Abstract
Klebsiella oxytoca M5a1 has the capacity to transport and to metabolize α-, β- and γ-cyclodextrins. Cyclodextrin transport is mediated by the products of the cymE, cymF, cymG, cymD, and cymA genes, which are functionally homologous to the malE, malF, malG, malK, and lamB gene products of Escherichia coli. CymE, which is the periplasmic binding protein, has been overproduced and purified. By substrate-induced fluorescence quenching, the binding of ligands was analyzed. CymE bound α-cyclodextrin, β-cyclodextrin, and γ-cyclodextrin, with dissociation constants (Kd) of 0.02, 0.14 and 0.30 μM, respectively, and linear maltoheptaose, with a Kd of 70 μM. In transport experiments, α-cyclodextrin was taken up by the cym system of K. oxytoca three to five times less efficiently than maltohexaose by the E. coli maltose system. Besides α-cyclodextrin, maltohexaose was also taken up by the K. oxytoca cym system, but because of the inability of maltodextrins to induce the cym system, growth of E. coli mal mutants on linear maltodextrin was not observed when the cells harbored only the cym uptake system. Strains which gained this capacity by mutation could easily be selected, however.
Cyclodextrins (CDs) are α-1,4-glycosidically linked, cyclic maltooligosaccharides. The main forms are α-, β-, and γ-CDs, which have 6, 7, and 8 glucose residues, respectively (5, 24). Their three-dimensional structure is torus shaped, with a hydrophilic outside ring and a interior hydrophobic cavity. Dependent on the size of the hydrophobic cavity of the respective CD, inclusion complexes with a variety of guest molecules can be formed, which is the basis of broad applications in industry (5, 24). CDs are formed enzymically from starch by CD-glucanotransferases (CGTases) a subgroup of the α-amylase class of enzymes (27, 35). One of the producers of CGTases is Klebsiella oxytoca M5a1 (1).
K. oxytoca can utilize starch as the sole carbon and energy source via two metabolic routes. The first one involves the extracellular degradation into linear maltodextrins by hydrolysis of the α-1,6-glycosidic bonds via the cell surface-associated pullulanase (1, 32) and the subsequent cleavage of the α-1,4-glycosidic linkages by the disproportionation activity of the extracellular α-CGTase (1). Escherichia coli (11, 15, 16, 36) and Klebsiella pneumoniae (7, 44) can then take up these linear maltodextrins (maltose up to maltoheptaose) via a binding protein-dependent ABC transporter (12) consisting of maltoporin (LamB), the maltodextrin-binding protein (MalE), the cytoplasmatic membrane proteins (MalF and MalG), and the ATP-binding protein (MalK). Intracellularly, the linear maltodextrins are degraded into glucose and glucose-1-phosphate by the enzymes amylomaltase (MalQ) (31), the maltodextrin phosphorylase (MalP) (43), and maltodextrin glucosidase (MalZ) (30).
In the second pathway, present only in K. oxytoca, starch is converted extracellularly into CDs by the cyclization activity of the α-CGTase, forming first and predominately α-CD, which later on is transformed into the thermodynamically favored β-CD (3, 35). The growth of K. oxytoca with CD as its sole carbon and energy source (2) is based on the uptake of the α- and β-CD via the cym system (17). Intracellularly, CDs are linearized by a cyclodextrinase (CymH) into linear maltooligosaccharides (14) which enter the maltose degradation pathway (17).
The genes responsible for CD metabolism were localized at the 5′ side of the gene coding for the α-CGTase (6, 17). Sequence analysis demonstrated the homology of the gene products of cymE, cymF, cymG, and cymD to MalE, MalF, MalG, and MalK, respectively (17). In addition, a functional homology could be shown between the gene product of cymA and the maltoporin LamB (29). A mutational analysis showed that cymE, cymF, cymD, and cymA were essential for growth at the expense of CDs (17, 29).
The existence of a specific transport system for CDs, which are rigid molecules of considerable size (ranging from 1.37 nm for α-CD to 1.69 nm for γ-CD [outer diameter]), was unexpected. Since their transfer through the outer membrane and the cytoplasmic membrane may involve novel mechanisms, we have initiated the biochemical analysis. Here we report on the biochemical characterization of CymE as a periplasmic CD binding protein as well as the kinetic analysis of the cym transport system.
MATERIALS AND METHODS
Bacterial strains and plasmids, media, and growth conditions.
The bacterial strains and the plasmids used in this study are listed in Table 1, together with their source or derivation.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Strains | ||
| Klebsiella oxytoca derivatives | ||
| M5a1 | Wild type | 1 |
| CYME | Like M5a1, Δ(cymE) | This work |
| Escherichia coli K-12 derivatives | ||
| JM109 | F′ (traD36 proAB+ lacIqZΔM15) recA1 endA1 gyrA96 thi-1 hsdR17 relA1 supE44 Δ(lac-proAB) λ− | 46 |
| MC4100 | F−, Δ(argF-lac) U169 araD139 ptsF25 deoC1 relA1 flbB5301 rpsL150 λ− | 13 |
| GM1 | Like MC4100, malE::Tn10 (Kanr) | 17 |
| JV2000 | Like MC4100, malS::Tn10 (Tetr) phoA8 rbsR | W. Boos |
| GM15 | Like GM1, malS::Tn10 (Tetr) | This work |
| Escherichia coli B derivatives; BL21 (DE-3) | F−hsdS gal λ(DE-3) | 41 |
| Plasmids | ||
| pT7-H6-TRXFUS | AprH6-trxA | 22 |
| pCYME2 (pT7-H6-TRXFUS derivative) | AprH6-trxA ′cymE | This work |
| pSU2719 | CmrlacZα | 25 |
| pGM200 (pSU2719 derivative) | CmrcymA-J | 17 |
| pCYMΔE (pSU2719 derivative) | CmrcymA-D Δ(cymE) cymF-J | 17 |
The rich medium used was Luria-Bertani (LB) medium (26). mal and cym mutants were discriminated on MacConkey agar plates enriched with 0.5% (wt/vol) maltose or α-CD. The minimal medium employed was salt solution P supplemented with 12 mM ammonium sulfate (18). If not indicated otherwise, particular carbon sources were added at concentrations of 0.2 or 0.5%. The concentrations of antibiotics added and growth conditions were described previously (17).
Standard genetic procedures.
Standard genetic procedures were adopted from Miller (26) and Sambrook et al. (33). Enzymes for recombinant DNA techniques were used according to the recommendations of the manufacturer.
Immunoblotting analysis.
Proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (23) and subjected to immunoblotting analysis (34) with rat anti-CymH monoclonal antibodies at a dilution of 1:10,000 as described previously (14).
Construction of a K. oxytoca cymE-deletion mutant.
The wild-type K. oxytoca strain was transformed with the plasmid pCYMΔE carrying an in-frame deletion in cymE (17). The transformants were grown overnight in LB medium without antibiotics, and the resulting cultures were diluted to cell densities of 1 × 103 to 2 × 103 cells/ml and plated on MacConkey plates supplemented with 0.5% α-CD. Colorless colonies (about 1%) were purified and tested for the inability to grow with α-CD as the sole carbon and energy source as well as for their sensitivity to the antibiotic marker of the plasmid used. The presence of the deletion was verified by PCR employing oligonucleotides priming up- and downstream of the deletion (MC7, 5′-CCCATTTCCGGAATAGAT-3′; MC15, 5′-CAGCCAGGTAAGATTTAC-3′). Finally, the strains were tested for complementation of the cym phenotype by transformation with a plasmid carrying a wild-type copy of the gene.
Construction of plasmid pCYME2.
Plasmid pCYME2 is a derivative of the plasmid pT7-H6-TRXFUS (22). It carries a translational fusion of the gene coding for a His-tagged thioredoxin, followed by an enterokinase cleavage site and the cymE gene lacking its signal sequence coding region (bp 1 to 60). For construction, the ′cymE part was amplified via PCR with the oligonucleotide primers CYME6 (5′-GCTTGGGGAGAGAGTATT-3′) and CYME7 (5′-TTTCCCGTCGACAATAACATAGTTACTCCT-3′). The resulting fragment was phosphorylated and cleaved with SalI. The pT7-H6-TRXFUS vector part was restricted with KpnI, treated with Klenow fragment, subsequently cleaved by SalI, and dephosphorylated. Finally, both fragments were ligated. All fusion joints and the entire ′cymE gene region were verified by sequencing.
Overproduction of the His-tag–thioredoxin–CymE fusion protein.
For overproduction and purification of the His-tag–thioredoxin–CymE fusion protein, strain BL21 (DE-3) was transformed with plasmid pCYME2. Cells were grown aerobically at 37°C in 200 ml of LB medium and inoculated 1:100 with an overnight culture. At an A600 of 1.0, cells were induced with isopropyl-β-d-thiogalactopyranoside (IPTG; 0.5 mM final concentration), harvested 2 h later by centrifugation, and stored at −80°C.
Purification of CymE.
The purification procedure was conducted at 4°C. The protein concentration was assayed during purification according to the method of Whitaker and Granum (45). The CymE content of the fractions was analyzed by SDS-PAGE (23) following the distribution of the 55-kDa band of the His-tag–thioredoxin–CymE fusion protein. After the enterokinase treatment, the appearance of the 39-kDa band of the mature form of CymE was monitored.
Cells were suspended in a 20-fold volume of buffer A (50 mM sodium phosphate [pH 7.4], 500 mM NaCl, 1 mM dithiothreitol) containing 1 mM phenylmethylsulfonyl fluoride and broken by three consecutive passages through a French pressure cell at 16,000 lb/in2. The extract was clarified by centrifugation for 10 min at 30,000 × g, resulting in the S30 fraction.
The S30 fraction was applied to a Zn2+-chelating Sepharose FF column (1-ml bed volume) that had been equilibrated in buffer A. After loading, the column was rinsed with buffer A and developed with a 30-ml linear gradient ranging from 10 to 200 mM imidazol. The flow rate was 0.5 ml/min. Fractions containing the His-tag–thioredoxin–CymE fusion protein were pooled and dialyzed against buffer B (20 mM Tris-HCl [pH 7.6], 0.5 mM EDTA).
For the cleavage of the fusion protein, the dialysate was incubated with enterokinase (Boehringer Mannheim GmbH) for 6 h at 37°C at a ratio between fusion protein and enterokinase of 1:20 (wt/wt). To separate the mature form of CymE from the His-tag–thioredoxin part of the fusion protein and from the noncleaved fusion protein and minor impurities, ion-exchange chromatography through a Mono Q-HR 5/5 column (1-ml bed volume) was performed. The column was equilibrated with buffer B; after the sample was loaded, the column was washed with 5 ml of buffer B, developed with a 20-ml linear gradient reaching from 0 to 1 M KCl, and finally washed with 5 ml (each) of buffer B and buffer C (buffer B plus 1 M KCl). The flow rate was 0.5 ml/min. Fractions containing the mature form of CymE were pooled and dialyzed against buffer B. The solution was frozen in liquid nitrogen and stored at −80°C.
Fluorescence spectroscopy.
Fluorescence spectroscopy was performed to measure the binding of substrates by the purified CymE (42). Fluorescence measurements were carried out in an LS50B Luminescence spectrometer (Perkin-Elmer, Norwalk, Conn.) with excitation and emission slits between 5 and 10 nm and at an excitation wavelength of 280 nm. The emission spectrum was monitored between 300 and 400 nm. Changes in the fluorescence intensity as a function of ligand concentrations were recorded at an emission wavelength of 355 nm. All measurements were performed at room temperature in 1 ml of 50 mM Tris-HCl (pH 7.6), which also served as a reference. Protein concentrations ranged from 0.8 to 8 μg/ml, and the substrates were added in small volumes. Changes in the intensity of fluorescence emission due to dilutions were corrected with a control containing the same concentration of protein and receiving buffer instead of substrate. For determination of the respective Kd values, a series of substrate concentrations were examined for the degree of quenching of fluorescence. The results reported are the mean values of three consecutive measurements for 5 min each. Maximal quenching was set at 1. The substrate concentration resulting in half-maximal quenching was defined as the Kd and was derived from Lineweaver-Burk plots.
Chromatographic techniques.
Linear and cyclic maltooligosaccharides were analyzed qualitatively by thin-layer chromatography (TLC) as described previously (14).
Transport assays.
In order to measure transport, the bacteria were first adapted in a preculture to the carbon source employed. For the characterization of CD uptake, 0.5% α-CD or a mixture of 1% succinate plus 0.1% α-CD was used as the carbon source, whereas for studying the maltose system, 0.2% maltose was used as the carbon source. After inoculation of the main culture, cells were grown to the late exponential growth phase, harvested by centrifugation, washed three times with minimal medium without a carbon source, resuspended in the same medium, and adjusted in terms of their cell density. For transport measurements, samples of 3 or 6 ml of this stock solution were adjusted to 25°C. To start the uptake reaction, a small volume of the labeled substrate was added (time zero). At different time intervals, samples of 500 or 1,000 μl were withdrawn, filtrated through Schleicher & Schuell (Dassel) NC45 membrane filters (0.45-μm pore size), washed with 10 ml of minimal medium and dried. Their radioactivity was determined in a TRI-CARB 2100 TR liquid scintillation analyzer (Packard, Dreieich, Germany) with the Ultima Gold scintillation cocktail (Packard). The rate of uptake was taken from the linear portion of the resulting curve (initial rate of uptake).
For the Vmax and Km determination of α-CD and maltohexaose uptake, [α-14C]CD (1,890 mCi/mmol) and [14C]maltohexaose (2 mCi/mmol) were synthesized as described by Pajatsch et al. (28) [α-14C]CD of low specific radioactivity (2 mCi/mmol) obtained from Wacker Chemicals (Munich, Germany) was also used. Km and Vmax values were deduced from Lineweaver-Burk plots. Competition of [α-14C]CD uptake by unlabeled cyclodextrins or linear maltodextrins was measured with a saturating concentration (20 μM) of the [α-14C]CD of the specific radioactivity (2 mCi/mmol) mixed with the indicated molar excess of unlabeled compounds.
To observe the metabolism of the α-CD taken up by the cells, chromatographic analysis of the intracellular low-molecular-weight compounds was performed. To this end, cells were incubated with a sufficient amount of the [α-14C]CD (2 mCi/mmol) to ensure maximal uptake rates over the entire incubation time. At the times indicated, samples were harvested by filtration, washed, frozen immediately in liquid nitrogen, and stored at −80°C. The cells were broken by resuspension in 1 ml of a solution containing 2.5% SDS and 1% chloroform. The solution was clarified by centrifugation, and the supernatant was desalted by the mixed bed ion-exchange material Serdolit MB-1 (Serva, Heidelberg, Germany). Finally, the solution was lyophilized, dissolved in a small volume of water, and separated by TLC. The TLC plate was then autoradiographed and developed.
Special chemicals.
The unlabeled α-, β-, and γ-CDs, as well as the [α-14C]CD of low specific radioactivity (2 mCi/mmol), were gifts from Wacker Chemicals. [α-14C]CD of high specific radioactivity (1,890 mCi/mmol) and [14C]maltohexaose (2 mCi/mmol) were synthesized as described previously (28). Nonradioactively labeled linear maltodextrins were purchased from Sigma Chemicals (Deisenhofen, Germany). [35S]dATP used for sequencing was delivered by NEN/DuPont (Dreieich, Germany). Oligonucleotide primers were synthesized by MWG (Ebersberg, Germany). Molecular biological reagents were from Boehringer Mannheim GmbH, Pharmacia (Freiburg, Germany). or New England Biolabs (Schwalbach, Germany).
RESULTS AND DISCUSSION
Overproduction and purification of CymE.
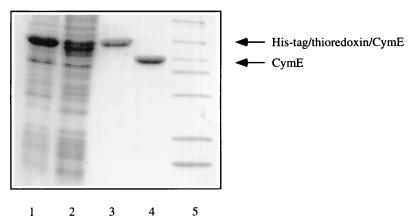
A fragment containing the part of the cymE gene corresponding to its mature (leaderless) form (17) plus 22 bp downstream of the coding region was amplified via PCR and cloned into the vector pT7-H6-TRXFUS to generate a translational fusion with the gene coding for a His-tagged thioredoxin, which at its C-terminus contains the cleavage site for enterokinase. Considerable attempts to overproduce native CymE from other vector systems and under many growth conditions only led to insoluble protein. For overproduction of the CymE fusion protein, the resulting plasmid, pCYME2, was transformed into E. coli BL21 (DE-3), and the expression was induced in LB medium at 37°C by the addition of IPTG. SDS-PAGE of a crude cellular extract showed that a protein of the expected molecular mass was overproduced (Fig. 1, lane 1), the major part in soluble form (data not shown). The purification involved the preparation of a 30,000 × g supernatant, affinity chromatography on a Zn2+-chelating Sepharose FF column, cleavage of the His-tag–thioredoxin–CymE fusion protein by enterokinase, and ion-exchange chromatography through a Mono-Q-HR5/5 column. A symmetrical peak consisting solely of the mature form of CymE was eluted from the final column (data not shown). Figure 1 demonstrates the course of purification as analyzed by SDS-PAGE of the pooled fractions from each step. From 200 ml of culture, 1 mg of purified CymE was obtained.
FIG. 1.
Course of purification of the mature form of CymE followed by SDS-PAGE of the respective fractions. Lanes: 1, crude extract; 2, S30 fraction; 3, eluate of the Zn2+-chelating Sepharose FF column; 4, eluate of the Mono-Q column; 5, molecular mass standard (97, 85, 55, 39, 33, 27, 19, and 14 kDa). Proteins were stained with Coomassie brilliant blue.
Specificity and affinity of substrate binding by CymE.
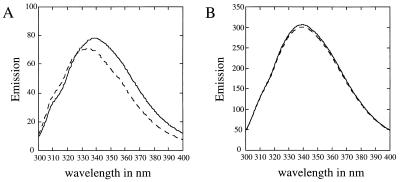
The CymE protein contains six tryptophan residues, which, when excited at a wavelength of 280 nm in the absence of ligand, induce a fluorescence emission spectrum with a maximum of 338 nm (Fig. 2). The addition of 0.1 mM substrate such as α-CD (Fig. 2A) decreased the fluorescence intensity and caused a shift of the emission spectrum to shorter wavelengths, in the case of α-CD by about 4 nm. Addition of 0.1 mM β- and γ-CD and of 0.1 mM maltodextrins also quenched fluorescence, with a shift of the emission peak to shorter wavelengths, although this shift becomes less prominent with increasing size of the CDs or decreasing chain length of the maltodextrins, respectively (data not shown). Addition of maltose (Fig. 2B) was without effect. Since the substrate-induced quenching was maximal at 355 nm (Fig. 2A), this wavelength was used in further experiments.
FIG. 2.
Fluorescence quenching of CymE induced by α-CD (A) and maltose (B). Excitation was at 280 nm. The emission spectrum of CymE (A, 0.8 μg/ml; B, 4 μg/ml) before (solid lines) and after (dotted lines) the addition of 0.1 mM substrate is shown.
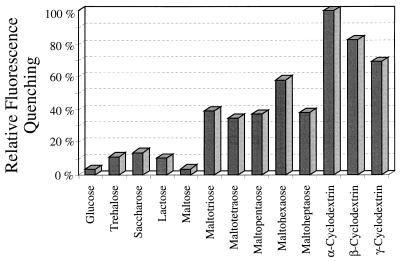
The substrate specificity of CymE was analyzed by measuring the effect of addition of 0.1 mM cyclic and linear maltodextrins and of several mono- and disaccharides (Fig. 3). The mono- and disaccharides, including maltose, only marginally quenched fluorescence, which may be due to maltodextrin impurities. Linear maltodextrins of a chain length of 3 glucose units and longer, however, had a significant effect. The CDs displayed the highest degree of quenching.
FIG. 3.
Decrease in fluorescence of CymE (4 μg/ml) induced by different substrates at a concentration of 0.1 mM. The value exhibited by α-CD is set at 100%.
The Kd values of CymE for the three CDs and for maltoheptaose were then determined by titrating the protein with a series of concentrations of the ligands and measuring the effect on fluorescence. As measured by fluorescence quenching, the affinity of CymE for the different sugars decreased with increasing size of the CDs as follows. The Kd values for α-CD, β-CD, and γ-CD were 0.02, 0.14, and 0.30 μM, respectively. The affinity for maltoheptaose (Kd, 70 μM) was much lower than those for the CDs.
CymE dependence of transport of CDs and maltodextrins into K. oxytoca and recombinant E. coli cells.
To gain information on whether the binding affinity of CymE determines the in vivo affinity for cellular uptake, transport experiments with radioactively labeled α-CD and maltohexaose were conducted at different substrate concentrations. Several strains were used: K. oxytoca (wild type and a cymE mutant), E. coli GM15/pGM200 harboring a plasmid that encodes the cym genes except that for the cyclodextrin glucanotransferase (cgt), and E. coli GM15/pCYMΔE with a deletion in cymE of pGM200 (both strains lack the E. coli maltose transport system because of a chromosomal mutation in malE). E. coli MC4100 served as control strain for the determination of the uptake of maltohexaose by the mal system. All strains, with the exception of MC4100, were grown under conditions used to induce the cym system. Table 2 gives the apparent Km and Vmax values for α-CD and maltohexaose. The results allow the following conclusions. (i) Under Vmax conditions, the recombinant E. coli strain transports the two substrates at nearly identical rates. (ii) The apparent Km for α-CD transport in K. oxytoca as well as in the recombinant E. coli strain is higher than the Kd of the binding protein (1 versus 0.02 μM). (iii) In the recombinant E. coli strain, the Km for α-CD is nearly 10-fold higher than that for maltohexaose. Also, transport of maltohexaose via the E. coli mal system is more efficient than that via the K. oxytoca cym system.
TABLE 2.
Kinetics of cym system-dependent uptake of α-CD and maltohexaose
| K. oxytoca or E. coli straina |
Vmax (nmol · min−1)b
|
Km (μM)
|
||
|---|---|---|---|---|
| α-CD | Maltohexaose | α-CD | Maltohexaose | |
| Wild type | 1.0 | NDc | 0.7–1.0 | ND |
| CYME | —d | ND | ND | ND |
| GM15/pGM200 | 1.5 | 1.2 | 0.7 | 6.5 |
| GM15/pCYMΔE | — | — | ND | ND |
| MC4100 | ND | 4.3 | ND | 0.33 |
K. oxytoca wild-type and CYME and E. coli GM15 strains were grown with a mixture of 1% succinate plus 0.1% α-CD as a carbon source, whereas for E. coli MC4100, the carbon source used was 0.2% maltose.
Values are for 109 cells.
ND, not determined.
—, from no uptake at all to about 3% of the Vmax of the respective cym+ strain.
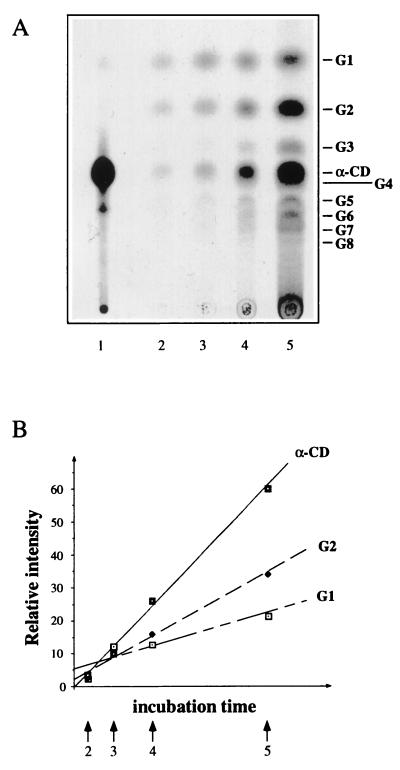
Since only the initial rate of uptake was used, the kinetic constants of Table 2 reflect the properties of the uptake system and are not influenced by subsequent metabolism or by a possible feedback inhibition due to the accumulating substrate. We also monitored the intracellular fate of radioactive α-CD during the course of an uptake experiment with the K. oxytoca wild type. Figure 4 shows that α-CD is accumulated more rapidly than it is hydrolyzed, which indicates that it is taken up in an unaltered form.
FIG. 4.
TLC analysis of the intracellular compounds of the K. oxytoca wild type during the uptake of [α-14C]CD. (A) Scanned autoradiograph. Lanes: 1, authentic [α-14C]CD applied as standard; 2 to 5, accumulated radioactivity after 20 s, 1 min, 2 min, and 5 min of incubation with [α-14C]CD, respectively. (B) Comparison of increases in spot intensities for α-CD, G2, and G1. Arrows correspond to the respective lanes of the autoradiograph. G1 and G2 to G8 denote glucose and maltose to maltooctaose, respectively. The spot for α-CD may be contaminated by a minor amount of G4.
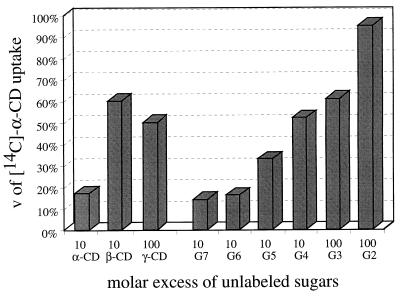
Finally, we have carried out α-CD uptake competition experiments in the absence and presence of competing substrates (Fig. 5). Competition decreased with increasing size of CDs and with decreasing chain length of linear maltooligosaccharides; maltose, again, was without any effect. This pattern roughly reflects the binding affinities of CymE towards these substrates.
FIG. 5.
Competition of the uptake of [α-14C]CD (20 μM) by the addition of unlabeled substrates. The % v (Vmax) values are relative to the rate of uptake of [α-14C]CD. G2 and G3 to G7 denote maltose and maltotriose to maltoheptaose, respectively. The competing substrates were added at the indicated fold concentration.
Can the cym system support growth at the expense of maltooligosaccharides?
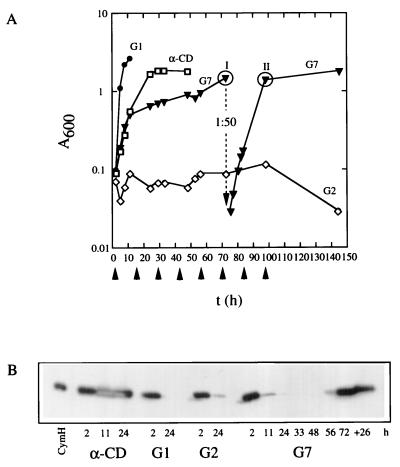
The results described above have shown that CymE binds maltooligosaccharides, that maltooligosaccharides can be transported in a CymE-dependent fashion, and that they compete with the uptake of α-CD. Therefore, one would assume that the cym system would allow the cells to grow on these linear maltooligosaccharides. This assumption was tested with strain GM15/pGM200 (Fig. 6A). It grows well in α-CD-containing minimal medium, with a mean doubling time of about 2 h. When subcultured into medium with linear maltodextrins from maltotetraose to maltoheptaose, growth continued with doubling times at least for maltohexaose and maltoheptaose comparable to those with α-CD, but decreased thereafter and finally stopped. However, after a considerable lag, growth resumed and a resubculture grew without a lag. For maltohexaose and maltoheptaose, this lag phase lasted at least 36 h, and it was even longer on maltopentaose and maltotetraose. No growth took place at all on maltose and maltotriose (not shown). Growth on maltodextrins from maltotetraose to maltoheptaose was completely blocked when a mutation was present in cymE (not shown).
FIG. 6.
(A) cym system-dependent growth of strain GM15/pGM200 with maltoheptaose (G7) as the carbon source. The preculture was grown with α-CD as the carbon source. Before being subcultured, the cells were harvested, washed, and resuspended in minimal medium. I and II indicate the time points at which samples were withdrawn from the culture to retrieve single colonies which consecutively were characterized. (B) Immunoblotting analysis of the expression of the cym system in strain GM15/pGM200 during growth with maltoheptaose (G7) as the carbon source. At the time points indicated, samples of the respective cultures were withdrawn, and a crude extract of the harvested cells was used for immunoblotting with an anti-CymH monoclonal antibody (14). G1 and G2 denote glucose and maltose, respectively. The amount of purified CymH (14) used was 40 ng.
To test whether the resumption of growth is the consequence of a selection of mutants, we have taken samples at the two time points indicated in Fig. 6. Only 20% of the colonies retrieved from the culture at time 1 were able to grow on α-CD and on maltoheptaose without a lag phase. This proportion increased to 80% at time 2.
Figure 6B shows that the ability to grow on α-CD correlates precisely with the expression pattern of cymH, an enzyme essential for CD metabolism (14, 17). The results of the experiment therefore show that the cym system can be used for the uptake of linear maltodextrins, but linear maltodextrins are unable to induce cym gene expression. Mutations, however, can be selected which lead to expression. The underlying mutations will be characterized in the future.
In conclusion, the results from previous work—that cym genes code for products of a transport system specific for CDs—have been established now by the in vitro analysis of CymE, the periplasmic binding protein, as well as by transport studies. Meanwhile, the gene product of cymA also has been identified as the functional homolog of the maltoporin LamB (29). CymE binds CDs, especially α-CD, with high affinity. It also binds linear maltooligosaccharides with a chain length of 3 and higher. However, binding appears to be productive and competent for transport only for α-CD and β-CD and for the longer linear maltooligosaccharides. The metabolism of γ-CD requires the activity of CGTase (17), which allows the conclusion that γ-CD has to be converted into α- or β-CD or linearized (3, 4, 29) by this enzyme to be transported. The fact that γ-CD does not support growth (17) could reside in the fact that its complex with CymE does not acquire the conformation required for docking to the membrane CymFG partner (37, 39, 40).
A similar situation has been described for MalE. MalE binds α-CD and β-CD with high affinity (Kd values of 4 and 1.8 μM, respectively) (38), but in the form of an unproductive complex (17, 19–21, 37, 38).
The Kd of CymE for α-CD binding is about 30-fold lower than the Km in the uptake reaction. Since it appears that the outer membrane does not limit the diffusion (10), the uptake process must be limited at some other step, most probably at the interaction of the substrate-loaded CymE with the membrane complex. Possibly, the amount of CymE is not large enough to ensure equality of Kd with Km (8, 9).
ACKNOWLEDGMENTS
We are greatly indebted to G. Wich from Wacker Chemicals, Munich, Germany, for the generous gift of CDs.
This work was supported by grants from the Deutsche Forschungsgemeinschaft (SFB156) to W.B. and the Fonds der Chemischen Industrie to A.B. and W.B.
REFERENCES
- 1.Bender H. Cyclodextrin-Glucanotransferase von Klebsiella pneumoniae. 1. Synthese, Reinigung und Eigenschaften des Enzyms von Klebsiella pneumoniae M5a1. Arch Microbiol. 1977;111:271–282. doi: 10.1007/BF00549366. [DOI] [PubMed] [Google Scholar]
- 2.Bender H. Cyclodextrin-Glucanotransferase von Klebsiella pneumoniae. 2. Bedeutung des Enzyms für den Metabolismus der Cyclodextrine bei Klebsiella pneumoniae M5a1. Arch Microbiol. 1977;113:49–66. doi: 10.1007/BF00428579. [DOI] [PubMed] [Google Scholar]
- 3.Bender H. Kinetische Untersuchungen der durch die Cyclodextrin-Glycosyltransferase katalysierten (1→4)-α-d-Glucopyranosyltransferreaktionen, insbesondere der Zyklisierungreaktion, mit (1→4)-α-d-Glucopyranosylketten (durchnittlicher Polymerisationsgrad von 16) als Substrat. Carbohydr Res. 1980;78:133–145. doi: 10.1016/s0008-6215(00)83667-8. [DOI] [PubMed] [Google Scholar]
- 4.Bender H. Proceedings of the First International Symposium on Cyclodextrins. D. Dordrecht, The Netherlands: Reichel Publishing Company; 1981. Enzymology of the cyclodextrins; pp. 77–88. [Google Scholar]
- 5.Bender H. Production, characterization, and application of cyclodextrins. Adv Biotechnol Proc. 1986;6:31–71. [Google Scholar]
- 6.Binder F, Huber O, Böck A. Cyclodextrin-glycosyltransferase from Klebsiella pneumoniae M5a1: cloning, nucleotide sequence and expression. Gene. 1986;47:269–277. doi: 10.1016/0378-1119(86)90070-3. [DOI] [PubMed] [Google Scholar]
- 7.Bloch M-A, Raibaud O. Comparison of the malA regions of Escherichia coli and Klebsiella pneumoniae. J Bacteriol. 1986;168:1220–1227. doi: 10.1128/jb.168.3.1220-1227.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bohl E, Shuman H A, Boos W. Mathematical treatment of the kinetics of binding protein-dependent transport systems reveals that both the substrate loaded and unloaded binding proteins interact with the membrane components. J Theor Biol. 1995;172:83–94. doi: 10.1006/jtbi.1995.0006. [DOI] [PubMed] [Google Scholar]
- 9.Bohl E, Boos W. Quantitative analysis of binding protein-mediated ABC transport systems. J Theor Biol. 1997;186:65–74. doi: 10.1006/jtbi.1996.0342. [DOI] [PubMed] [Google Scholar]
- 10.Bohl, E., M. Pajatsch, and W. Boos. Unpublished data.
- 11.Boos W, Peist R, Decker K, Zdych E. The maltose system. In: Lin E C C, Lynch A S, editors. Regulation of gene expression in Escherichia coli. R. G. Georgetown, Tex: Landes Company; 1996. pp. 201–229. [Google Scholar]
- 12.Boos W, Lucht J M. Periplasmic binding protein-dependent ABC transporters. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: Cellular and molecular biology. Vol. 1. Washington, D.C: American Society for Microbiology; 1996. pp. 1175–1209. [Google Scholar]
- 13.Casadaban M J, Cohen S N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci USA. 1979;76:4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feederle R, Pajatsch M, Kremmer E, Böck A. Metabolism of cyclodextrins by Klebsiella oxytoca M5a1: purification and characterisation of a cytoplasmically located cyclodextrinase. Arch Microbiol. 1996;165:206–212. doi: 10.1007/BF01692863. [DOI] [PubMed] [Google Scholar]
- 15.Ferenci T. The recognition of maltodextrins by Escherichia coli. Eur J Biochem. 1980;108:631–636. doi: 10.1111/j.1432-1033.1980.tb04758.x. [DOI] [PubMed] [Google Scholar]
- 16.Ferenci T, Muir M, Lee K-S, Maris D. Substrate specificity of the Escherichia coli maltodextrin transport system and its component proteins. Biochim Biophys Acta. 1986;860:44–50. doi: 10.1016/0005-2736(86)90496-7. [DOI] [PubMed] [Google Scholar]
- 17.Fiedler G, Pajatsch M, Böck A. Genetics of a novel starch utilisation pathway in Klebsiella oxytoca. J Mol Biol. 1996;256:279–291. doi: 10.1006/jmbi.1996.0085. [DOI] [PubMed] [Google Scholar]
- 18.Fraenkel D G, Neidhardt F C. Use of chloramphenicol to study control of RNA synthesis in bacteria. Biochim Biophys Acta. 1961;53:96–100. doi: 10.1016/0006-3002(61)90797-1. [DOI] [PubMed] [Google Scholar]
- 19.Hall J A, Gehring K, Nikaido H. Two modes of ligand binding in maltose-binding protein of Escherichia coli: correlation with the structure of ligands and the structure of binding protein. J Biol Chem. 1997;272:17605–17609. doi: 10.1074/jbc.272.28.17605. [DOI] [PubMed] [Google Scholar]
- 20.Hall J A, Thorgeirsson T E, Liu J, Shin Y-K, Nikaido H. Two modes of ligand binding in maltose-binding protein of Escherichia coli: electron paramagnetic resonance study of ligand-induced global conformational changes by site-directed spin labeling. J Biol Chem. 1997;272:17610–17614. doi: 10.1074/jbc.272.28.17610. [DOI] [PubMed] [Google Scholar]
- 21.Hall J A, Ganesan A K, Chen J, Nikaido H. Two modes of ligand binding in maltose-binding protein of Escherichia coli: functional significance in active transport. J Biol Chem. 1997;272:17615–17622. doi: 10.1074/jbc.272.28.17615. [DOI] [PubMed] [Google Scholar]
- 22.Kromayer M, Wilting R, Tormay P, Böck A. Domain structure of the procaryotic selenocysteine-specific elongation factor SelB. J Mol Biol. 1996;262:413–420. doi: 10.1006/jmbi.1996.0525. [DOI] [PubMed] [Google Scholar]
- 23.Laemmli U K. Cleavage of structural proteins during the assembly of the head of the bacteriophage T4. Nature (London) 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 24.Loftsson T, Brewster M E. Pharmaceutical applications of cyclodextrins. 1. Drug solubilisation and stabilization. J Pharm Sci. 1996;85:1017–1025. doi: 10.1021/js950534b. [DOI] [PubMed] [Google Scholar]
- 25.Martinez E, Bartolomé B, de la Cruz F. pACYC184-derived cloning vectors containing the multiple cloning site and lacZa reporter gene of pUC8/9 and pUC18/19 plasmids. Gene. 1988;68:159–162. doi: 10.1016/0378-1119(88)90608-7. [DOI] [PubMed] [Google Scholar]
- 26.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1992. [Google Scholar]
- 27.Nakamura A, Haga K, Yamane K. Three histidine residues in the active center of cyclodextrin glucanotransferase from alkalophilic Bacillus sp. 1011: effects of the replacement on pH dependence and transition-state stabilisation. Biochemistry. 1993;32:6624–6631. doi: 10.1021/bi00077a015. [DOI] [PubMed] [Google Scholar]
- 28.Pajatsch, M., A. Böck, and W. Boos. Enzymatic preparation of radioactively labeled linear maltodextrins and cyclodextrins of high specific activity from [14C]maltose using amylomaltase, cyclodextrin glucanotransferase and cyclodextrinase. Carbohydr. Res., in press. [DOI] [PubMed]
- 29.Pajatsch, M., G. Fiedler, and A. Böck. Unpublished data.
- 30.Peist R, Schneider-Fresenius C, Boos W. The MalT-dependent and malZ-encoded maltodextrin glucosidase of Escherichia coli can be converted into a dextrinyltransferase by a single mutation. J Biol Chem. 1996;271:10681–10689. doi: 10.1074/jbc.271.18.10681. [DOI] [PubMed] [Google Scholar]
- 31.Pugsley A P, Dubreuil C. Molecular characterisation of malQ, the structural gene for the Escherichia coli enzyme amylomaltase. Mol Microbiol. 1988;2:473–479. doi: 10.1111/j.1365-2958.1988.tb00053.x. [DOI] [PubMed] [Google Scholar]
- 32.Pugsley A P. The complete general secretory pathway in gram-negative bacteria. Microbiol Rev. 1993;57:50–108. doi: 10.1128/mr.57.1.50-108.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 34.Schmid G, Böck A. Immunoblotting analysis of ribosomal proteins from archaebacteria. Syst Appl Microbiol. 1984;5:1–10. [Google Scholar]
- 35.Schmid G. Enzymology of cyclodextrins. In: Atwood J L, Davies J E D, MacNicol D D, Vögtle F, editors. Comprehensive supramolecular chemistry. 3. Cyclodextrins. Oxford, United Kingdom: Elsevier Science, Ltd.; 1996. pp. 615–626. [Google Scholar]
- 36.Schwartz M. The Maltose regulon. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Vol. 2. Washington, D.C: American Society for Microbiology; 1987. pp. 1482–1502. [Google Scholar]
- 37.Sharff A J, Rodseth L E, Spurlino J C, Quiocho F A. Crystallographic evidence of a large ligand-induced hinge-twist motion between the two domains of the maltodextrin binding protein involved in active transport and chemotaxis. Biochemistry. 1992;31:10657–10663. doi: 10.1021/bi00159a003. [DOI] [PubMed] [Google Scholar]
- 38.Sharff A J, Rodseth L E, Quiocho F A. Refined 1.8 Å structure reveals the mode of binding of β-cyclodextrin to the maltodextrin binding protein. Biochemistry. 1993;32:10553–10559. doi: 10.1021/bi00091a004. [DOI] [PubMed] [Google Scholar]
- 39.Shilton B H, Shuman H A, Mowbray S L. Crystal structures and solution conformations of a dominant-negative mutant of Escherichia coli maltose-binding protein. J Mol Biol. 1996;264:364–376. doi: 10.1006/jmbi.1996.0646. [DOI] [PubMed] [Google Scholar]
- 40.Spurlino J C, Lu G-Y, Quiocho F A. The 2.3-Å resolution structure of the maltose- or maltodextrin-binding protein, a primary receptor of bacterial active transport and chemotaxis. J Biol Chem. 1991;266:5202–5219. doi: 10.2210/pdb1mbp/pdb. [DOI] [PubMed] [Google Scholar]
- 41.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 42.Szmelcman S, Schwartz M, Silhavy T J, Boos W. Maltose transport in Escherichia coli K12. A comparison of transport kinetics in wild-type and λ-resistant mutants with the dissociation constants of the maltose binding protein as measured by fluorescence quenching. Eur J Biochem. 1976;65:13–19. doi: 10.1111/j.1432-1033.1976.tb10383.x. [DOI] [PubMed] [Google Scholar]
- 43.Watson K A, Schinzel R, Palm D, Johnson L N. The structure of Escherichia coli maltodextrin phosphorylase provides an explanation for the activity without control in this basic archetype of a phosphorylase. EMBO J. 1997;16:1–14. doi: 10.1093/emboj/16.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Werts C, Charbit A, Bachellier S, Hofnung M. DNA sequence analysis of the lamB gene from Klebsiella pneumoniae: implications for the topology and the pore functions in maltoporin. Mol Gen Genet. 1992;233:372–378. doi: 10.1007/BF00265433. [DOI] [PubMed] [Google Scholar]
- 45.Whitaker J R, Granum P E. An absolute method for protein determination based on difference in absorbance at 235 and 280 nm. Anal Biochem. 1980;109:156–159. doi: 10.1016/0003-2697(80)90024-x. [DOI] [PubMed] [Google Scholar]
- 46.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;133:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]