Abstract
Staphylococcus aureus KSI9051 has a complex mutation that was associated with the aberrant expression of cell surface and extracellular proteins (M. S. Smeltzer, M. E. Hart, and J. J. Iandolo, J. Bacteriol. 61:919–925, 1993). This mutation was named xpr, although no specific gene was identified. Here this mutation is referred to as Δ1058::Tn551. In this study, we show that in strain KSI9051, the Δ1058::Tn551 mutation occurred coincidentally with a frameshift in agrC that is expected to truncate the sensor component of the known staphylococcal global regulatory locus agr. Remarkably, pleiotropic mutations affecting cell surface and extracellular proteins are generated at frequencies approaching 50% upon the transduction of erythromycin resistance (Emr) encoded by Δ1058::Tn551 from S. aureus KSI905 back to its parental strain, S6C. Three independent isolates created in the manner of KSI9051 contained mutations within agrC. Each isolate had different mutations, suggesting that the transduction of Emr encoded by Δ1058::Tn551 affects the stability of agrC in S6C. In similar experiments with strains from an S. aureus 8325 genetic background, a mutant AgrC phenotype could not be isolated, implying that strain S6 has aberrant genetic behavior. A comparison of the nucleotide sequences of AgrC from several strains revealed seven errors in the GenBank entry for agr (X52543); these data were confirmed with plasmid pRN6650, the original wild-type clone of agr.
The virulence of Staphylococcus aureus is dependent on the expression of cell surface and extracellular proteins (16). The current understanding is that many of these proteins are under the coordinate control of three interacting genetic loci known as agr, sar, and xpr (5, 31, 36). The regulatory loci function to repress the transcription of genes encoding cell surface proteins and to activate the transcription and, at least in one case, the translation of genes encoding extracellular proteins beginning at the transition from the exponential phase to the stationary phase of bacterial growth (14, 18, 24). The importance of these regulatory loci to the pathogenesis of S. aureus has been demonstrated in studies that show mutant strains are less virulent upon intraperitoneal challenge of mice or in animal models of staphylococcal disease (1, 6, 13, 36).
The agr locus consists of divergent messages transcribed from adjacent promoters (22). One promoter, P2, transcribes an ≈3.5-kb message (RNAII) that encodes four proteins ordered AgrB, AgrD, AgrC, and AgrA (27). Based on DNA sequence homology, the activity of the isolated components, or phenotypic complementation, the RNAII products have been shown to function in an autocatalytic signal transduction system (18, 26, 27). AgrC shows homology to signal transducers, and AgrA shows homology to the response regulators found in other bacterial signaling systems (27). The two remaining open reading frames complete the system, with agrD encoding the activating signal and agrB encoding a putative processing enzyme that is required for AgrD activity (19).
In addition to stimulation of transcription from the P2 promoter, the gene products of RNAII enhance transcription of an ≈0.5-kb message (RNAIII) from a second promoter, P3 (26). Synthesis of RNAIII from a heterologous promoter in an agr-null mutant of S. aureus 8325-4 returns a wild-type pattern of virulence factor messages repressing transcription of cell surface protein genes and activating transcription of extracellular-protein genes (26, 38). Mutational analysis has ruled out the involvement of any RNAIII translation product, leaving the transcript itself as the effector molecule of the agr system (18, 26).
A second locus, sar, both independently and through interactions with agr regulates the expression of virulence factors (8). Minimally, the sar locus consists of a single gene product, SarA, that is encoded by three messages ranging in size from 0.58 to 1 kb (3, 7). The sar messages are thought to provide distinct factors, since the attenuation of protein A transcription requires different messages in either sar or agr mutant strains of S. aureus (9). Cross-complementation in S. aureus has shown that sar independently regulates protein A expression in an agr mutant strain, although the expression of the other regulated virulence factors is apparently controlled by an agr-dependent mechanism. This mechanism involves the binding of SarA to a site within the region encoding the divergent promoters of agr. The binding of SarA to this region is required for the expression of RNAIII (24). Consistent with this finding, sar mutants have an agr-like phenotype in strains with an 8325-4 genetic background (7, 15).
Smeltzer et al. reported a chromosomal transposon insertion in S. aureus KSI9051 that defined a third global regulator, named xpr (36). KSI9051 is a derivative of S. aureus S6, the archetype enterotoxin-B-producing strain that is a hyperproducer of many extracellular proteins. To create KSI9051, strain S6 was first cured of a large penicillin resistance plasmid. The plasmid-cured strain, S6C, was then subjected to transposon Tn551 mutagenesis. One resulting strain, KSI905, was characterized as deficient only in the production of lipase (34). Upon transduction of erythromycin resistance from the transposon in KSI905 back to S6C, KSI9051 was isolated. This strain has an agr-like pattern of transcription and expression of cell surface and extracellular proteins and, like agr and sar mutants, has reduced levels of agr mRNA (14, 34, 36).
A comparison of the genome of S. aureus KSI905 and KSI9051 and that of their parental strain, S6C, indicated that the genetic lesion in both the mutant strains was an 18.7-kb chromosomal deletion that accompanied the insertion of transposon Tn551 (17, 34). The transposon insertion and the chromosomal deletion in KSI9051 were reported as the genetic locus xpr, although it is more appropriately designated Δ1058::Tn551, since the coding capacity of the region associated with the transposon-induced deletion is unknown and the transposon insertion has been mapped to position Ω1058 on the staphylococcal chromosome (17).
In the present study, we show that Δ1058::Tn551 does not confer the expected phenotype upon transduction into a derivative of S. aureus 8325. Given the similarity between the phenotype of S. aureus KSI9051 and agr mutant strains and reports of the genetic instability of agr in the S. aureus 8325-4 background, we attempted to complement the mutation in KSI9051 with DNA encoding the agr locus. We found that the P2 operon completely restored a wild-type phenotype to KSI9051. Consistent with this finding, we show that KSI9051 has a frameshift mutation that is expected to truncate AgrC. Transduction of erythromycin resistance (Δ1058::Tn551) from S. aureus KSI905 into strain S6C was used to create independent extracellular-protein-deficient mutants. A wild-type phenotype was restored to three of these strains with DNA that encodes AgrC. Sequence analysis of this region from the newly created strains revealed that each isolate had a different mutation or mutations within agrC, indicating the inherent instability of this gene under the conditions used to generate KSI9051.
MATERIALS AND METHODS
Bacterial strains, phage, media, and growth conditions.
Bacteria and phage relevant to this study are summarized in Table 1. S. aureus was routinely cultivated in Trypticase soy broth (TSB; Difco Laboratories, Detroit, Mich.) incubated at 37°C with rotary agitation or on Trypticase soy agar (TSA) plates. S. aureus was also cultivated on chemically defined synthetic medium without lysine or supplemented with erythromycin (29). Escherichia coli was grown at 37°C in Luria-Bertani (LB) broth with agitation or on LB agar plates (LB plus 1.5% agar). Antibiotic-resistant staphylococci were selected and maintained at 5 μg of tetracycline, erythromycin, or chloramphenicol ml−1. Antibiotic-resistant E. coli were grown in media augmented with 100 μg of ampicillin ml−1.
TABLE 1.
Summary of bacterial strains
| Strain or phage | Genotype and/or relevant phenotypea | Reference or source |
|---|---|---|
| E. coli DH5α | F− φ80lacZDM15 D(lacZYA-argF)U196 endA1 recA1 hsdR17(rK− mK+) deoR thi-1 supE44 λ− gyrA96 relA1 | BRL |
| S. aureus | ||
| ISP32 | 8325, lys-115 pig-131 | 29 |
| DU1090 | 8325-4, pig-131 hla-421::erm Hlb+ Hld+ | 28 |
| ISP2094 | 8325-4, pig-131 hlb::gen Hla+ Hld+ | Gift from T. J. Foster |
| KSI2111 | 80α/KSI9051 × ISP32b, Emr Geh+ Hla+ Hlb+ Lys+ Prt+ Nuc+ | This work |
| KSI2116 | KSI9051 with pIM34 integrated and resolved from the chromosome, Emr Geh+ Hla+ Hlb− Nuc+ Prt+ Sak+ Tcs | This work |
| KSI2121 | KSI9051 with pIM35 integrated and resolved from the chromosome, Emr Geh+ Hla+ Hlb− Nuc+ Prt+ Sak+ Tcs | This work |
| KSI2300 | 80α/DU1090 × ISP2094, pig-131 hla-421::erm hlb::gen, Hld+ | This work |
| KSI2401 | 80α/KSI905 × S6C, Geh− Hla− Hlb− Nuc− Prt− Sak− | This work |
| KSI2402 | 80α/KSI905 × S6C, Geh− Hla− Hlb− Nuc− Prt− Sak− | This work |
| KSI2403 | 80α/KSI905 × S6C, Geh− Hla− Hlb− Nuc− Prt− Sak− | This work |
| KSI905 | S6C, Ω[chr::Tn551]1058, Emr Geh− Hla+ Hlb− Nuc+ Prt+ Sak+ | 33 |
| KSI9051 | S6C, Ω[chr::Tn551]1058, Emr Geh− Hla− Hlb− Nuc− Prt− Sak− | 33 |
| 8325 | PS47, φ11, φ12, and φ13 lysogen | National Culture Type Collection |
| 8325-4 | 8325, UV cured of prophages | 25 |
| RN4220 | 8325-4, nitrosoguanidine-induced restriction mutant used as primary recipient for plasmids propagated in E. coli | 27 |
| RN4282 | Naturally occurring TSST-1-producing strain | 27, 30 |
| RN6390 | 8325-4, Hla+ Prt+ | 30 |
| S6 | Hyperproducer of extracellular proteins Geh+ Hla+ Hlb− Nuc+ Pcr Prt+ Sak+ | 34 |
| S6C | S6, plasmid free, φ42d and φ15 lysogen, Geh+ Hla+ Hlb− Nuc+ PCs Prt+ Sak+ | 34 |
| Phage 80α | S. aureus lytic group I phage 80 selected on lytic group III, propagated in 8325-4 | Gift from P. Pattee |
Abbreviations: Emr, erythromycin resistance; Geh, lipase; Hla, α-toxin; Hlb, β-toxin; Hld, δ-toxin; Lys, lysine; Pcr, penicillin resistant; PCs, penicillin sensitive; Prt, protease; Tcs, tetracycline sensitive; Nuc, nuclease; Sak, staphylokinase.
Transduction with phage from donor × recipient.
Indicator plates and extracellular protein assays.
Individual hemolysins were differentiated by examining the pattern of hemolysis on sheep blood agar following cross-streaking of the strain to be tested with S. aureus DU1090, ISP2094, or KSI2300. Observations for α-toxin or δ-toxin activity were made following 24 h of growth at 37°C. β-Toxin activity (hot-cold hemolysin) was visualized on the same plates following 8 h of refrigeration at 4°C. The pattern used to distinguish the hemolysins has been described previously by Elek and Levy (12). Protease activity was visualized as clear zones surrounding single colonies grown 12 h on nutrient agar plates (37) supplemented with 5% skim milk (Difco Laboratories). Nuclease activity was determined on DNase test agar (Difco Laboratories) as described by the manufacturer. Staphylokinase was visualized after 12 h of growth as halos surrounding single colonies on TSA containing 1% horse serum.
Quantitative measurements of α-toxin activity were determined from culture supernatant fluids by a method modified from that described by Peng et al. (30). Filtered culture supernatant fluids (100 μl) were added to 900 μl of 0.5% defibrinated rabbit blood washed three times in Tris-saline buffer (50 mM Tris, 100 mM NaCl [pH 7.0]). Samples were incubated for 30 min and centrifuged at 800 × g for 6 min, and activity from the supernatant fluids was determined spectrophotometrically at A410. Units of activity are expressed as the reciprocal of the dilution giving 50% lysis. Measurements of total lysis were determined from an average of 10 samples lysed with sodium dodecyl sulfate (SDS) at a final concentration of 1%. Lipase activity was measured as a decrease in optical density over time due to the hydrolysis of emulsified tributyrin (35). Units of lipase activity are reported as the linear slope of a plot of optical density at 540 nm versus time.
DNA isolation.
Staphylococcal chromosomal DNA was isolated by the method of Dyer and Iandolo (11). Plasmid DNA was purified from 3 ml of cultures of S. aureus with a kit from Qiagen (Chatsworth, Calif.). The plasmid isolation procedure was modified by incubating the cell suspension in P1 buffer containing 100 μg of recombinant lysostaphin ml−1 (AMBI UK, Trowbridge, United Kingdom) for 30 min at 37°C. The procedure was further modified by removal of the precipitate that formed after the addition of neutralization buffer by ultracentrifugation for 30 min at 1.5 × 105 × g. DNA was isolated from E. coli by standard methods (2).
PCR and plasmid constructs.
DNA was amplified by the method of Coen (10) with a range of MgCl2 concentrations from 0 to 9 mM, 1 μg of chromosomal DNA template, nucleotides (Promega Biotec, Madison, Wis.), Mg2+-free reaction buffer, and Advantage Tth polymerase (Clontech, Palo Alto, Calif.). Standard recombinant DNA techniques (2) were used for cloning DNA in E. coli DH5α. Restriction endonucleases, calf intestinal phosphatase, and T4 DNA ligase were obtained from Promega Biotec and used as recommended by the manufacturer.
Plasmid pIM29 contains the P2 operon of the agr locus (agrBDCA) from S. aureus RN6390, amplified with the primers 5′-GCATGCGTACTAAATCGTATAATGAC and 5′-TCTAGAGAATACGTCGTTAACTGAC, t-tail cloned into pT7Blue (R) (Novagen, Madison, Wis.). The underlined nucleotides highlight SphI and XbaI sites added to the 5′ ends of the primers. Plasmid pIM34 was constructed by transferring the ≈2.9-kb SacI-SalI fragment containing agrBDCA from pIM29 to identically cleaved sites in pSPT181 (18). Plasmid pIM35 consists of the ≈2.0-kb PstI-PvuII fragment from the insert in pIM34 and contains the 3′ end of agrB, all of agrD and agrC, and the 5′ end of agrA (agrB′DCA′) cloned into PstI-SmaI sites in pSPT181. Plasmids pIM30, pIM31, and pIM32 are pT7Blue (R) containing the entire agr locus amplified from strain RN6390, KSI9051, or S6C with primers 5′-ATGACTAAACATAGATTTATGAG and 5′-TCTAGAGAATACGTCGTTAACTGAC. Plasmids pIM49, pIM50, pIM51, pIM52, and pIM62 are pCR-Script (Stratagene, La Jolla, Calif.) with agrB′DCA′ from strain KSI2401, KSI2402, KSI2403, RN6390, or KSI2121 amplified with primers 5′-CACTCATAAGGATTATCAGTTGCGAGGGC and 5′-GAGCCATTTGCCCAATTCATTCAATTAGGC. Plasmid pIM54 contains the insert from pIM52 cloned into the SmaI site of pSPT181. Plasmid pIM36 is an ≈1.2-kb HindIII fragment from pKUS535 that encodes the transposase of Tn551 (see GenBank U75387 and U75368) cloned into pBluescript II KS(+) (Stratagene). Plasmid pIM40 is pT7Blue (R) with the structural gene for lipase (GenBank M12715) from S6C amplified with primers 5′-CCATGCACTTGGTTGTTGTG and 5′-TTCAACACTGTGAGATGCTAAC. Plasmid pIM42 is pT7Blue (R) containing DNA amplified from the P3 operon of RN6390 with primers 5′-CATGACTAAACATAGATTTATGAG and 5′-CACAGAGATGTGATGGAAAATAG.
Staphylococcal transduction, transformation, and allele exchange.
Bacteriophage 80α lysates were obtained from infected strains growing in top agar (TSB, 0.5 mM CaCl2, 0.5% agar), sterilized by passage through 0.2-μm-pore-size filters, and titered on S. aureus 8325-4. Transduction of erythromycin from S. aureus DU1090, KSI9051, or KSI905 to KSI2300, KSI2111, or KSI2401 through KSI2403 involved the use of 5 × 1010 CFU of exponentially growing bacteria per ml of TSB containing 0.5 mM CaCl2 and 5 × 109 PFU of the bacteriophage (multiplicity of infection = 0.1) per ml in a total of 0.6 ml. After 5 min at room temperature, 1.5 ml of TSB containing 0.5 mM CaCl2 was added, and tubes were incubated for 20 min at 37°C. Following the addition of 1 ml of 0.2 mM sodium citrate, the cells were harvested by centrifugation at 4 × 103 × g for 20 min, suspended in 1 ml of 0.2 mM sodium citrate, and plated on TSA supplemented with 2 mM sodium citrate and the appropriate antibiotic or modified chemically defined synthetic medium. Transductional frequencies, when reported, were based on scores of 100 colonies.
Transformation of S. aureus was by the electroporation procedure described by Kraemer and Iandolo (23). Plasmid DNA initially isolated from E. coli was introduced into S. aureus RN4220 prior to transformation of strain KSI9051 or KSI2401 through KSI2403. For allele replacement experiments, we used plasmid pIM34, pIM35, or pIM54. The conditions for plasmid integration and cointegrate resolution have been described in detail by Janzon and Arvidson (18).
SDS-polyacrylamide gel electrophoresis (PAGE) analysis.
Filtered culture supernatant fluids from bacterial cultures grown to an optical density at 540 nm of 5 were concentrated 40-fold by lyophilization, and 1.5 μl of the material was subjected to electrophoresis through a 4% stacking, 10 to 20% separating Tris-glycine gel (Bio-Rad, Hercules, Calif.) as described by the manufacturer. Broad-range weight standards were purchased from Bio-Rad. Protein bands were visualized following staining with Coomassie blue.
DNA and RNA hybridizations.
Staphylococcal chromosomal DNA was digested with restriction endonucleases. The digested DNA was subjected to electrophoresis through 0.7% agarose gels and transferred to nylon membranes (MagnaGraph; Fisher Scientific, Pittsburgh, Pa.) and probed with the Genius system (Boehringer Mannheim, Indianapolis, Ind.) as instructed by the manufacturer. Hybridization was done with randomly primed digoxigenin-labeled probes and the standard buffer plus 50% formamide for prehybridization and hybridization, and stringent washes were performed at 68°C. Detection was done with the chemiluminescent substrate disodium 3-{4-methoxyspiro[1,2-dioxatane-3,2′-tricyclo-(3.3.1.13,7)decan]-4yl}phenyl phosphate (AMPPD).
Total cellular RNA was isolated by the method of Hart et al. (14) and purified with RNAeasy (Qiagen). Electrophoresis of RNA was conducted in 1% SeaKem LE agarose-glyoxal gels. The RNA was transferred to a nylon membrane (MagnaGraph) and probed with the Genius system. Probes included a 718-bp ClaI fragment internal to lipase from pIM40, an ≈2-kb AatII-SmaI fragment encoding most of protein A from pRIT5 (Pharmacia Biotech, Piscataway, N.J.), and a ClaI-XbaI fragment from pIM42 encoding part of RNAIII. The probes were digoxigenin labeled and hybridized with high-SDS buffer at 50°C. Stringent washes were performed at 65°C, and detection was done with AMPPD.
Sequencing.
The nucleotide sequence for the 3′ end of agrB, agrD, and agrC and the 5′ end of agrA were determined for plasmids pIM29, pIM31, pIM32, pIN62, and pRN6650. In addition, the entire inserts from plasmids pIM49, pIM50, and pIM51 were sequenced. Sequence data were obtained with an Applied Biosystems 373A or 377 DNA sequencer with dye terminator cycle sequencing chemistry (Perkin-Elmer, Foster City, Calif.) on Qiagen or CsCl2-purified plasmid templates. Sequencing primers are listed in Table 2. For plasmids pIM29 through pIM32 and pRN6650, the insert sequence was determined for both strands up to base 1690. The remaining sequence was verified with only the forward primer within agrA. The region of conflict in pRN6650, positions 3555 to 3011 (GenBank X52543), was sequenced six times with the three different flanking primers (26, 31). Plasmids pIM49 through pIM51 were sequenced in both directions with primers within the T7 and reverse primers and the primers within agrC. Data obtained from cloned PCR products were confirmed by sequencing two independently amplified templates that were purified with QIAquick (Qiagen) and primers adjacent to the suspected mutation. Sequence data were analyzed with Sequencher version 3.0 (Gene Codes, Ann Arbor, Mich.), SeqEd version 1 (Applied Biosystems, Foster City, Calif.), and the Genetics Computer Group (Madison, Wis.) sequence analysis software package version 8.1.
TABLE 2.
Oligonucleotide primers used to sequence agrB′DCA′
| Forward | Reverse |
|---|---|
| GCTCCTGCAGCAACTAAAAAGa | ATAAGTGTGATAATGAAAAGGG |
| TAAARAGAGAGTGTGATAGTAG | ATTAAGGACGCGCTATCAAAC |
| GATTATTCTATACTGTGCTAAC | AGTCACATAAACTACAAAAAAGC |
| AAGAAGAAATTGAAACCTATTATG | TATATTCATAATAGGTTTCAATTTC |
| GCTATAAAATTAAATGGTATCGAG | CTTTTGTTTTGGATCGTCTTC |
| AATACCCGATGAAGTAAGTAG | ACGATACCATTTAATTTTATAGCA |
| CGAAGACGATCCAAAACAAAG |
Primers are written 5′ to 3′.
RESULTS
Cotransduction of Tn551::Δ1058 and Lys from S. aureus KSI9051 to S. aureus ISP32.
Mutations in sar and agr have been described in the defined genetic background of S. aureus 8325-4 (7, 30). Transduction of erythromycin resistance (Emr) encoded by Tn551 resident in KSI9051 to 8325-4 results in the generation of an extracellular-protein-deficient phenotype. When the location of the transposon in these strains was examined by Southern analysis, multiple sites of insertion were found (data not shown). Given this lack of success, we cotransduced Δ1058::Tn551 and a nearby prototrophic marker into a derivative of the parental strain of 8425-4.
The Δ1058::Tn551 mutation in KSI9051 was introduced into the lysine auxotroph ISP32 by transduction. When either erythromycin resistance (Emr) or the ability to synthesize lysine (Lys+) was selected for, 98 or 97%, respectively, of each transductant class was converted to the nonselected phenotype, confirming the linkage of Δ1058::Tn551 with lysine prototrophy. Cotransductants generated by selecting for both an Emr and an Lys+ phenotype produced α-toxin as indicated by the cross-streaking of the transductants against S. aureus DU1090, ISP2094, and KSI2300. This result was confirmed by assaying culture supernatant fluids from ISP32 and the five cotransductants for the ability to lyse rabbit erythrocytes. Each of the transductant strains produced 1,100 ± 150 (mean ± standard deviation) U of activity, which was equal to that seen in the recipient strain. The cotransductants and ISP32 generated approximately ≈0.34 U of lipase activity compared to an undetectable level in KSI9051 and 1.4 U of activity for S. aureus S6C. Further phenotypic examination of the cotransductants on indicator plates showed that β-toxin, protease, and nuclease activities were similar to those for ISP32 (data not shown). The transfer of Δ1058::Tn551 from KSI9051 to ISP32 was confirmed in the Emr Lys+ transductants by demonstration of the predicted ≈12.6-kb EcoRI fragment that hybridizes to the insert from pIM36 (data not shown; 34).
Phenotypic restoration of S. aureus KSI9051.
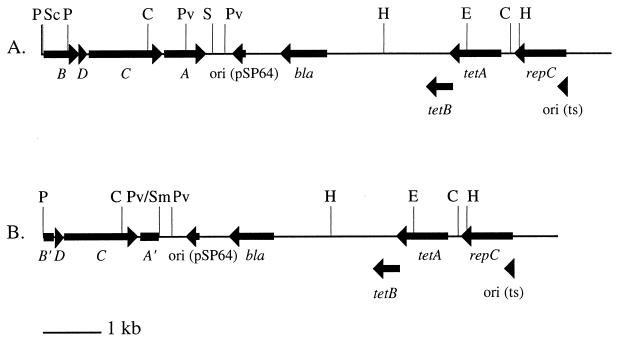
Reports of spontaneous mutations occurring within agr (4, 18, 30) and the similarity between transcript and protein profiles from KSI9051 and those of agr mutant strains (14, 36) suggested that KSI9051 may contain a mutation within agr. We attempted to restore a wild-type phenotype to KSI9051 by allele replacement with plasmids pIM34 and pIM35. Plasmid pIM34 (Fig. 1A) contains the entire P2 operon (agrBDCA) cloned into pSPT181, a shuttle vector containing a temperature-sensitive staphylococcal replicon. Unlike the vector alone, allele replacement with agrBDCA resulted in the isolation of colonies that produced α-toxin, staphylokinase, and lipase (data not shown). Similar colonies were obtained with pIM35, pSPT181 carrying the 3′ end of agrB, all of agrD and agrC, and the 5′ end of agrA (agrB′DCA′) (Fig. 1B). Resolution of plasmid cointegrates was determined by Southern analysis of strains created by allele replacement with pIM35. The expected pattern of hybridizing EcoRV DNA fragments, approximately 4.2, 1.0, and 0.24 kb in size (data not shown), was found and corresponds to those seen in S. aureus S6C and those predicted from the sequence of agr (22, 34).
FIG. 1.
Maps of complementing plasmids pIM34 (A) and pIM35 (B). The locations of relevant restriction sites (C, ClaI; E, EcoRV; H, HindIII; P, PstI; Pv, PvuI; S, SalI; Sc, SacI; Sm, SmaI), genes (B, agrB; D, agrD; C, agrC; A, agrA; B′, truncated agrB; A′, truncated agrA), and origins of replication are shown. Details of the construction are described in Materials and Methods.
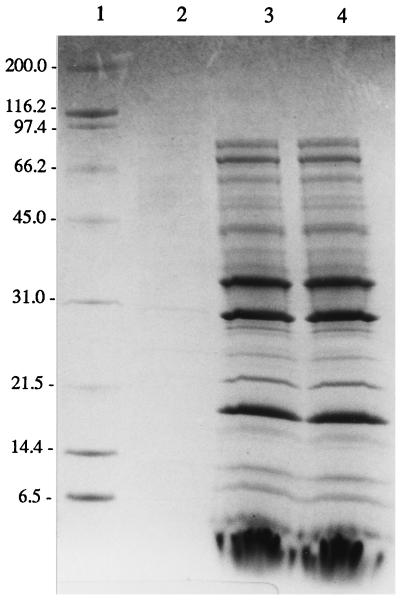
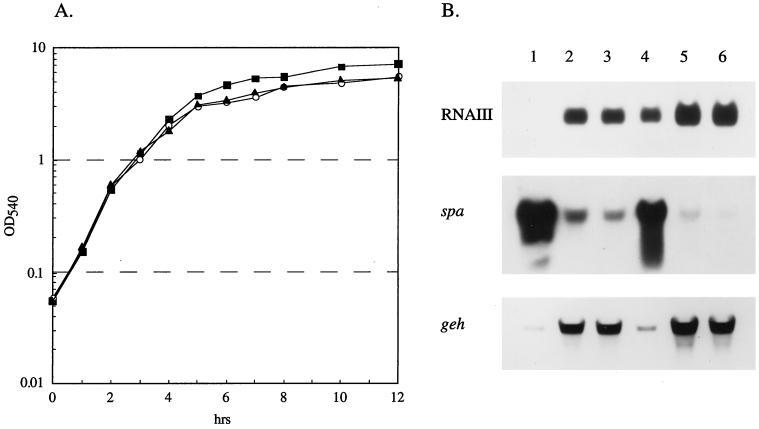
S. aureus KSI2121 was further characterized by examining total extracellular protein production, α-toxin, and lipase activity, and the transcript levels of the lipase, protein A, and RNAIII. SDS-PAGE analysis of equilibrated culture supernatant fluids from strains KSI9051, KSI2121, and S6C revealed that while KSI9051 does not produce an appreciable amount of extracellular proteins, KSI2121 and S6C have indistinguishable protein profiles (Fig. 2). Hemolytic and enzymatic activities are consistent with this result, with both producing 3,540 ± 200 U of α-toxin and ≈0.98 U of lipase activity. Northern analysis of select messages from KSI2121 showed the return of an S6C-like level and pattern of transcription (Fig. 3).
FIG. 2.
SDS-PAGE analysis of cell-free culture supernatant fluids from S. aureus KSI9051 (lane 2), KSI2110 (lane 3), and S6C (lane 4) stained with Coomassie blue. Molecular mass standards (in kilodaltons) are in lane 1.
FIG. 3.
Northern analysis of strains KSI9051, KSI2121, and S6C. (A) Growth curves of S. aureus KSI9051 (▴), KSI2121 (○), and S6C (▪) indicate the growth phase during which RNA was isolated. OD540, optical density at 540 nm. (B) RNA probed with DNA encoding protein A (spa) and lipase (geh) and RNA from the P3 (RNAIII) operon of agr. Each lane contains 10 μg of total cellular RNA isolated at either 3 h (lanes 1 to 3) or 12 h (lanes 4 to 6). Samples from strains KSI9051 (lanes 1 and 4), KSI2121 (lanes 2 and 5), and S6C (lanes 3 and 6) demonstrate the return of reciprocal regulation of surface and extracellular proteins and levels of agr transcripts to KSI2121.
Sequence analysis of agrB′DCA′ from S. aureus KSI9051, S6C, and KSI2121 and plasmid pRN6650.
The nucleotide sequence and a conceptual translation of the agrB′DCA′ region from KSI9051 (GenBank U85095), S6C (GenBank U85096), and KSI2121 were determined. A comparison of this region from strains KSI9051 and S6C revealed four differences in the nucleotide sequence. The first was a frameshift mutation in KSI9051 caused by the insertion of a seventh thymidine residue in a string of six thymidines at positions 469 to 474. Upon translation, the frameshift would alter 12 contiguous amino acids, beginning at amino acid 39, before truncating AgrC at amino acid 51. Other mutations specific to the cloned PCR product included two thymidine-to-cytidine transitions at positions 622 and 1783 and a thymidine-to-adenosine transversion at position 1133. As expected, the sequence of agrC from KSI2121 corresponds to that from S6C, indicating that the mutations found in KSI9051 are corrected upon allele replacement (data not shown).
In a comparison of agrC from S. aureus S6C and RN4282 (GenBank X52543), seven differences were noted. Six of the conflicts are additional thymidine residues in strings of thymidine residues that begin at positions 3555, 3058, 3078, 3095, 3098, and 3011 of the RN4282 sequence. These bases are all within AgrC and, upon translation, result in nine substituted and two added amino acids. The seventh difference is a single base deletion in a string of thymidine residues that begins at position 3761 of the RN4282 sequence. This deletion is predicted to alter four amino acids before truncation of the AgrC five amino acids before the predicted stop in the S6C allele. We determined the sequence of agrC from plasmid pRN6650 (GenBank U85097), which is Novick and coworkers’ original agr clone. Its sequence is identical to agrC from S6C, suggesting that the reported differences are errors in the GenBank entry. Finally, the sequence of agrC from S. aureus 8325-4 was determined from plasmid pIM29. Sequence from this plasmid was found to be identical to that obtained from strain S6C (data not shown). Blast searches with the AgrC from S6C identify, in addition to the histidine kinase sensor proteins previously reported by Novick et al. (27), similarities to BlaR1, the sensor component of two β-lactamase operons (Fig. 4).
FIG. 4.
Blast alignment of AgrC from S6C and BlaR1 from Tn552 (PIR S34445) showing amino acid identity (|) and similarity (+) over the range presented. The underlined sequence represents corrected amino acids in AgrC.
Phenotypic restoration and sequence analysis of S. aureus KSI2401, KSI2402, and KSI2403.
Extracellular-protein-deficient strains were independently generated by the transduction of erythromycin resistance from S. aureus KSI905 to S6C. Between 43 and 48% of the resulting transductants were α-toxin- and protease-deficient mutants. Three mutants, one from each transduction, were chosen for further study. They were phenotypically rescued for the missing enzymatic activities by allele replacement with pIM54 (data not shown), and the nucleotide sequence that corresponds to the insert from pIM54 was determined for each isolate.
Sequence data from three strains revealed that the isolates had mutations only within agrC. Here nucleotide positions refer to the insert sequence, and numbered amino acids refer to positions within AgrC. Strain KSI2401 (GenBank AF026120) has a thymidine-to-cytidine transition at position 483. Upon translation, this first mutation is predicted to substitute proline for serine-18. The second isolate, KSI2402 (GenBank AF026121), has two adenine-to-guanidine transitions, at positions 294 and 1245, that are expected to substitute alanine for threonine-15 and glycine for serine-332, respectively. The final isolate, KSI2403 (GenBank AF026122), has a thymidine-to-cytidine transition at position 961 and an adenine-to-thymidine transversion at position 1528. These mutations are respectively predicted to substitute serine for phenylalanine-237 and valine for glutamic acid-426. Additional mutations were found in the cloned PCR products; however, these mutations could not be verified by directly sequencing amplified DNA.
DISCUSSION
We show that the aberrant expression of proteins in S. aureus KSI9051 originally ascribed to a mutation known as xpr is actually due to a frameshift mutation within the gene encoding the sensor component of the global regulator agr. Remarkably, creation of KSI9051 by the transduction of Tn551::Δ1058 from the chromosome of S. aureus KSI905 to its parental strain, S6C, results in the isolation of agr mutants at frequencies approaching 50%. In contrast, the inability to isolate similar mutations in the 8325 genetic background indicates that strain S6 is genetically less stable. Sequence analysis of the 5′ end of agrB, agrD, and agrC and the 5′ end of agrA DNA that restores a wild-type phenotype to three strains isolated in the manner of strain KSI9051 suggests that transduction of erythromycin resistance from KSI905 to strain S6C generates independent mutations in agrC.
The Δ1058::Tn551 mutation is part of a linkage group containing the lysine biosynthetic operon (Lys), allowing the cotransduction of Emr and Lys+ from KSI9051 into ISP32. Emr Lys+ cotransductants were wild type for the production of both α-toxin and lipase, indicating that the Δ1058::Tn551 mutation is not linked to the loss of either phenotype. These results contrast with the association of Δ1058::Tn551 with the loss of lipase activity in KSI905 and the pleiotropic phenotype seen in strain KSI9051 (34, 36).
With agr mutants, trans-complementation has been complicated by both the instability of plasmids containing agr sequences and the ability of stable plasmids to only partially return a wild-type phenotype (27, 30). Additionally, with the possible exception of agrA, RNAII-encoded genes require the P2 promoter for expression (22, 27). To circumvent these problems, allele replacement with plasmid pIM34 or pIM35 was used to screen for a wild-type pattern of protein expression in KSI9051. The mutation responsible for the phenotype of KSI9051 was found to reside within the 3′ end of agrB, agrD, or agrC, or the 5′ end of agrA (agrB′DCA′). This is clearly indicated by the fact that DNA from the P2 operon of agr returns a S6C-like pattern of proteins to strain KSI2121. Many agr-regulated extracellular proteins are known only as bands on stained gels, and the data in Fig. 2 are consistent with previously published results (31). Northern analysis confirmed phenotypic restoration of protein expression in strain KSI2121 and corresponded with the return of growth-phase-dependent transcription of mRNA for both a negatively regulated surface protein and positively regulated extracellular protein. Unlike in the agrA or agr-null mutants, RNAIII transcription is evident in KSI9051 in the postexponential phase of growth (18, 38). These data are consistent with previous findings and may be due to the nature of the agr mutation or may represent differences between the genetic backgrounds of S. aureus 8325-4 and S6 (14).
To determine the location of the mutation within KSI9051, we sequenced agrB′DCA′ from strains S6C, KSI9051, and KSI2121. The sequence data suggest that a frameshift mutation within agrC, the sensor component of the agr system, is responsible for the mutant phenotype of KSI9051. The truncated allele of agrC is not predicted to be functional, since conserved regions found in bacterial sensor proteins are not expected to be present. A requirement for a functional agrC gene product for the agr response has previously been inferred from complementation studies (27).
In a comparison of the sequence of agrC from RN4282—the wild-type locus described by Novick et al. (27)—and strain S6C, several differences are evident. The sequence of agrC from RN4282 (GenBank X52543) could not be verified in plasmid pRN6650, suggesting that the reported differences are errors in the GenBank entry (27, 32). The sequences of agrC from S. aureus 8325, 8325-4, and S6C are all identical, suggesting that the sequence represents the wild-type allele of this gene. A Blast search with our wild-type AgrC sequence identifies similarity to BlaR1 in addition to the histidine kinase sensor proteins previously described by Novick et al. (27). BlaR1 is the sensor protein encoded by the penicillinase operons from S. aureus and Bacillus licheniformis (21, 33). Like AgrC, BlaR1 is a membrane-bound sensor protein that acts as a positive activator of transcription. Two of the identified regions are presumptive transmembrane domains, and while the function of the third region is unknown, it is flanked by conserved amino acids that define the active site of the penicillin-binding proteins (33).
To demonstrate that agr is an unstable locus, we re-created strain KSI9051 and sequenced three independently derived isolates. S. aureus KSI2401 through KSI2403 were generated by the transduction of erythromycin resistance from Δ1058::Tn551 in KSI905 back to S6C. Screening for loss of α-toxin and protease activities, we confirmed that extracellular-toxin-deficient strains could be obtained at frequencies approaching 50%. Why the transduction of Δ1058::Tn551, a marker that is unlinked to agr, produces agrC mutants remains unknown. In general, the transduction of Tn551 or other chromosomal markers does not generate mutations in agrC. Moreover, with strain KSI905, extracellular-protein-deficient mutants are created at frequencies of approximately 0.6% when a chromosomal copy of a tetracycline resistance marker at the bacteriophage φ11 att site or the chloramphenicol-resistant plasmid pC194 is transduced to strain S6C (data not shown). The generation of agrC mutants by the transduction of Δ1058::Tn551 appears to be specific to the S6 genetic background, since similar experiments with KSI2111 as the donor and ISP32 as the recipient strains did not produce extracellular-protein-deficient transductants (data not shown).
A wild-type phenotype could be restored to strains KSI2401 through KSI2403 with DNA that included agrB′DCA′ from RN6390 (data not shown). This region was cloned from the three strains, and the sequence of the insert DNA was determined. Each of the three strains had point mutations within agrC. Only the agrC sequence from each of the strains differed, confirming that this gene varies at a high frequency. Recently, the sequences of agrC and the agrBD complex have been shown to diverge in staphylococci from different phage groups (20). Despite this divergence, the receptor-ligand interaction between AgrC and AgrD is maintained among each phage grouping. Our data suggest that the differences in the agr loci may involve a hypervariability-generating mechanism, rather than a cassette-switching model.
ACKNOWLEDGMENTS
We thank Allen Gies and Caroline Thompson for technical support in sequencing and Duane Kerr for photographic services. We also thank Mark Smeltzer for helpful discussions and George Stewart for discussing this work, critical reading of the manuscript, and the gift of plasmid pKUS535.
This work was supported by grant no. AI17474 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Abdelnour A, Arvidson S, Bremell T, Ryden C, Tarkowski A. The accessory gene regulator (agr) controls Staphylococcus aureusvirulence in a murine model. Infect Immun. 1993;61:3879–3895. doi: 10.1128/iai.61.9.3879-3885.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1996. [Google Scholar]
- 3.Bayer M G, Heinrichs J H, Cheung A L. The molecular architecture of the sar locus in Staphylococcus aureus. J Bacteriol. 1996;178:4563–4570. doi: 10.1128/jb.178.15.4563-4570.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Björklind A, Arvidson S. Mutants of Staphylococcus aureus affected in the regulation of exoprotein synthesis. FEMS Microbiol Lett. 1980;7:203–206. [Google Scholar]
- 5.Cheung A L, Koomey J M, Butler C A, Projan S J, Fischetti V A. Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc Natl Acad Sci USA. 1992;89:6462–6466. doi: 10.1073/pnas.89.14.6462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cheung A L, Eberhardt K J, Chung E, Yeaman M R, Sullam P M, Ramos M, Bayer A S. Diminished virulence of a sar−/agr− mutant of Staphylococcus aureusin the rabbit model of endocarditis. J Clin Invest. 1994;94:1815–1822. doi: 10.1172/JCI117530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cheung A L, Projan S J. Cloning and sequencing of sarA of Staphylococcus aureus, a gene required for the expression of agr. J Bacteriol. 1994;176:4168–4172. doi: 10.1128/jb.176.13.4168-4172.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheung A L, Woltz C, Yeaman M R, Bayer A S. Insertional inactivation of a chromosomal locus that modulates expression of potential virulence determinants in Staphylococcus aureus. J Bacteriol. 1995;177:3220–3226. doi: 10.1128/jb.177.11.3220-3226.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cheung A L, Eberhardt K, Heinrichs J H. Regulation of protein A synthesis by the sar and agr loci of Staphylococcus aureus. Infect Immun. 1997;65:2243–2249. doi: 10.1128/iai.65.6.2243-2249.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Coen D. Enzymatic amplification of DNA by PCR: standard procedures and optimization. In: Ausubel F A, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1992. pp. 15.1.1–15.1.7. [Google Scholar]
- 11.Dyer D W, Iandolo J J. Rapid isolation of DNA from Staphylococcus aureus. Appl Environ Microbiol. 1983;46:283–285. doi: 10.1128/aem.46.1.283-285.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elek S, Levy E. Distribution of haemolysin in pathogenic and non-pathogenic staphylococci. J Pathol Bacteriol. 1950;62:541–554. doi: 10.1002/path.1700620405. [DOI] [PubMed] [Google Scholar]
- 13.Gillaspy A F, Hickmon S G, Skinner R A, Thomas J R, Nelson C L, Smeltzer M S. Role of the accessory gene regulator (agr) in pathogenesis of staphylococcal osteomyelitis. Infect Immun. 1995;63:3373–3380. doi: 10.1128/iai.63.9.3373-3380.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hart M E, Smeltzer M S, Iandolo J J. The extracellular protein regulator (xpr) affects exoprotein and agr mRNA levels in Staphylococcus aureus. J Bacteriol. 1993;175:7875–7879. doi: 10.1128/jb.175.24.7875-7879.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heinrichs J, Bayer M, Cheung A. Characterization of the sar locus and its interaction with agr in Staphylococcus aureus. J Bacteriol. 1996;178:418–423. doi: 10.1128/jb.178.2.418-423.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iandolo J J. Genetic analysis of extracellular toxins of Staphylococcus aureus. Annu Rev Microbiol. 1989;43:375–402. doi: 10.1146/annurev.mi.43.100189.002111. [DOI] [PubMed] [Google Scholar]
- 17.Iandolo J J, Bannantine J P, Stewart G C. Genetic and physical map of the chromosome of Staphylococcus aureus. In: Crossley K B, Archer G L, editors. The staphylococci in human disease. New York, N.Y: Churchill Livingstone; 1997. pp. 39–53. [Google Scholar]
- 18.Janzon L, Arvidson S. The role of the delta-lysin gene (hld) in the regulation of virulence genes by the accessory gene regulator (agr) in Staphylococcus aureus. EMBO J. 1990;9:1391–1399. doi: 10.1002/j.1460-2075.1990.tb08254.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ji G, Beavis R C, Novick R P. Cell density control of staphylococcal virulence mediated by an octapeptide pheromone. Proc Natl Acad Sci USA. 1995;92:12055–12059. doi: 10.1073/pnas.92.26.12055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ji G, Beavis R C, Novick R P. Bacterial interference caused by autoinducing peptide variants. Science. 1997;276:2027–2030. doi: 10.1126/science.276.5321.2027. [DOI] [PubMed] [Google Scholar]
- 21.Kobayashi T, Zhu Y F, Nicholls N J, Lampen J O. A second regulatory gene, blaR1, encoding a potential penicillin-binding protein required for induction of beta-lactamase in Bacillus licheniformis. J Bacteriol. 1987;169:3873–3878. doi: 10.1128/jb.169.9.3873-3878.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kornblum J, Kreiswirth N, Projan S J, Ross H, Novick R P. agr: a polycistronic locus regulating exoprotein synthesis in Staphylococcus aureus. In: Novick R P, editor. Molecular biology of the staphylococci. New York, N.Y: VCTL Publishers; 1990. pp. 373–402. [Google Scholar]
- 23.Kraemer G R, Iandolo J J. High-frequency transformation of Staphylococcus aureusby electroporation. Curr Microbiol. 1990;21:373–376. [Google Scholar]
- 24.Morfeldt E, Tegmark K, Arvidson S. Transcriptional control of the agr-dependent virulence gene regulator, RNAIII, in Staphylococcus aureus. Mol Microbiol. 1996;21:1227–1237. doi: 10.1046/j.1365-2958.1996.751447.x. [DOI] [PubMed] [Google Scholar]
- 25.Novick R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967;33:155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 26.Novick R P, Ross H F, Projan S J, Kornblum J, Kreiswirth B, Moghazeh S. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 1993;12:3967–3975. doi: 10.1002/j.1460-2075.1993.tb06074.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Novick R P, Projan S J, Kornblum J, Ross H F, Ji G, Kreiswirth B, Vandenesch F, Moghazeh S. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet. 1995;248:446–458. doi: 10.1007/BF02191645. [DOI] [PubMed] [Google Scholar]
- 28.Pattee P A. Chromosomal map location of the alpha-hemolysin structural gene in Staphylococcus aureus. Infect Immun. 1986;54:593–596. doi: 10.1128/iai.54.2.593-596.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pattee P A, Neveln D S. Transformation analysis of three linkage groups in Staphylococcus aureus. J Bacteriol. 1975;124:201–211. doi: 10.1128/jb.124.1.201-211.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peng H-L, Novick R P, Kreiswirth B, Kornblum J, Schlievert P. Cloning, characterization, and sequence of an accessory gene regulator (agr) in Staphylococcus aureus. J Bacteriol. 1988;170:4365–4372. doi: 10.1128/jb.170.9.4365-4372.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Recsei P, Kreiswirth B, O’Reilly M, Schlievert P, Gross A, Novick R P. Regulation of exoprotein expression in Staphylococcus aureus by agr. Mol Gen Genet. 1986;202:58–61. doi: 10.1007/BF00330517. [DOI] [PubMed] [Google Scholar]
- 32.Regassa L B, Novick R P, Betley M J. Glucose and nonmaintained pH decrease expression of the accessory gene regulator (agr) in Staphylococcus aureus. Infect Immun. 1992;60:3381–3388. doi: 10.1128/iai.60.8.3381-3388.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rowland S-J, Dyke G H. Tn552, a novel transposable element from Staphylococcus aureus. Mol Microbiol. 1990;4:961–975. doi: 10.1111/j.1365-2958.1990.tb00669.x. [DOI] [PubMed] [Google Scholar]
- 34.Smeltzer M S, Gill R, Iandolo J J. Localization of a chromosomal mutation affecting expression of extracellular lipase in Staphylococcus aureus. J Bacteriol. 1992;174:4000–4006. doi: 10.1128/jb.174.12.4000-4006.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Smeltzer M S, Hart M E, Iandolo J J. Quantitative spectrophotometric assay for staphylococcal lipase. Appl Environ Microbiol. 1992;58:2815–2819. doi: 10.1128/aem.58.9.2815-2819.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smeltzer M S, Hart M E, Iandolo J J. Phenotypic characterization of xpr, a global regulator of extracellular virulence factors in Staphylococcus aureus. Infect Immun. 1993;61:919–925. doi: 10.1128/iai.61.3.919-925.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smibert R M, Krieg N R. General characterization. In: Kreig N, editor. Manual of methods for general bacteriology. Washington, D.C: American Society for Microbiology; 1981. p. 435. [Google Scholar]
- 38.Vandenesch F, Kornblum J, Novick R P. A temporal signal, independent of agr, is required for hla but not spa transcription in Staphylococcus aureus. J Bacteriol. 1991;173:6313–6320. doi: 10.1128/jb.173.20.6313-6320.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]