on the entire cohort, that is, the performance of distinguishing normal from abnormal on the entire cohort.
Robustness to variation in image quality and class imbalances
We evaluated the performance of the proposed method in real-world scenarios, which typically include many variations. Table 3 compares the performance of DBA+DRP and the naive method in the presence of blur, high contrast, and low contrast corruption. This experiment was run on only single-fold data and showed that DBA+DRP outperformed the naive method on all the metrics and almost all backbones. Table 4 shows the results of solving the class imbalance problem using data augmentation. The performance of DBA+DRP improved with augmentation, which suggests that our method is effective with large amounts of data.
Table 3.
Robustness to distorted images that occur in the real-world.
| Corruption | Backbone | Methods | Accuracy (%) | Specificity (%) | Sensitivity (%) | AUROC |
|---|---|---|---|---|---|---|
| Blur | ResNet | Naive | 67.53 | 83.66 | 66.92 | 0.878 |
| DBA+DRP | 76.62 | 88.31 | 76.46 | 0.891 | ||
| ResNext | Naive | 62.34 | 81.15 | 62.00 | 0.826 | |
| DBA+DRP | 72.73 | 86.43 | 72.72 | 0.917 | ||
| WideResNet | Naive | 66.23 | 83.01 | 65.38 | 0.852 | |
| DBA+DRP | 71.43 | 85.73 | 71.23 | 0.873 | ||
| DenseNet | Naive | 72.73 | 86.34 | 72.72 | 0.858 | |
| DBA+DRP | 64.94 | 82.42 | 64.67 | 0.848 | ||
| EfficientNet | Naive | 51.95 | 75.83 | 51.49 | 0.708 | |
| DBA+DRP | 51.95 | 75.87 | 51.59 | 0.714 | ||
| High Contrast | ResNet | Naive | 65.38 | 82.69 | 65.38 | 0.880 |
| DBA+DRP | 78.21 | 89.10 | 78.21 | 0.926 | ||
| ResNext | Naive | 73.08 | 86.54 | 73.08 | 0.874 | |
| DBA+DRP | 74.36 | 87.18 | 74.36 | 0.855 | ||
| WideResNet | Naive | 66.67 | 83.33 | 66.67 | 0.821 | |
| DBA+DRP | 70.51 | 85.26 | 70.51 | 0.868 | ||
| DenseNet | Naive | 69.23 | 84.62 | 69.23 | 0.848 | |
| DBA+DRP | 73.08 | 86.54 | 73.08 | 0.885 | ||
| EfficientNet | Naive | 56.41 | 78.21 | 56.41 | 0.585 | |
| DBA+DRP | 66.67 | 83.33 | 66.67 | 0.827 | ||
| Low Contrast | ResNet | Naive | 73.08 | 86.54 | 73.08 | 0.880 |
| DBA+DRP | 80.77 | 90.38 | 80.77 | 0.927 | ||
| ResNext | Naive | 74.36 | 87.18 | 74.36 | 0.889 | |
| DBA+DRP | 79.49 | 89.74 | 79.49 | 0.919 | ||
| WideResNet | Naive | 62.82 | 81.41 | 62.82 | 0.856 | |
| DBA+DRP | 75.64 | 87.82 | 75.64 | 0.887 | ||
| DenseNet | Naive | 71.79 | 85.90 | 71.79 | 0.890 | |
| DBA+DRP | 78.21 | 89.10 | 78.21 | 0.905 | ||
| EfficientNet | Naive | 71.79 | 85.90 | 71.79 | 0.896 | |
| DBA+DRP | 80.77 | 90.38 | 80.77 | 0.906 |
For each corruption type and backbone, DBA+DRP outperformed the naive method.
Table 4.
Augmentation to deal with class imbalance: performance improves when augmentation is applied with the backbones regardless of the methods.
| Backbone | Methods | Augmentation | w/o Augmentation |
|---|---|---|---|
| ResNet | Naive | 0.848 | 0.823 |
| DBA+DRP | 0.876 | 0.841 | |
| ResNext | Naive | 0.874 | 0.831 |
| DBA+DRP | 0.883 | 0.845 | |
| WideResNet | Naive | 0.861 | 0.833 |
| DBA+DRP | 0.896 | 0.839 | |
| DenseNet | Naive | 0.868 | 0.854 |
| DBA+DRP | 0.873 | 0.865 | |
| EfficientNet | Naive | 0.841 | 0.856 |
| DBA+DRP | 0.868 | 0.859 |
All experiments were performed using fivefold cross-validation.
DBA, dual-branch architecture; DRP, depth retention pooling.
Resource overhead of the proposed method
We evaluated the proposed method in terms of computational complexity (i.e. FLOPs), number of parameters, and throughput by comparing it with the backbone models. Our method had a relatively minor loss in parameters and throughput but significantly improved model performance. As shown in Table 5, some of the proposed method’s parameters improved and the throughput was only 0.6% lower. The FLOPs remained almost constant.
Table 5.
Resource overhead of proposed methods.
| Backbone | Method | Throughput (img/s) | Param. (M) | FLOPs (G) |
|---|---|---|---|---|
| ResNet | Naive | 42.65 | 46.17 | 220.14 |
| DBA+DRP | 42.43 (−0.2) | 48.29 (+2.12) | 220.14 (+0) | |
| DenseNet | Naïve | 28.71 | 11.24 | 229.924 |
| DBA+DRP | 28.51 (−0.2) | 11.79 (+0.55) | 229.928 (+0.004) | |
| EfficientNet | Naïve | 41.16 | 4.69 | 13.985 |
| DBA+DRP | 40.91 (−0.25) | 5.53 (+0.84) | 13.991 (+0.006) |
We report here the throughput, number of parameters, and floating-point operations (FLOPs) of each backbone when applying DBA and DRP. Throughput refers to the number of images that can be inferred per second, and parameter means to the size of the model. In general, the more parameters model has, the slower the speed and the better the performance. FLOPs are the number of floating-point operations. Operations include root square, log, exponential, and arithmetic, each of which counts as a single operation.
DBA, dual-branch architecture; DRP, depth retention pooling.
Qualitative result
We visualized gradient-weighted class activation mapping (Grad-CAM)38 as a qualitative empirical study. For this visualization, the feature map was extracted using WideResNet. After collecting five CT images per patient in each class, we constructed a feature map with a middle depth to compare the Grad-CAM images. In Figure 4, the number of titles in each image is the prediction probability, and true or false indicates whether the prediction is accurate. Figure 4 shows the Grad-CAM images of the naive model and our proposed model. The first line of the figures is the Grad-CAM of the normal data, and the second and third lines are abnormal values. Figure 4A shows the Grad-CAM of the naive model, where a heatmap can be observed stamped on a similar part in all classes; it does not correctly predict HGSBO because the heatmap is not well captured. However, in Figure 4B, the heatmap is taken differently for each row, and in one row, that is, in the same class, a heatmap is taken in a uniform form. From this, we can conclude that the model reasonably predicts the CT class. Figure 5 shows a generally correct example of model classification in which both high-grade and low-grade obstacles are classified accordingly.
Figure 4.
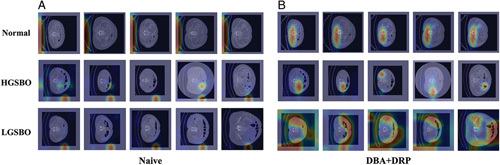
Gradient-weighted class activation mapping (Grad-CAM) for CT in multiple patients. (A) Grad-CAM of naive WideResNet and (B) Grad-CAM of DBA+DRP. The first row is the normal class, and the second and third are HGSBO and LGSBO, respectively. Figure 4(A) represents the naive model, in which the heat maps are activated on similar regions across all classes. This indicates that the model fails to accurately classify each individual class. Figure 4(B) illustrates the DBA+DRP model. In the normal class, the heat maps predominantly show activity in the lower center of the abdomen. However, for the HGSBO class, activity is concentrated in specific areas of the abdomen. In the case of LGSBO, the heatmap exhibits widespread activity across various regions of the abdomen, implying that the model distinguishes each class differently. DBA, dual-branch architecture; DRP, depth retention pooling; HGSBO, high-grade small bowel obstruction; LGSBO, low-grade small bowel obstruction.
Figure 5.
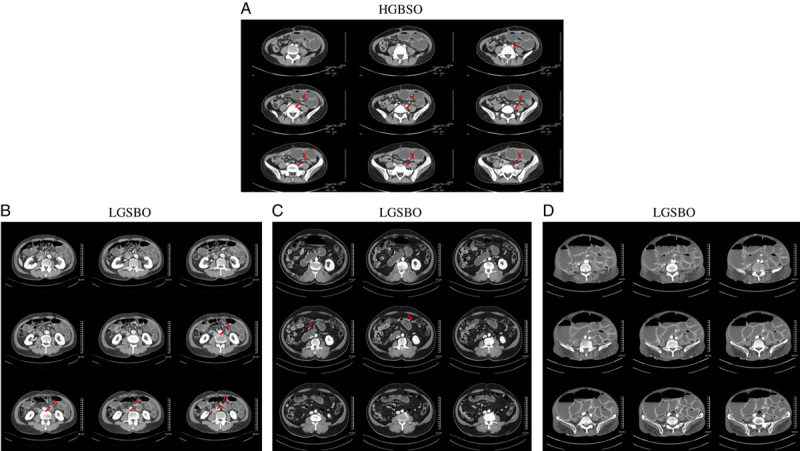
High-grade small bowel obstruction and low-grade small bowel obstruction (LGSBO) images. (A) a case of high-grade small bowel obstruction that shows a bird beak sign with each leading point close to the other. (B)–(D) represent LGSBO cases. Both are typical cases of small bowel obstruction and were correctly classified by our model. Especially, (D) is a case of severe paralytic ileus, and our classifier correctly classified it as LGSBO.
Discussion
In this study, we developed a novel classifier to identify high-risk patients with intestinal obstruction and verified its validity through fivefold cross-validation for a cohort including those who underwent surgery. In addition, this novel diagnostic tool can identify patients at risk by classifying them as normal or abnormal and effectively detect high-risk patients who may require surgery for ASBO. High-risk patients, such as older, sarcopenic, or diabetic patients, are more vulnerable to progressive bowel ischemia caused by high-grade obstruction, which may cause major morbidities such as AKI, cardiac injury, cardiovascular events, or even mortality39,40. In addition, if appropriately operated in the early stages, bowel resection can be avoided. Short bowel syndrome, which can occur if a large portion of the small bowel is resected, can also be prevented.
The accuracy of multidetector computed tomography (MDCT) in predicting SBO varied significantly and ranged from as low as 65%41 to as high as 100%. Most studies are based on small sample sizes and heterogeneous cohorts42,43. Thompson et al.44 reported an accuracy of 83% for MDCT and Maglinte et al.41 reported that MDCT can correctly identify 81% of HGSBOs and 48% of LGSBOs, and the overall accuracy is 65%. Our model is highly accurate in detecting SBO in a normal cohort. Therefore, it can be used as a screening tool for initial assessments. Our model’s overall accuracy is 78%, which is slightly better than that previously reported. In addition, compared to a previous machine learning model, our model is satisfactory in predicting HGSBO13.
We demonstrated that the proposed DBA and DRP methods improve classification performance. In future studies, we will apply DBA to multiple-branch architectures if diseases are analyzed hierarchically and several categories can be created. Furthermore, instead of simply flattening the features after DRP, we present a research direction to enrich the information by applying a new feature recalibration method.
However, the following limitations of this study should be noted. First, because this study was based on data from a single tertiary center, there could be a potential selection bias or the patient population could be poorly represented. However, our dataset contains more than 500 cases and shows a higher class imbalance, such as variations in BMI or image quality. We demonstrated this aspect using our model via further analysis, as detailed in Appendix 3 (Supplemental Digital Content 5, http://links.lww.com/JS9/B27). Second, all high-grade patients in the nonsurgical group did not undergo surgery and were diagnosed only by imaging findings. This could raise the question of whether the nonsurgical group patients were high-risk. It has been reported that predicting the high-risk group with only CT scans is difficult, and interobserver variation among radiologists is also high45. A case in which the distance between two transition points is less than 8 mm was recently reported as an important finding. Still, this finding also requires prospective validation, and 19 out of 62 patients showed success in NOM Tx, which could potentially be false-positive cases7. Although the final reports of the high-grade group included in this study were confirmed by a radiologist with more than 10 years of experience, there could be a potential risk of bias and subjective error. In terms of ground truth, it is necessary to design a prospective study that confirms high-risk findings, such as strangulation, closed-loop, or band adhesion by surgery7–10. However, we retrospectively identified ground truths in the surgical records, and Grad-CAM showed consistent results, supporting our findings. Third, because our model lacks external validation, our findings are relatively weak in terms of usability and generalizability. Further prospective validation or external validation studies are required to confirm the reliability and generalizability of the proposed classifier. For external validation, the model weights are available online. (link) Another limitation is that this classifier was trained to only classify cases of adhesive SBO and many clinical scenarios of ASBO were excluded. The reason for excluding these other cases from the analysis is as follows: First, there are fewer cases compared to adhesive SBOs. Second, the treatment strategies vary for each case, such as malignancy, which can potentially degrade the classifier’s performance. However, if sufficient cases are secured, hierarchical classification will be possible according to the proposed method. Fourth, because the clinical information of patients was excluded, and the classification was based only on CT images, future studies are required to improve performance. Although the regions activated in Grad-CAM are apparently activated in the small intestine, a few regions exist that are not46. These results are expected to be improved through further modifications of the model or through segmentation in future studies. Fifth, overall performance was slightly degraded despite the better distinction between HGSBO and LGSBO. In future studies, these issues could be solved with hyperparameter tuning (e.g. by selecting an appropriate optimizer or learning rate scheduler). In addition, we did not perform any tests to determine the clinical impact of our model. Therefore, before applying this classifier in clinical settings, further assessments are required, for example, the potential clinical impact and cost-effectiveness of implementing the proposed approach in real-world clinical settings should be evaluated. Finally, both accurate readings from an experienced radiologist and careful clinical consideration by the clinician in charge of patient care should be prioritized.
Conclusion
This work introduced a significant development in predicting high-risk patients with ASBO by applying a novel DBA using only CT images. The AI model efficiently classifies the abnormal group and guides radiologists and surgeons to identify high-risk patients requiring surgery. Further prospective validation studies are required to confirm the efficacy of this model.
Ethical approval
This study received ethical approval from the Ajou University Hospital IRB on 9 February 2023 (Approval # AJOUIRB-DB-2023-070).
Sources of funding
This research was supported by a grant from the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (grant no. NRF-2022R1A2C1012113) to Dr Shin, and supported in part by the National Research Foundation of Korea (NRF) of the Korea Government (MSIT) under grant no. NRF-2021R1F1A1062807, and Institute of Information and Communications Technology Planning and Evaluation (IITP) grant funded by the Korean Government (MSIT) (Artificial Intelligence Innovation Hub) under grant no 2021-0-02068.
Author contribution
S.R.: had full access to all data in the study and take responsibility for the integrity of the data and accuracy of data analysis, concept, and design; O.S., R.: acquisition, analysis, or interpretation of data; O.S.: statistical analysis; S.R.: obtained funding and supervision. All authors contributed in administrative, technical, material support, drafting of the manuscript, and critical revision of the manuscript for important intellectual content.
Conflicts of interest disclosures
All authors declare no conflicts of interest. No support from any organization has been received for the submitted work; there are no financial relationships with organizations that might be interested in the submitted work. There are no other relationships or activities that have influenced the submitted work.
Research registration unique identifying number (UIN)
Name of the registry : CRIS(Clinical Research Information Service).
Unique identifying number or registration ID: KCT0008330.
Hyperlink to your specific registration (must be publicly accessible and will be checked): https://cris.nih.go.kr/cris/search/detailSearch.do?all_type=Y&search_page=NU&search_lang=&pageSize=10&seq=24386&searchGubun=1&page=1.
Guarantor
Ho-Jung Shin, MD, MS.
Data availability statement
The datasets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request. Anonymized data is available on the local server at Ajou University.
Provenance and peer review
Not commissioned, externally peer reviewed.
Supplementary Material
Footnotes
Sponsorships or competing interests that may be relevant to content are disclosed at the end of this article.
Supplemental Digital Content is available for this article. Direct URL citations are provided in the HTML and PDF versions of this article on the journal’s website, www.lww.com/international-journal-of-surgery.
Published online 14 September 2023
Contributor Information
Seungmin Oh, Email: mangusn1@ajou.ac.kr.
Jongbin Ryu, Email: jongbin.ryu@gmail.com.
Ho-Jung Shin, Email: agnusdei13@gmail.com.
Jeong Ho Song, Email: songs1226@hanmail.net.
Sang-Yong Son, Email: spy798@gmail.com.
Hoon Hur, Email: hhcmc75@naver.com.
Sang-Uk Han, Email: hansu@ajou.ac.kr.
References
- 1.Foster NM, McGory ML, Zingmond DS, et al. Small bowel obstruction: a population-based appraisal. J Am Coll Surg 2006;203:170–176. [DOI] [PubMed] [Google Scholar]
- 2.Scott JW, Olufajo OA, Brat GA, et al. Use of national burden to define operative emergency general surgery. JAMA Surg 2016;151:e160480–e160480. [DOI] [PubMed] [Google Scholar]
- 3.Zielinski MD, Bannon MP. Current management of small bowel obstruction. Adv Surg 2011;45:1–29. [DOI] [PubMed] [Google Scholar]
- 4.Long B, Robertson J, Koyfman A. Emergency medicine evaluation and management of small bowel obstruction: evidence-based recommendations. J Emerg Med 2019;56:166–176. [DOI] [PubMed] [Google Scholar]
- 5.Rami Reddy SR, Cappell MS. A systematic review of the clinical presentation, diagnosis, and treatment of small bowel obstruction. Curr Gastroenterol Rep 2017;19:28. [DOI] [PubMed] [Google Scholar]
- 6.Fung BSC, Behman R, Nguyen M-A, et al. Longer trials of non-operative management for adhesive small bowel obstruction are associated with increased complications. J Gastrointest Surg 2020;24:890–898. [DOI] [PubMed] [Google Scholar]
- 7.Rondenet C, Millet I, Corno L, et al. CT diagnosis of closed loop bowel obstruction mechanism is not sufficient to indicate emergent surgery. Eur Radiol 2020;30:1105–1112. [DOI] [PubMed] [Google Scholar]
- 8.Zins M, Millet I, Taourel P. Adhesive small bowel obstruction: predictive radiology to improve patient management. Radiology 2020;296:480–492. [DOI] [PubMed] [Google Scholar]
- 9.Millet I, Boutot D, Faget C, et al. Assessment of strangulation in adhesive small bowel obstruction on the basis of combined CT findings: implications for clinical care. Radiology 2017;285:798–808. [DOI] [PubMed] [Google Scholar]
- 10.Millet I, Ruyer A, Alili C, et al. Adhesive small-bowel obstruction: value of CT in identifying findings associated with the effectiveness of nonsurgical treatment. Radiology 2014;273:425–432. [DOI] [PubMed] [Google Scholar]
- 11.Cheng PM, Tejura TK, Tran KN, et al. Detection of high-grade small bowel obstruction on conventional radiography with convolutional neural networks. Abdom Radiol 2018;43:1120–1127. [DOI] [PubMed] [Google Scholar]
- 12.Daugaard Jørgensen M, Antulov R, Hess S, et al. Convolutional neural network performance compared to radiologists in detecting intracranial hemorrhage from brain computed tomography: a systematic review and meta-analysis. Eur J Radiol 2022;146:110073. [DOI] [PubMed] [Google Scholar]
- 13.Goyal R, Mui LW, Riyahi S, et al. Machine learning based prediction model for closed-loop small bowel obstruction using computed tomography and clinical findings. J Comput Assist Tomogr 2022;46:169–174. [DOI] [PubMed] [Google Scholar]
- 14.Vanderbecq Q, Ardon R, De Reviers A, et al. Adhesion-related small bowel obstruction: deep learning for automatic transition-zone detection by CT. Insights Imaging 2022;13:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mathew G, Agha R, Albrecht J, et al. STROCSS 2021: strengthening the reporting of cohort, cross-sectional and case-control studies in surgery. Int J Surg 2021;96:106165. [DOI] [PubMed] [Google Scholar]
- 16.Collins GS, Reitsma JB, Altman DG, et al. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD Statement. BMC Med 2015;13:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mongan J, Moy L, Kahn CE, Jr. Checklist for Artificial Intelligence in Medical Imaging (CLAIM): a guide for authors and reviewers. Radiol Artif Intell 2020;2:e200029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen JF, Korevaar DA, Altman DG, et al. STARD 2015 guidelines for reporting diagnostic accuracy studies: explanation and elaboration. BMJ Open 2016;6:e012799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng C, Elazab A, Yang P, et al. Deep Learning Framework for Alzheimer’s Disease Diagnosis via 3D-CNN and FSBi-LSTM IEEE Access 2019;7:63605–63618. [Google Scholar]
- 20.Kruthika KR, Rajeswari, Maheshappa HD. CBIR system using Capsule Networks and 3D CNN for Alzheimer’s disease diagnosis. Inform Med Unlocked 2019;14:59–68. [Google Scholar]
- 21.Oh K, Chung YC, Kim KW, et al. Classification and visualization of alzheimer’s disease using volumetric convolutional neural network and transfer learning. Sci Rep 2019;9:18150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parmar HS, Nutter B, Long R, et al. Deep learning of volumetric 3D CNN for fMRI in Alzheimer’s disease classification. Medical Imaging 2020: Biomedical Applications in Molecular, Structural, and Functional Imaging; 2020 February 01, 2020.
- 23.Yang C, Rangarajan A, Ranka S. Visual explanations from deep 3D convolutional neural networks for Alzheimer’s disease classification. AMIA Annu Symp Proc 2018;2018:1571–1580. [PMC free article] [PubMed] [Google Scholar]
- 24.Kitaguchi D, Takeshita N, Matsuzaki H, et al. Development and validation of a 3-dimensional convolutional neural network for automatic surgical skill assessment based on spatiotemporal video analysis. JAMA Netw Open 2021;4:e2120786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nie D, Zhang H, Adeli E, et al. 3D Deep Learning for Multi-modal Imaging-Guided Survival Time Prediction of Brain Tumor Patients. Med Image Comput Comput Assist Interv 2016;9901:212–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wegmayr V, Aitharaju S, Buhmann J. Classification of brain MRI with big data and deep 3D convolutional neural networks. Medical Imaging 2018: Computer-Aided Diagnosis; 2018 February 01, 2018.
- 27.Schraufnagel D, Rajaee S, Millham FH. How many sunsets? Timing of surgery in adhesive small bowel obstruction: a study of the Nationwide Inpatient Sample. J Trauma Acute Care Surg 2013;74:181–187. [DOI] [PubMed] [Google Scholar]
- 28.Szegedy C, Vanhoucke V, Ioffe S, et al. Rethinking the Inception Architecture for Computer Vision. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 27-30 June 2016.
- 29.Wu W, Zhang Y, Wang D, et al. SK-Net: deep learning on point cloud via end-to-end discovery of spatial keypoints. Proc AAAI Conf Artif Intel 2020;34:6422–6429. [Google Scholar]
- 30.Xie S, Girshick R, Dollár P, et al. Aggregated residual transformations for deep neural networks. Proceedings of the IEEE conference on computer vision and pattern recognition; 2017.
- 31.Zhang H, Wu C, Zhang Z, et al. ResNeSt: Split-Attention Networks. 2022 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW); 2022 19-20 June 2022.
- 32.Nillmani, Jain PK, Sharma N, et al. Four types of multiclass frameworks for pneumonia classification and its validation in X-ray scans using seven types of deep learning artificial intelligence models. Diagnostics (Basel, Switzerland) 2022;12:652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nirthika R, Manivannan S, Ramanan A, et al. Pooling in convolutional neural networks for medical image analysis: a survey and an empirical study. Neural Comput Appl 2022;34:5321–5347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He K, Zhang X, Ren S, et al. Deep Residual Learning for Image Recognition. 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR); 2016 27-30 June 2016.
- 35.Huang G, Liu Z, Van Der Maaten L, et al. Densely connected convolutional networks. Proceedings of the IEEE conference on computer vision and pattern recognition; 2017.
- 36.Tan M, Le Q. Efficientnet: Rethinking model scaling for convolutional neural networks. International conference on machine learning; 2019: PMLR.
- 37.Zagoruyko S, Komodakis NJapa. Wide residual networks. 2016.
- 38.Selvaraju RR, Cogswell M, Das A, et al. Grad-cam: Visual explanations from deep networks via gradient-based localization. Proceedings of the IEEE international conference on computer vision; 2017.
- 39.Fevang BT, Fevang J, Stangeland L, et al. Complications and death after surgical treatment of small bowel obstruction: a 35-year institutional experience. Ann Surg 2000;231:529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Karamanos E, Dulchavsky S, Beale E, et al. Diabetes mellitus in patients presenting with adhesive small bowel obstruction: delaying surgical intervention results in worse outcomes. World J Surg 2016;40:863–869. [DOI] [PubMed] [Google Scholar]
- 41.Maglinte DD, Gage SN, Harmon BH, et al. Obstruction of the small intestine: accuracy and role of CT in diagnosis. Radiology 1993;188:61–64. [DOI] [PubMed] [Google Scholar]
- 42.Fukuya T, Hawes DR, Lu CC, et al. CT diagnosis of small-bowel obstruction: efficacy in 60 patients. AJR Am J Roentgenol 1992;158:765–769. [DOI] [PubMed] [Google Scholar]
- 43.Megibow A, Balthazar E, Cho K, et al. Bowel obstruction: evaluation with CT. Radiology 1991;180:313–318. [DOI] [PubMed] [Google Scholar]
- 44.Thompson WM, Kilani RK, Smith BB, et al. Accuracy of abdominal radiography in acute small-bowel obstruction: does reviewer experience matter? AJR Am J Roentgenol 2007;188:W233–W238. [DOI] [PubMed] [Google Scholar]
- 45.Makar RA, Bashir MR, Haystead CM, et al. Diagnostic performance of MDCT in identifying closed loop small bowel obstruction. Abdom Radiol 2016;41:1253–1260. [DOI] [PubMed] [Google Scholar]
- 46.Draelos RL, Carin L. Explainable multiple abnormality classification of chest CT volumes. Artif Intell Med 2022;132:102372. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analyzed during the current study are not publicly available but are available from the corresponding author on reasonable request. Anonymized data is available on the local server at Ajou University.


