Abstract
The COVID-19 pandemic, caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has infected more than 160 million people worldwide. Researchers have targeted the SARS-CoV-2 structural proteins to better combat the pandemic. Of the four structural proteins, spike (S), membrane (M), envelope (E) and nucleocapsid (N), the S, M and E proteins are glycosylated whereas the N protein is phosphorylated. The glycosylation of the S protein has been reported previously by multiple research groups, and this knowledge has assisted the pharmaceutical industry in developing vaccines and treatment options. In the United States, there are currently three approved COVID-19 vaccines. All three of these vaccines use the S protein to teach host cells how to react when SARS-CoV-2 particles are present. Treatment options utilizing antivirals and immunosuppressants are being developed in addition to vaccines. Different treatment approaches are needed based on the severity of COVID-19 infection. The therapeutic options currently available are not derived through the direct knowledge on SARS-CoV-2 glycosylation. However, more research on the glycosylation of the structural proteins and how this effects SARS-CoV-2 and host cell binding could lead to new and more effective therapeutics. Herein we outline the current vaccine and therapeutic options against COVID-19 available to the public, as well as those still in development.
Introduction
The pharmaceutical industry has a key role in the development of vaccines and therapeutics, which became even more critical after the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in late 2019 and quickly caused a global pandemic. SARS-CoV-2 infection results in COVID-19, a respiratory disease that can lead to severe illness and death.1,2 The COVID-19 pandemic demanded a rapid re-direction of pharmaceutical research and development efforts towards the development of vaccines as well as drugs. Additionally, the scientific community expended great effort in compiling data regarding the virus and COVID-19 to accelerate time to market during this worldwide public health crisis.
To combat the virus, scientists have studied the structural proteins of SARS-CoV-2 and host cell receptors that assist in viral entry to host cells. SARS-CoV-2 has four key structural proteins: spike (S), membrane (M), envelope (E) and nucleocapsid (N) proteins (Figure 1). The S protein is highly glycosylated with 22 potential glycosylation sites. In silico experiments paired with knowledge about the glycosylation of previous human coronaviruses support that both the M and E proteins are likely N-glycosylated.3–9 The M protein has eight N-glycosylation sites, six of which are in common with the M protein of SARS-CoV-1. The M protein is utilized to evade host cell immune response and is important for the coronavirus budding process.10 The E protein has two possible N-glycosylation sites. The E protein induces retention of the S protein, and both the E and M proteins regulate intracellular trafficking and processing of the S protein.11 The N protein is located in the nucleocapsid of the virus and does not enter the secretory pathway like the other structural proteins. Therefore, the native N protein has been found to be phosphorylated but not glycosylated.12 Functionally, the N protein facilitates entering the host cell, binding to viral RNA genome and forms the ribonucleoprotein core.13,14
Figure 1.
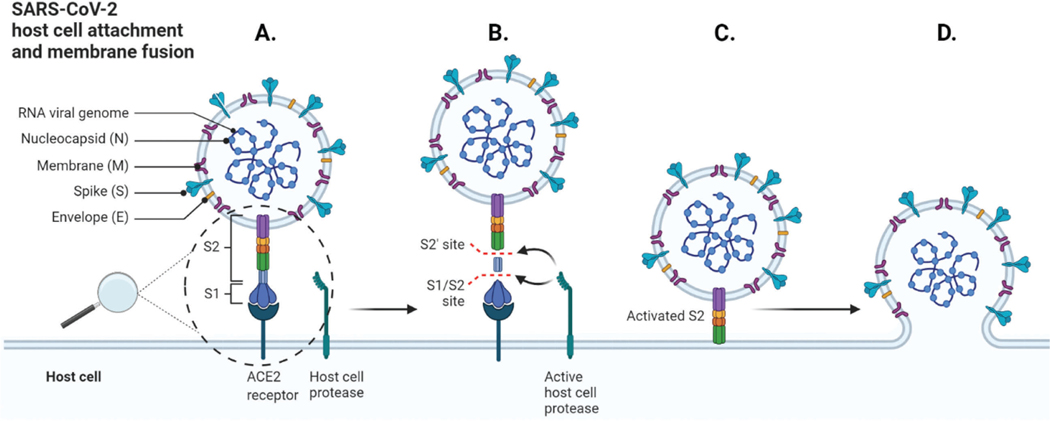
Mechanisms for SARS-CoV-2 attachment to the host cell leading to membrane fusion between the viral membrane with the host cell membrane. A. Attachment of a viral SARS-CoV-2 particle via the S1 subunit. B. Cleavage of SARS-CoV-2 S protein. C. Activation of the S2 domain and attachment to the host cell membrane. D. Fusion of viral and host membranes, which releases the viral components into the host cell and leads to infection. (created with BioRender.com).
The viral S protein has been studied in great detail given its key role; it binds to host cell receptors, and becomes enzymatically activated by host proteases resulting in viral fusion and endocytosis.15–17 The S protein is highly glycosylated, and attaches to human angiotensin-converting enzyme 2 (hACE2) and cell surface glycosaminoglycans (GAGs), which are independent processes and, in combination, facilitate the viral attachment to the host cell surface (Figure 1).18–25 All four structural proteins are ideally co-expressed for the efficient generation of virus-like particles (VLPs). VLPs are nanostructures that resemble the structure of the virus and are promising tools for studying vaccines.11
Since working with the native virus requires significantly more precautions, e. g. high-cost equipment and work at a higher biosafety level facility, it was difficult to generate this data at the beginning of the pandemic. Therefore, most research has so far been conducted on recombinant SARS-CoV-2 proteins and their glycosylation patterns. For example, recent work by Watanabe et al., Zhang et al., and Shajahan et al. outlined the glycosylation of the S protein, however the three results showed differences.18,21,26 This variance was due to all three research groups using different recombinant versions of the S protein. Manufacturers of the recombinant S protein may use amino acid sequence substitutions to effectively stabilize the recombinant protein, which may result in differing N-glycosylation.18,27 Additionally, the expression of full-length S protein versus its subunits can result in glycosylation diff erences.18,28 Hence, the differences in glycosylation should be considered when developing vaccines and drugs that aim for glycosylated structures of SARS-CoV-2 or its binding targets in the host.
COVID-19 Prevention
To combat the pandemic, there are governmental measures as well as public health advice in place.29 Pharmaceutical interventions are another option, with two categories of medical intervention that are important to consider for infectious diseases: prophylaxis, which is a protective measure or process that is created to guard against the onset or development of a specific disease, and treatment, which supports the clearance of an infectious disease so that the individual’s health is restored.30,31
To date, the vaccines that have been developed against COVID-19 are prophylactic, available to a broad population, and trigger the individual’s immune response to produce antibodies against antigens present on SARS-CoV-2. Antigen targets that have been used for SARS-CoV-2 vaccines are the structural proteins mentioned earlier – mainly the S protein.31–33
In the United States, there are currently three COVID-19 vaccines with an Emergency Use Authorization (EUA) status from the U.S. Food and Drug Administration (FDA). They have been authorized for active immunization against COVID-19, to both reduce the likelihood of an infection and to prevent a severe course of disease in the event an infection with SARS-CoV-2 should occur. These approved vaccines utilize either messenger RNA (mRNA) or viral vectors.33–35
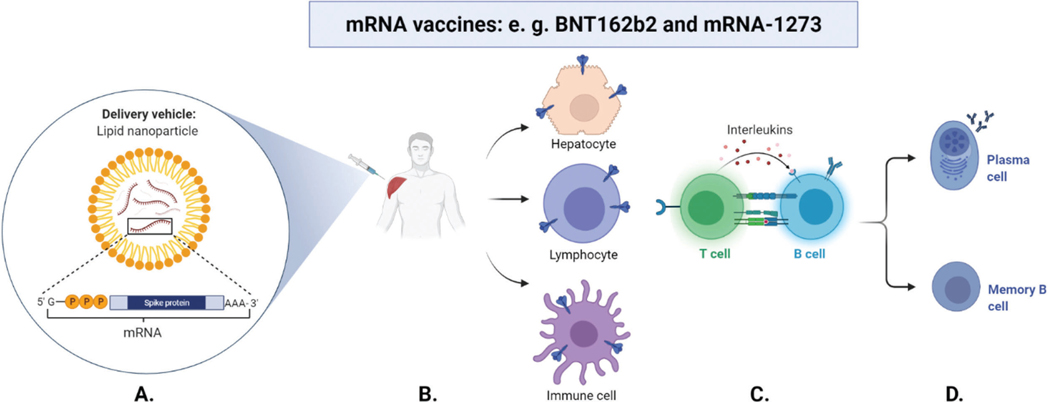
mRNA is a template for protein generation and in the case of mRNA COVID-19 vaccines, they teach cells how to make copies of the SARS-CoV-2 S protein.36 The advantage is that the individual’s cells will produce and glycosylate the S protein exactly how it would happen during an actual infection. This triggers an immune response, which produces antibodies to protect the individual from becoming infected if exposed to the virus later on (Figure 2).32–34, 36–38
Figure 2.
Schematic overview of the working principle of mRNA vaccines to protect against COVID-19. A. The mRNA encoding the S protein is encapsulated in a lipid nanoparticle and injected intramuscular in the deltoid muscle of the shoulder. B. Production of the S protein antigen: The mRNA provides the template to produce the S protein in cells that were reached by lipid nanoparticles, which are likely muscle tissue (site of injection), nearby lymphatic tissue and spleen tissue (with immune cells) as well as liver tissue. C. Activation of the immune response: Antigen (S protein) recognition on cell surfaces induces expression of effector molecules (interleukins) by T cells, which activates B cells, leads to B cell proliferation and subsequent B cell differentiation. D. Antibody production and long-term immunization: B cells differentiate to antibody-secreting plasma cells (antibodies target the S protein) and resting memory B cells, which enable a faster antibody production in future infections. (created with BioRender.com)
BNT162b2, better known as Pfizer-BioNTech COVID-19 vaccine, is an mRNA-based vaccine that encodes the SARS-CoV-2 S protein in full-length and is modified by two proline mutations, which lock the S protein in a conformation right before fusing with the host’s cell.39 Another mRNA vaccine authorized for emergency use in the US is mRNA-1273, better known as Moderna vaccine. Similar to BNT162b2, it encodes the S protein from SARS-CoV-2 in its stabilized prefusion conformation via two proline substitutions.33 Both Pfizer-BioNTech and Moderna vaccines require the administration of two doses, and the effectiveness is determined after receiving both doses (efficacies: BNT162b2 95%, mRNA 1273 94.1%).32,34 Current clinical trials of the mRNA vaccines are based on the patient receiving both vaccine doses. It was found that after the initial dose, a weak immune response is noted within the first few weeks. A much stronger immune response is noted following the second dose of the vaccine.38 A negative side effect to only receiving the first dose is partial immunity, which could result in the emergence of vaccine-resistant variants of SARS-CoV-2, similar to what can occur with improper administration of antibiotics. Additionally, there are no results that indicate long-term immunity to COVID-19 infection with only the initial dose.38
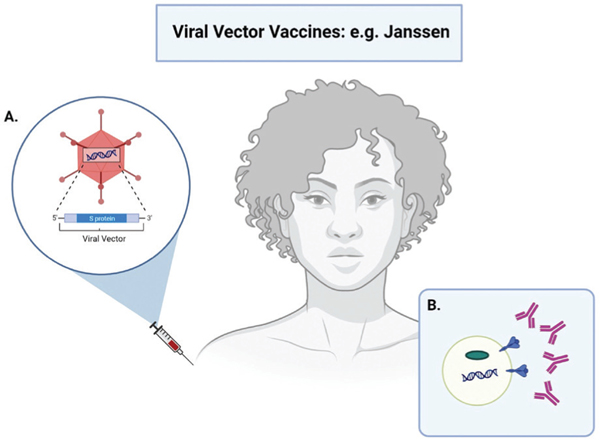
Viral vector vaccines utilize an established viral system, also known as the vector, to deliver instructions to the cell.31,36 The vector is not the SARS-CoV-2 virus, but a different, harmless virus that has been modified by stripping of disease-causing genes. The genetic instructions for producing the S protein of the SARS-CoV-2 virus are then encoded in the vector.40 Once these vectors enter the host cells, the genetic material is converted by the host cell machinery, resulting in host cells displaying the S protein on their surface.37,41 This allows the immune system to recognize the S protein and trigger an immune response and antibody production. Vector vaccines, like mRNA vaccines, cannot result in COVID-19 infection (Figure 3).36,40
Figure 3.
Schematic overview of the working principle of viral vector vaccines to protect against COVID-19. A. A harmless non-coronavirus viral particle known as a vector is modified with the SARS-CoV-2 S protein DNA and injected intramuscular in the deltoid muscle of the shoulder. B. Production of the S protein antigen: the viral vector teaches the host cells to make copies of the S protein. This allows the host cell antibodies to recognize the spike protein if exposed to the virus. The host’s immune response is then similar to that of the mRNA vaccine, and is outlined in the caption of Figure 2. (created with BioRender.com)
The third COVID-19 vaccine that has been authorized under EUA in the United States is JNJ-78436735, manufactured by Johnson & Johnson’s Janssen. It belongs to the viral vector vaccine category and only requires administration of a single dose. The efficacy of preventing COVID-19 has been reported with 66.3% but it has shown that the prevention of severe courses of COVID-19 that required hospitalization was even higher – with estimated efficacies between 93.1% and 100%.35,42
Aside from mRNA and viral vector vaccines, other vaccine types are currently in clinical trials and/or approved in other countries. Unlike viral vector vaccines, attenuated microbes are weakened versions of the virus and able to replicate in vivo which can result in a limited disease. This allows the immune system to induce an immune memory.37 Another option is vaccines based on chemically inactivated viruses. The immune response in this case would be against a multitude of SARS-CoV-2 antigens, and not just the S protein.37 An additional vaccine type is based on the SARS-CoV-2 proteins, specifically the S protein. These vaccines use the S protein or their fragments and adjuvants to elicit an immune response.37 DNA-based vaccines are commonly used in veterinary medicine and currently not used in humans. However, this vaccine type is also being investigated as a potential strategy for SARS-CoV-2 vaccine development in humans. DNA-based vaccines enter the host cell and induce the cell to produce the target protein temporarily. This simulates the production of antibodies.37
COVID-19 Treatment
Whilst prophylaxis is ideally available to a major proportion of individuals at risk, statistically, a smaller percentage of patients with an infectious disease like COVID-19 require additional treatment options due to a severe course. In the early stages of COVID-19, the disease is driven by replication of SARS CoV 2. In the later stages, a dysregulated and heightened inflammatory response leads to tissue damage.43 Hence, antiviral therapies are thought to have the greatest eff ect at early stages in the disease, while pharmacological interventions with an immunosuppressive characteristic should have the greatest effect at later stages in the disease. As a result, the more severe the symptoms of an individual with COVID-19, the more specific treatment options are available, which we outline below.
Orally administered antivirals have characteristics of both prophylaxis and treatment. Antivirals target properties that, ideally, are only presented by the pathogen and not the host, such as the S protein for the coronaviruses. These drugs are administered after a suspected or confirmed exposure to the virus to prevent the onset of the disease or minimize the viral load. Soluble hACE2 in combination with human antibodies are being developed to prevent SARS-CoV-2 infection.44 Soluble ACE2 as a means to prevent acute lung injury was fi rst conceived during the first SARS-CoV-1 epidemics.45 Coronaviruses have evolved to utilize cellular ACE2 for S protein binding and resultant infection. Introducing soluble ACE2 results in the SARS-CoV-2 S protein binding to soluble ACE2, which can then block SARS-CoV-2 from binding to cellular ACE2.45 Additionally, studies have shown that host cell surface heparin (Hp) and heparin sulfate (HS) are crucial for S protein binding.22,24,46 Hp and HS are a class of GAGs with antithrombotic characteristics. GAGs are linear polysaccharides covalently bound to a protein backbone on the surface of cells and in the extracellular matrix.24,47 When testing a variety of GAG and GAG-like structures, heparin, heparin sulfate and fucoidan-like structures were able to compete with cell surface GAGs for SARS-CoV-2 binding with fucoidan-like structures being the most effective.23,24 Hp/HS are highly sulfated, heterogenous polysaccharides with anticoagulant properties. Currently Hp is being utilized to treat thrombosis caused by COVID-19. Fucoidan also has antithrombotic properties, and like Hp/HS, is sulfated.23 Unlike GAGs which are linear, fucoidan can be branched. It is possible that the effectiveness of fucoidan-like structures is due to these branches, which would result in more multivalent interaction possibilities than linear structures.23 These findings still need to be tested in a clinical setting.48
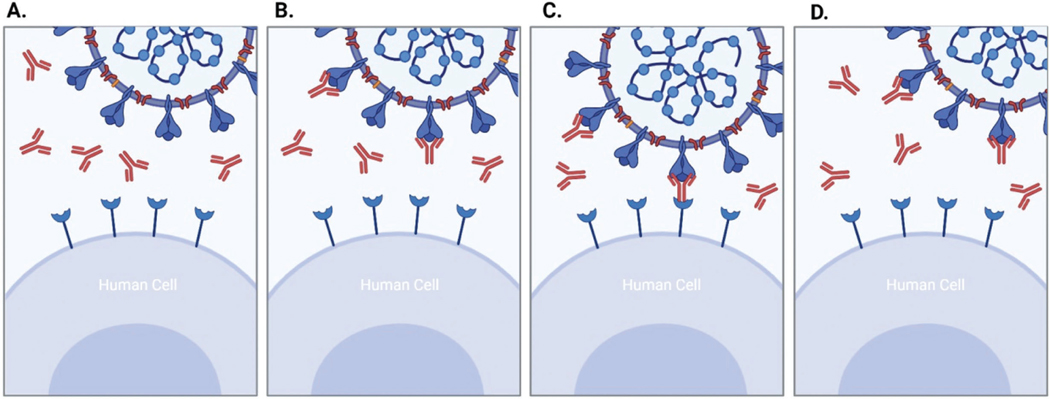
Another option for the treatment of COVID-19 is the administration of neutralizing antibodies (nAbs). They can block interactions between the virus and the host cell receptor and support the clearance of the viruses, similar to antivirals. However, nAbs are only used if the severity of the disease has increased and, therefore, are only available to a smaller number of patients. The FDA has issued EUA for several monoclonal antibody treatments for patients at a higher risk of developing severe COVID-19. Monoclonal antibodies (mAbs) are laboratory-made molecules which respond more effectively to the virus (Figure 4).49 mAbs are highly specifi c and selective, and are often glycosylated in the fragment crystallizable (Fc) region. The glycosylation modulates the binding of the virus to the Fc region of the mAbs.50 Understanding glycosylation patterns of the target virus is crucial for developing effective antibody treatment methods.50
Figure 4.
Mechanisms for SARS-CoV-2 attachment to the host cell leading to membrane fusion between the viral membrane with the host cell membrane. A. Attachment of a viral SARS-CoV-2 particle via the S1 subunit. B. Cleavage of SARS-CoV-2 S protein. C. Activation of the S2 domain and attachment to the host cell membrane. D. Fusion of viral and host membranes, which releases the viral components into the host cell and leads to infection. (created with BioRender.com).
If the patient develops even more severe symptoms of COVID-19 and hospitalization is required, the next level of treatment options includes intravenous antivirals.43 Considering the disease severity, the requirements for licensed personnel to administer drugs intravenously as well as the possibility of more significant adverse effects, fewer patients receive this treatment. Currently, there has been only one intravenous antiviral approved by the FDA for COVID-19 treatment; Remdesivir is an inhibitor of RNA-dependent, RNA polymerase with known inhibitory activity against SARS-CoV-1 and Middle East respiratory syndrome (MERS-CoV).51 Remdesivir treatment results in shorter recovery time for patients with COVID-19 compared to patients who did not receive treatment (10 days versus 15 days respectively). The mortality rate of those treated with Remdesivir also decreased compared to patients who did not receive treatment (6.7% and 11.9% at day 15, respectively).51
On the next level of pharmacological intervention, hospitalized patients with an overreacting immune system also referred to as ‘cytokine storm’, can be treated with immune modulating drugs. Because only a fraction of COVID-19 patients develops this immune reaction, only a very narrow population of patients receives this treatment.
Since the systemic inflammation in the most severe cases of COVID-19 is associated with heightened cytokines, such as interleukins (IL; Figure 2C), the pharmacological modulation of IL levels is considered a promising route for reducing the duration and severity of COVID-19. The FDA has approved treatment with monoclonal antibodies that either bind to the IL itself, such as Interleukin-6 (IL-6), or to the IL receptor, therefore blocking the interaction. The treatment guidelines currently suggest the administration of monoclonal antibodies targeting the IL-6 receptor in combination with dexamethasone.43,52 Dexamethasone is a corticosteroid, which reduces the immune system response to disease, resulting in reduced symptoms. When treating hospitalized COVID-19 patients receiving invasive mechanical ventilation with dexamethasone, there was a decrease in mortality rate; however, this was not seen in cases where a ventilator was not used.52 This indicates that dexamethasone provides benefit to patients with severe COVID-19 symptoms who are in need of respiratory support, but the medication provides no benefit to those not receiving respiratory support.43,52
Outlook and Concluding Remarks
Since the onset of the COVID-19 pandemic, researchers have been working to learn as much as possible about the SARS-CoV-2 virus and the resultant COVID-19 disease to develop preventative and treatment options. Vaccines are the preferred pharmaceutical tool, as they can prevent infection in a broad population. The S protein of SARS-CoV-2 is the focus for the majority of the developed vaccines as it plays a crucial role in SARS-CoV-2 infection of host cells. Currently approved vaccines in the U.S. utilize a template for the S protein to teach host cells temporarily how to generate the protein so that the host’s immune system is prepared should an infection occur. Although currently available vaccines were developed without considering the deeper roles of glycosylation, recent advances in the glycosylation study on SARS-CoV-2 would pave way for the development of more robust therapeutic possibilities.
In addition to preventative measures, treatment options are necessary to help those who either could not receive a vaccine or developed COVID-19 despite vaccination. For early onset COVID-19 cases, treatment with antivirals is effective, whereas for severe and critical COVID-19 cases, an immunosuppressive agent is more effective. ACE2, which is glycosylated, facilitates SARS-CoV-2 viral entry by binding to the viral S protein. The S protein also binds to cell surface GAGs, which further facilitates viral entry to the host cell. Treatment with soluble ACE2 can reduce viral infection, preventing the S protein from binding to cellular ACE2. Similarly, treatment with Hp/HS or fucoidan-like structures can act as competitive binders, preventing the S protein from binding to cell surface GAGs. Majority of SARS-CoV-2 and COVID-19 therapeutics do not require a background knowledge of SARS-CoV-2 glycosylation. However, understanding the glycosylation of the SARS-CoV-2 structural proteins and the host cell receptors could result in more effective therapeutics.
Funding
The US National Institutes of Health (R24GM137782, S10OD018530); U.S. Department of Energy, Office of Science, Basic Energy Sciences (under Award DE-SC0015662 to DOE—Center for Plant and Microbial Complex Carbohydrates at the Complex Carbohydrate Research Center, USA).
Biographies
 Dr. Anne Gleinich received her M.Sc. in Biochemistry in 2014 from the Goethe University Frankfurt am Main, Germany, with a focus on Biophysical Chemistry. The Ph.D. in Medical Sciences was conferred on her by the University of Warwick, United Kingdom, in 2019 and she joined the Analytical Service and Training Laboratory at the Complex Carbohydrate Research Center in the same year. Her research centers around the glycomic and glycoproteomic characterization of N- and O-linked glycans using mass spectrometry techniques (MALDI-MS, ESI-MS and LC-MS), the characterization of composition and structure of biopolymers, such as peptidoglycans via GC-MS and LC-MS, as well as biomolecular interaction/kinetic studies using Surface Plasmon Resonance (SPR). LinkedIn: www.linkedin.com/in/annegleinich
Dr. Anne Gleinich received her M.Sc. in Biochemistry in 2014 from the Goethe University Frankfurt am Main, Germany, with a focus on Biophysical Chemistry. The Ph.D. in Medical Sciences was conferred on her by the University of Warwick, United Kingdom, in 2019 and she joined the Analytical Service and Training Laboratory at the Complex Carbohydrate Research Center in the same year. Her research centers around the glycomic and glycoproteomic characterization of N- and O-linked glycans using mass spectrometry techniques (MALDI-MS, ESI-MS and LC-MS), the characterization of composition and structure of biopolymers, such as peptidoglycans via GC-MS and LC-MS, as well as biomolecular interaction/kinetic studies using Surface Plasmon Resonance (SPR). LinkedIn: www.linkedin.com/in/annegleinich
 Dr. Lauren Pepi received her B.A. (ACS certified) in Chemistry and Biology in 2015 from Assumption University (Worcester, MA) and her Ph.D. in Analytical Chemistry in 2020 from the University of Georgia under the direction of Dr. Jon Amster. Her Ph.D. work focused on fundamental studies of tandem mass spectrometry of glycosaminoglycans. Lauren joined the Analytical Service and Training team at the end of 2020, where her research focuses on glycomic and glycoproteomic characterization of N- and O-glycans using mass spectrometry techniques. Lauren has experience using a wide range of mass spectrometers, including FT-ICR MS, Orbitrap MS, MALDI MS and Ion trap MS. LinkedIn: www.linkedin.com/in/laurenpepi
Dr. Lauren Pepi received her B.A. (ACS certified) in Chemistry and Biology in 2015 from Assumption University (Worcester, MA) and her Ph.D. in Analytical Chemistry in 2020 from the University of Georgia under the direction of Dr. Jon Amster. Her Ph.D. work focused on fundamental studies of tandem mass spectrometry of glycosaminoglycans. Lauren joined the Analytical Service and Training team at the end of 2020, where her research focuses on glycomic and glycoproteomic characterization of N- and O-glycans using mass spectrometry techniques. Lauren has experience using a wide range of mass spectrometers, including FT-ICR MS, Orbitrap MS, MALDI MS and Ion trap MS. LinkedIn: www.linkedin.com/in/laurenpepi
 Dr. Asif Shajahan received his master’s degree in Pharmaceutical Chemistry at Hamdard University, India, and his Ph.D. at the National Institute of Immunology, India in 2014. His doctorate study was in the interface of chemistry and biology, where he synthesized various glycoconjugates for the glycoengineering of brain glycans (across BBB) in mice models. He studied glycoengineering in both in vitro and in vivo models using molecular biological and glycoproteomics techniques. He is now an assistant research scientist at the Analytical Service and Training Laboratory, Complex Carbohydrate Research Center (University of Georgia), as a part of glycoprotein analysis team. His research focuses on method development for glycoprotein characterizations by both glycomics and glycoproteomics. LinkedIn: www.linkedin.com/in/asifklm
Dr. Asif Shajahan received his master’s degree in Pharmaceutical Chemistry at Hamdard University, India, and his Ph.D. at the National Institute of Immunology, India in 2014. His doctorate study was in the interface of chemistry and biology, where he synthesized various glycoconjugates for the glycoengineering of brain glycans (across BBB) in mice models. He studied glycoengineering in both in vitro and in vivo models using molecular biological and glycoproteomics techniques. He is now an assistant research scientist at the Analytical Service and Training Laboratory, Complex Carbohydrate Research Center (University of Georgia), as a part of glycoprotein analysis team. His research focuses on method development for glycoprotein characterizations by both glycomics and glycoproteomics. LinkedIn: www.linkedin.com/in/asifklm
 Dr. Christian Heiss received his B.Sc. in Chemistry in 1991 from the University of Erlangen, Germany, and his Ph.D. in Organic Chemistry in 1999 from the University of Georgia. He serves as the Assistant Technical Director of Analytical Service and Training Laboratory at the Complex Carbohydrate Center and since then has focused on the structural characterization of glycoconjugates and polysaccharides by GC-MS, ESI- and MALDI-MS, and NMR. He has written multiple papers on the analysis of carbohydrates, and established the expansion of the CCRC’s analysis to glycosaminoglycans in 2006. LinkedIn: www.linkedin.com/in/christian-heiss-14781028
Dr. Christian Heiss received his B.Sc. in Chemistry in 1991 from the University of Erlangen, Germany, and his Ph.D. in Organic Chemistry in 1999 from the University of Georgia. He serves as the Assistant Technical Director of Analytical Service and Training Laboratory at the Complex Carbohydrate Center and since then has focused on the structural characterization of glycoconjugates and polysaccharides by GC-MS, ESI- and MALDI-MS, and NMR. He has written multiple papers on the analysis of carbohydrates, and established the expansion of the CCRC’s analysis to glycosaminoglycans in 2006. LinkedIn: www.linkedin.com/in/christian-heiss-14781028
 Dr. Parastoo Azadi received her B.Sc. in Chemistry in 1987 from University of North London, UK and her Ph.D. degree in Biochemistry in 1991 from Imperial College of Science and Technology, University of London, studying structural characterization of carbohydrates and glycoproteins by mass spectrometry under the supervision of Profs. A. Dell and H.R. Morris. Since 2001, Dr. Parastoo Azadi has been the Executive Technical Director of Analytical Service and Training at the Complex Carbohydrate Research Center. The samples submitted for these types of analyses come from academic, government, non-profit organizations, and private companies throughout the United States and internationally. LinkedIn: https://www.linkedin.com/in/parastoo-azadi-57a06133
Dr. Parastoo Azadi received her B.Sc. in Chemistry in 1987 from University of North London, UK and her Ph.D. degree in Biochemistry in 1991 from Imperial College of Science and Technology, University of London, studying structural characterization of carbohydrates and glycoproteins by mass spectrometry under the supervision of Profs. A. Dell and H.R. Morris. Since 2001, Dr. Parastoo Azadi has been the Executive Technical Director of Analytical Service and Training at the Complex Carbohydrate Research Center. The samples submitted for these types of analyses come from academic, government, non-profit organizations, and private companies throughout the United States and internationally. LinkedIn: https://www.linkedin.com/in/parastoo-azadi-57a06133
Footnotes
Conflicts of Interest:
The authors declare no conflict of interest.
References
- 1.Ge HP, Wang XF, Yuan XN, et al. The epidemiology and clinical information about COVID-19. Eur J Clin Microbiol. 2020;39(6):1011–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibson PG, Qin L, Puah SH. COVID-19 acute respiratory distress syndrome (ARDS): clinical features and differences from typical pre-COVID-19 ARDS. Med J Aust. 2020;213(2):54–56 e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duart G, García-Murria MJ, Grau B, et al. SARS-CoV-2 envelope protein topology in eukaryotic membranes. Open biology. 2020;10(9):200209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas S.The structure of the membrane protein of SARS-CoV-2 resembles the sugar transporter semiSWEET. Pathog Dis. 2020;5(1):342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dawood AA. Glycosylation, ligand binding sites and antigenic variations between membrane glycoprotein of COVID-19 and related coronaviruses. Vacunas. 2021;22(1):1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.De Haan CA, De Wit M, Kuo L, et al. The glycosylation status of the murine hepatitis coronavirus M protein affects the interferogenic capacity of the virus in vitro and its ability to replicate in the liver but not the brain. 2003;312(2):395–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Locker JK, Griffiths G, Horzinek M, Rottier PJJoBC. O-glycosylation of the coronavirus M protein. Differential localization of sialyltransferases in N-and O-linked glycosylation. 1992;267(20):14094–14101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oostra M, De Haan C, De Groot R, Rottier PJJov. Glycosylation of the severe acute respiratory syndrome coronavirus triple-spanning membrane proteins 3a and M. 2006;80(5):2326–2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yokomori K, La Monica N, Makino S, Shieh C-K, Lai MMC. Biosynthesis, structure, and biological activities of envelope protein gp65 of murine coronavirus. Virology. 1989;173(2):683–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fu Y-Z, Wang S-Y, Zheng Z-Q, et al. SARS-CoV-2 membrane glycoprotein M antagonizes the MAVS-mediated innate antiviral response. Cellular & Molecular Immunology. 2021;18(3):613–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boson B, Legros V, Zhou B, et al. The SARS-CoV-2 envelope and membrane proteins modulate maturation and retention of the spike protein, allowing assembly of virus-like particles. J Biol Chem. 2021;296:100111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Supekar NT, Shajahan A, Gleinich AS, et al. Variable post-translational modifications of SARS-CoV-2 nucleocapsid protein. Glycobiology. 2021;In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rahman MS, Islam MR, Alam ASMRU, et al. Evolutionary dynamics of SARS-CoV-2 nucleocapsid protein and its consequences. 10.1002/jmv.26626. J Med Virol. 2021;93(4):2177–2195. [DOI] [PubMed] [Google Scholar]
- 14.Zeng W, Liu G, Ma H, et al. Biochemical characterization of SARS-CoV-2 nucleocapsid protein. Biochem Biophys Res Commun. 2020;527(3):618–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morse JS, Lalonde T, Xu S, Liu WR. Learning from the Past: Possible Urgent Prevention and Treatment Options for Severe Acute Respiratory Infections Caused by 2019-nCoV. ChemBioChem. 2020;21(5):730–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhou P, Yang XL, Wang XG, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoffmann M, Kleine-Weber H, Schroeder S, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181(2):271–280 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shajahan A, Supekar NT, Gleinich AS, Azadi P. Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology. 2020;30(12):981–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Watanabe Y, Berndsen ZT, Raghwani J, et al. Vulnerabilities in coronavirus glycan shields despite extensive glycosylation. Nat Commun. 2020;11(1):2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walls AC, Park YJ, Tortorici MA, et al. Structure, Function, and Antigenicity of the SARS-CoV-2 Spike Glycoprotein. Cell. 2020;183(6):1735–1735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Watanabe Y, Allen JD, Wrapp D, McLellan JS, Crispin M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science. 2020;369(6501):330−+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Clausen TM, Sandoval DR, Spliid CB, et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell. 2020;183(4):1043–1057.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kwon PS, Oh H, Kwon S-J, et al. Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro. Cell Discov. 2020;6(1):1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim SY, Jin W, Sood A, et al. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antivir Res. 2020;181:104873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shajahan A, Archer-Hartmann S, Supekar NT, et al. Comprehensive characterization of N- and O- glycosylation of SARS-CoV-2 human receptor angiotensin converting enzyme 2. Glycobiology. 2021;31(4) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang Y, Zhao W, Mao Y, et al. Site-specific N-glycosylation characterization of recombinant SARS-CoV-2 spike proteins. Molecular & Cellular Proteomics. 2021;20:100058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pritchard LK, Vasiljevic S, Ozorowski G, et al. Structural constraints determine the glycosylation of HIV-1 envelope trimers. Cell reports. 2015;11(10):1604–1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shrimal S, Trueman SF, Gilmore R. Extreme C-terminal sites are posttranslocationally glycosylated by the STT3B isoform of the OST. Journal of Cell Biology. 2013;201(1):81–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.World Health Organization. Coronavirus disease (COVID-19) advice for the public. Available at: https://www.who.int/emergencies/diseases/novel-coronavirus-2019/advice-for-public. Accessed May 10, 2021.
- 30.Nolte E.Disease Prevention. In: Heggenhougen HK, ed. International Encyclopedia of Public Health. Academic Press; 2008:222–234. [Google Scholar]
- 31.Centers for Disease Control and Prevention. Different COVID-19 Vaccines. Available at: https://www.cdc.gov/coronavirus/2019-ncov/vaccines/different-vaccines.html. Accessed May 05, 2021.
- 32.Baden LR, El Sahly HM, Essink B, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med. 2021;384(5):403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corbett KS, Edwards DK, Leist SR, et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586(7830):567–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Polack FP, Thomas SJ, Kitchin N, et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med. 2020;383(27):2603–2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sadoff J, Gray G, Vandebosch A, et al. Safety and Efficacy of Single-Dose Ad26.COV2.S Vaccine against Covid-19. N Engl J Med. 2021;In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Centers for Disease Control and Prevention. Understanding Viral Vector COVID-19 Vaccines. Available at: https://www.cdc.gov/coronavirus/2019-ncov/vaccines/different-vaccines/viralvector.html#:~:text=Some%20vaccines%20recently%20used%20for,more%20about%20getting%20your%20vaccine. Accessed April 30, 2020.
- 37.Forni G, Mantovani A, Forni G, et al. COVID-19 vaccines: where we stand and challenges ahead. Cell Death Differ. 2021;28(2):626–639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Livingston EH. Necessity of 2 Doses of the Pfizer and Moderna COVID-19 Vaccines. JAMA. 2021;325(9):898–898. [DOI] [PubMed] [Google Scholar]
- 39.Wrapp D, Wang N, Corbett KS, et al. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020;367(6483):1260–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gavi The Vaccine A. What are viral vector-based vaccines and how could they be used against COVID-19? Available at: https://www.gavi.org/vaccineswork/what-are-viral-vector-basedvaccines-and-how-could-they-be-used-against-covid-19. Accessed May 3, 2021.
- 41.Gavi TVA. What are viral vector-based vaccines and how could they be used against COVID-19? Available at: https://www.gavi.org/vaccineswork/what-are-viral-vector-based-vaccines-and-how-could-they-be-used-against-covid-19 Accessed May 3, 2021.
- 42.Oliver SE GJ, Scobie H, et al. Interim Recommendation for Use of Janssen COVID-19 Vaccine. Available at: https://www.cdc.gov/mmwr/volumes/70/wr/mm7009e4.htm?s_cid=mm7009e4_w. Accessed 2021, 5 May. [DOI] [PMC free article] [PubMed]
- 43.National Institutes of Health. Therapeutic Management of Adults With COVID-19. Available at: https://www.covid19treatmentguidelines.nih.gov/therapeutic-management/. Accessed April 30, 2021.
- 44.Formycon. Innovative SARS-CoV-2 blocker. Available at: https://www.formycon.com/en/pipeline/fyb207/. Accessed April 30, 2021.
- 45.Sokolowska M.Outsmarting SARS-CoV-2 by empowering a decoy ACE2. Signal Transduct Tar. 2020;5(1):1–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tandon R, Sharp JS, Zhang F, et al. Effective inhibition of SARS-CoV-2 entry by heparin and enoxaparin derivatives. J Virol. 2021;95(3) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pepi LE, Sanderson P, Stickney M, Amster IJ. Developments in Mass Spectrometry for Glycosaminoglycan Analysis: A Review. Molecular & Cellular Proteomics. 2021:100025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ardestani A, Azizi Z. Targeting glucose metabolism for treatment of COVID-19. Signal Transduct Tar. 2021;6(1):1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.U.S. Food and Drug Administration. Know Your Treatment Options for COVID-19. Available at: https://www.fda.gov/consumers/consumer-updates/know-your-treatment-options-covid-19. Accessed April 30, 2021.
- 50.Zheng K, Bantog C, Bayer R. The impact of glycosylation on monoclonal antibody conformation and stability. MAbs. 2011;3(6):568–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Beigel JH, Tomashek KM, Dodd LE, et al. Remdesivir for the treatment of Covid-19. New England Journal of Medicine. 2020;383(19):1813–1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Horby P, Lim WS, Emberson JR, et al. Dexamethasone in Hospitalized Patients with Covid-19. N Engl J Med. 2021;384(8):693–704. [DOI] [PMC free article] [PubMed] [Google Scholar]