Abstract
Neuropeptide Y targets the Y1 receptor (Y1) in the spinal dorsal horn (DH) to produce endogenous and exogenous analgesia. DH interneurons that express Y1 (Y1-INs; encoded by Npy1r) are necessary and sufficient for neuropathic hypersensitivity after peripheral nerve injury. However, as Y1-INs are heterogenous in composition in terms of morphology, neurophysiological characteristics, and gene expression, we hypothesized that a more precisely defined subpopulation mediates neuropathic hypersensitivity. Using fluorescence in situ hybridization, we found that Y1-INs segregate into 3 largely nonoverlapping subpopulations defined by the coexpression of Npy1r with gastrin-releasing peptide (Grp/Npy1r), neuropeptide FF (Npff/Npy1r), and cholecystokinin (Cck/Npy1r) in the superficial DH of mice, nonhuman primates, and humans. Next, we analyzed the functional significance of Grp/Npy1r, Npff/Npy1r, and Cck/Npy1r INs to neuropathic pain using a mouse model of peripheral nerve injury. We found that chemogenetic inhibition of Npff/Npy1r-INs did not change the behavioral signs of neuropathic pain. Further, inhibition of Y1-INs with an intrathecal Y1 agonist, [Leu31, Pro34]-NPY, reduced neuropathic hypersensitivity in mice with conditional deletion of Npy1r from CCK-INs and NPFF-INs but not from GRP-INs. We conclude that Grp/Npy1r-INs are conserved in higher order mammalian species and represent a promising and precise pharmacotherapeutic target for the treatment of neuropathic pain.
Keywords: Neuroscience
Keywords: NPY, Pain, Pharmacology
Introduction
Pain protects organisms from injury, and neurons in the superficial spinal cord dorsal horn (DH) are fundamental for the processing of harmful or potentially harmful nociceptive input (1). Most DH interneurons (INs) are glutamatergic with marked functional and chemical heterogeneity (2–4). These glutamatergic DH neurons mediate the appropriate and maladaptive expression of pain behavior (2, 5–7). For example, peripheral nerve damage can lead to pathological allodynia in which normally innocuous sensory input is amplified by excitatory DH neurons and conveyed as painful (7–10). The molecular identification of the subpopulations of excitatory DH neurons that mediate allodynia is an intense area of investigation (9), including those that express protein kinase C γ (PKCγ) (11–18), somatostatin (SST) (19, 20), cholecystokinin (CCK) (14, 21), neurokinin 1 receptor (NK1R) (22), mu opiate receptor (OPRM1) (23), and neuropeptide Y Y1 receptor (NPY1R) (24–26). Among these, Npy1r-expressing interneurons (Y1-INs) represent a particularly promising, druggable, pharmacotherapeutic target for the treatment of neuropathic pain (24, 26). We recently demonstrated that Y1-INs are both necessary and sufficient for the manifestation of neuropathic pain-like behavior using cell type–specific modulation/ablation studies (24, 25). Importantly, the Y1 receptor is coupled to inhibitory G proteins (Gi/o), and application of NPY Y1–selective agonists reduces the excitability of Y1-INs and decreases pro-nociceptive signaling both in vitro and in vivo (27–30). Markedly, intrathecal administration of Y1 agonist efficaciously reduces behavioral signs of neuropathic pain, and this can be reversed with Y1-selective antagonists (24, 31–33). These findings establish Y1 on Y1-INs as the general target for the antihyperalgesic effects of NPY.
However, further questions are raised by the highly heterogenous composition of Y1-INs in terms of morphology, neurophysiological characteristics, and gene expression (25, 26, 34, 35). For example, Y1 immunoreactivity in the rat revealed distinctive morphological and neurochemical Y1-IN subpopulations (25, 34, 35), our ex vivo lumbar spinal cord slice recordings from a reliable Npy1reGFP mouse line identified 4 distinct neurophysiological firing patterns of Y1-INs (24, 29), and unbiased single-cell transcriptomics in the mouse identified Npy1r expression in 3 excitatory DH neuron clusters (36). The heterogeneity of Y1-INs may suggest the existence of subpopulations with distinct physiological roles. Of particular interest are recent results that segregate excitatory interneurons into largely nonoverlapping populations defined by the expression of CCK, neurotensin, neurokinin B (NKB), neuropeptide FF (NPFF), substance P (SP; encoded by Tac1), enhanced green fluorescent protein (EGFP) or Cre recombinase under control of the gastrin-releasing peptide (Grp) promoter in BAC transgenic mice from the GENSAT project (GrpeGFP and GrpCre), and gastrin-releasing peptide receptor (GRPR) (37–40). Here, we performed fluorescence in situ hybridization (FISH) to characterize the coexpression of these markers with the Npy1r gene. We found that Y1-INs largely segregated into 3 subpopulations (demarcated by the coexpression of Npy1r with Cck, Grp, and Npff, respectively), which are conserved in the nonhuman primate and human DH. We then determined the functional contribution of the Npy1r/Npff population to the behavioral manifestation of mechanical and cold allodynia after peripheral nerve injury. Further, we asked whether pharmacological inhibition with a NPY Y1 agonist eliminates the signs of neuropathic pain in mice with conditional deletion of Npy1r in GRP-INs, CCK-INs, or NPFF-INs. Together, our results elucidate the molecular makeup and functional role of Y1-IN subpopulations and support Grp/Npy1r-INs as an optimal, evolutionarily conserved, and relatively precise pharmacotherapeutic target for the treatment of neuropathic pain.
Results
Y1-INs are excitatory neurons that segregate into 3 distinct subpopulations.
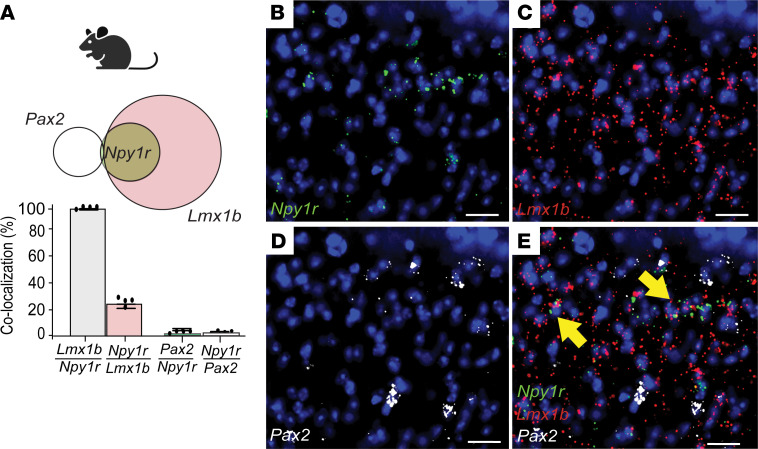
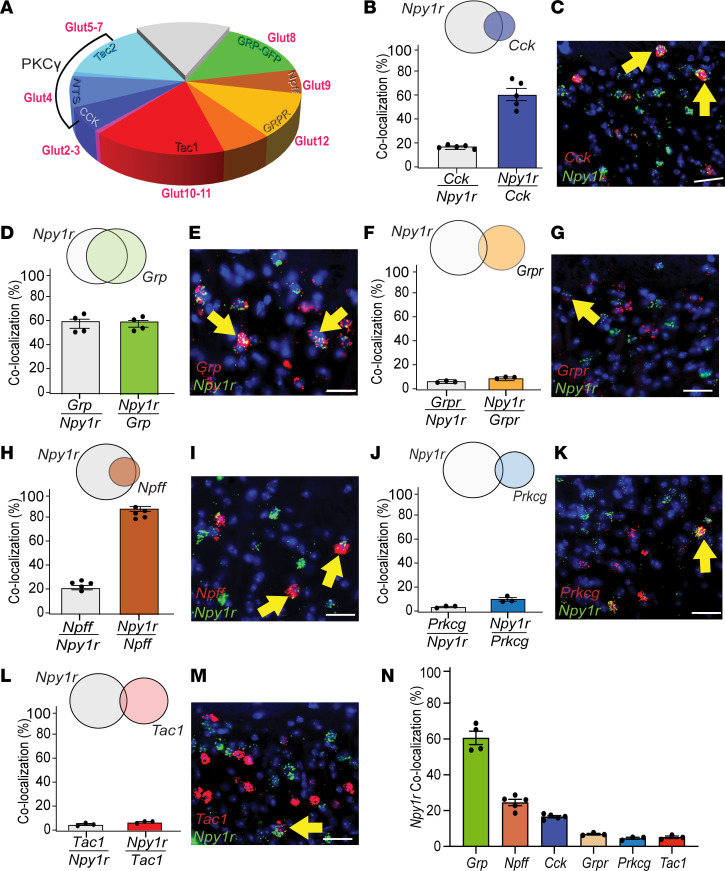
We used multilabel FISH to neurochemically characterize L4 Y1-INs in superficial DH. First, in mouse, we found Npy1r extensively colocalized with the excitatory marker LIM homeobox transcription factor 1-β (Lmx1b) (96.91% ± 0.49%) but not the inhibitory marker paired box 2 (Pax2) (1.39% ± 0.47%) (Figure 1, A–E) (41–44), indicating that Y1-INs are glutamatergic. Next, we identified putative Y1-IN subpopulation(s) using recent data that segregate approximately 90% of DH INs into largely nonoverlapping excitatory interneuron subpopulations based on the expression of neuropeptides/receptors (36–40, 45, 46) (Figure 2A). We found that Npy1r colocalized with Cck (16.60% ± 0.63%), Grp (60.61% ± 3.78%), and Npff (24.55% ± 1.75%) but minimally with Grpr (6.85% ± 0.45%), protein kinase C γ (Prkcg) (4.55% ± 0.55%), or Tac1 (5.13% ± 0.68%) (Figure 2, B–N). Note that Prkcg was used as a marker to broadly encompass both the neurotensin and NKB subpopulations (Figure 2A). Next, we characterized DH Npy1r expression with canonical genes implicated in neuropathic and inflammatory mechanical allodynia (9, 14, 19, 20, 22, 47) and found that Npy1r colocalized extensively with Sst (49.26% ± 0.98%) and Calb2 (33.96% ± 2.42%) but not Tacr1 (6.94% ± 1.11%) (Supplemental Figure 1, A–F; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.169554DS1). Last, recent single-cell RNA-sequencing data sets have suggested 2 genes, Nmur2 and Car12, have significant overlap with Npy1r (36, 45). Nmur2 encodes the neuromedin U receptor type 2 and contributes to nociceptive processing (48), and Car12 encodes the enzyme carbonic anhydrase 12 that is highly enriched in dorsal horn somatostatin neurons (49) but has no identified role in pain processing. Here, we verified the colocalization of Npy1r with both Nmur2 (37.04% ± 0.94%) and Car12 (32.83% ± 6.33%) (Supplemental Figure 1, G–J). Together, these results indicate that Y1-INs are excitatory DH interneurons and largely segregate into 3 largely nonoverlapping subpopulations demarcated by coexpression of Npy1r with the genes Cck, Npff, or Grp. Further, many Y1-INs colocalize with Sst, Calb2, Car12, and Nmur2.
Figure 1. Y1-INs are a glutamatergic subpopulation in mouse lumbar superficial dorsal horn.
(A–E) Npy1r in laminae I-II extensively colocalizes with Lmx1b (Lmx1b/Npy1r 96.91% ± 0.49%; Npy1r/Lmx1b 25.18% ± 1.59%) but not Pax2 (Pax2/Npy1r 1.39% ± 0.47%; Npy1r/Pax2 2.17% ± 0.81%) (n = 4–5 mice/group). Each data point indicates the average of 2–4 quantified sections/mouse. Scale bars: 25 μm. Yellow arrows indicate colocalization. Data shown as mean ± SEM.
Figure 2. Identification of major subpopulations of Y1-INs in mouse lumbar superficial dorsal horn.
(A) Excitatory interneurons segregate into largely nonoverlapping subpopulations defined by expression of cholecystokinin (CCK), neurotensin, neurokinin B (NKB), neuropeptide FF (NPFF), substance P (SP; encoded by Tac1), enhanced green fluorescent protein (EGFP) or Cre recombinase under control of the gastrin-releasing peptide (Grp) promoter in BAC transgenic mice from the GENSAT project (GrpeGFP and GrpCre), and gastrin-releasing peptide receptor (GRPR). (B–N) Npy1r colocalizes with superficial Cck (Cck/Npy1r 16.60% ± 0.63%; Npy1r/Cck 56.98% ± 4.73%), Grp (Grp/Npy1r 60.61% ± 3.78%; Npy1r/Grp 60.58% ± 2.78%), and Npff (Npff/Npy1r 24.55% ± 1.75%; Npy1r/Npff 91.21% ± 2.00%) but not Grpr (Grpr/Npy1r 6.85% ± 0.45%; Npy1r/Grpr 10.16% ± 0.43%), Prkcg (Prkcg/Npy1r 4.55% ± 0.55%; Npy1r/Prkcg 10.59% ± 1.50%), or Tac1 (Tac1/Npy1r 5.13% ± 0.68%; Npy1r/Tac1 7.09% ± 0.55%) (n = 3–5 mice/group). Each data point indicates the average of 2–4 quantified sections/mouse. Scale bars: 25 μm. Yellow arrows indicate colocalization. Data shown as mean ± SEM.
Y1-IN subpopulations are conserved across higher order mammalian species.
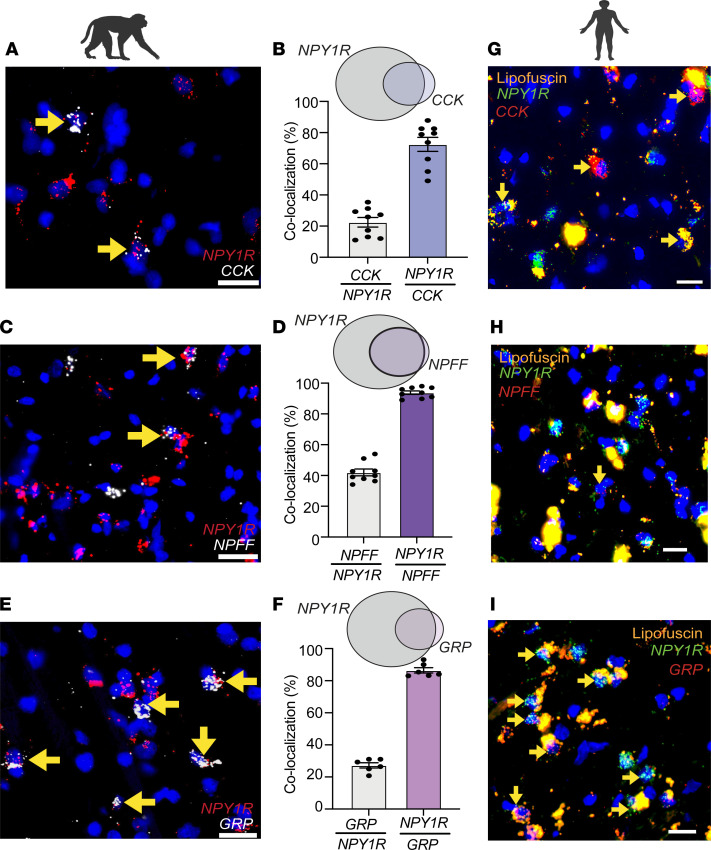
To determine the evolutionary conservation of Y1-IN subpopulations, we evaluated the expression of NPY1R, CCK, NPFF, and GRP in spinal cord tissue from rhesus macaque and human organ donors. As in the mouse, NPY1R-expressing cells were abundantly expressed in the superficial DH of macaque and extensively colocalized with CCK (22.49% ± 3.08%), GRP (27.26% ± 1.63%), and NPFF (41.98% ± 2.20%) (Figure 3, A–F). Similarly, NPY1R-expressing cells were plentiful in the superficial DH of human spinal cord and coexpressed CCK, GRP, and NPFF (acknowledging that NPFF expression is sparse in human DH as compared with mouse or macaque) (Figure 3, G–I). Intense lipofuscin prevented accurate quantification in the human spinal cord.
Figure 3. Conservation of NPY1R/CCK, NPY1R/GRP, and NPY1R/NPFF subpopulations in the superficial dorsal horn of nonhuman primates and humans.
(A–F) NPY1R in the substantia gelatinosa extensively colocalizes with superficial CCK (CCK/NPY1R 22.49% ± 3.08%; NPY1R/CCK 72.45% ± 4.53%), GRP (GRP/NPY1R 27.26% ± 1.63%; NPY1R/GRP 86.46% ± 1.70%), and NPFF (NPFF/NPY1R 41.98% ± 2.20%; NPY1R/NPFF 93.82% ± 1.12%) in the rhesus macaque (n = 2 macaques). Individual data points represent 1 single quantified section. Scale bars: 25 μm. (G–I) NPY1R colocalizes with superficial CCK, NPFF, and GRP in the human spinal cord dorsal horns (n = 2 human organ donors). Scale bars: 20 μm. Yellow arrows indicate colocalization. Data shown as mean ± SEM.
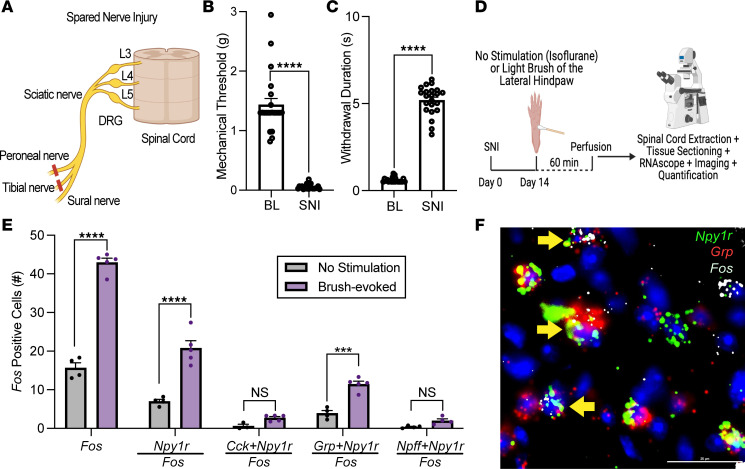
Light brush of the hind paw in mice with neuropathic hypersensitivity evokes Fos in DH Grp/Npy1r-INs.
Next, we determined whether the primary Y1-IN subpopulation(s) are sensitized by nerve injury. We performed spared nerve injury (SNI), a model of peripheral neuropathic pain (50) (Figure 4A), which produced robust mechanical and cold hypersensitivity in the spared nerve (sural) innervation territory (lateral aspect) of the injured hind paw (Figure 4, B and C) (24, 25). Two weeks after SNI, we applied light brush stimulation to the plantar hind paw (Figure 4D), and as predicted, we found increased Fos expression (using the immediate early gene Fos as a correlate for neuronal activity) in the ipsilateral superficial DH in Npy1r-expressing neurons (Figure 4E). Fos expression was principally expressed in the Grp/Npy1r subpopulation rather than the Npff/Npy1r or Cck/Npy1r subpopulations (Figure 4, E and F). These results suggest that in the context of peripheral nerve injury, mechanical stimulation predominately activates Y1-INs that coexpress Grp.
Figure 4. Non-noxious mechanical stimulation increases neuronal activity in Grp/Npy1r-INs.
(A) Schematic representation of the spared nerve injury (SNI) model of neuropathic pain. (B and C) SNI produces mechanical (****P < 0.0001) and cold hypersensitivity (****P < 0.0001) in the ipsilateral hind paw when assessed 14 days after injury. Unpaired 2-tailed t test (n = 24 mice/group). (D) Experimental timeline for SNI, light brush of the lateral hind paw, and FISH labeling for Fos in Y1-IN subpopulations in the lumbar dorsal horn of SNI mice. (E) Light brush increases Fos-positive cells in the superficial dorsal horn (****P < 0.0001) primarily in Npy1r neurons (****P < 0.0001) and Grp/Npy1r neurons (***P = 0.0001) but not Cck/Npy1r neurons (P = 0.7371) or Npff/Npy1r neurons (P = 0.8095) (2-way ANOVA: mRNA expression × stimulation, F4, 33 = 1.215, P < 0.0001, Holm-Šídák post hoc tests) (n = 3–5 mice/group). Each data point indicates the average of 2–4 quantified sections/mouse. (F) Representative example of Grp/Npy1r-INs coexpressing light brush–evoked Fos. Scale bars: 25 μm. Yellow arrows indicate colocalization. Data shown as mean ± SEM.
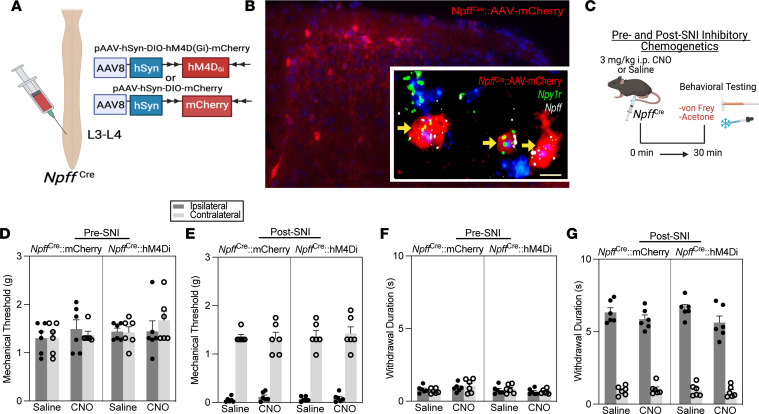
Inhibition of Npff/Npy1r-INs does not change neuropathic hypersensitivity.
Previously, we demonstrated that chemogenetic inhibition of Y1-INs with designer receptors exclusively activated by designer drugs (DREADDs) abolished SNI-induced neuropathic hypersensitivity (24). Similarly, we sought to determine the functional contribution of the Npff/Npy1r, Grp/Npy1r, and Cck/Npy1r subpopulations to neuropathic hypersensitivity. First, we took advantage of the fact that virtually all Npff-expressing DH interneurons coexpress Npy1r (Figure 2H). This feature of NPFF-INs allowed us to intraspinally administer Cre-dependent inhibitory DREADDs into the left lumbar (targeting L3–L4) DH of our developed NpffCre mouse line to inhibit the activity of Npff/Npy1r-INs (Figure 5A). As predicted, AAV-mCherry transfection expression with immunohistochemistry and FISH was detected ipsilateral to viral injection (left but not right DH) in superficial DH cells that coexpressed Npff and Npy1r (Figure 5B). We then intraperitoneally administered a 3 mg/kg dose of clozapine N-oxide (CNO, TOCRIS); this dose is the lowest recommended dose to behave as a DREADD agonist in the absence of off-target effects (51) and is commonly utilized for spinal cord chemogenetics (13, 24, 52). Chemogenetic inhibition of Npff/Npy1r-INs did not change mechanical or cold responses before or after SNI (Figure 5, C–G). These findings indicate that spinal Npff/Npy1r-INs are not necessary for the behavioral manifestation of mechanical or cold neuropathic hypersensitivity.
Figure 5. Npff/Npy1r-INs do not contribute to nerve injury–induced mechanical and cold allodynia.
(A) Strategy for selectively targeting Npff/Npy1r-INs in NpffCre mice with intraspinal injections of the Cre-dependent virus AAV8-hSyn-DIO-hM4DGi-mCherry. (B) AAV transfection in NpffCre mice (large image) and FISH confirmation of Npy1r and Npff expression in transfected cells (insert). Scale bar: 10 μm. (C) Experimental timeline of chemogenetic reflexive behavioral testing at both pre- and post-SNI time points. (D and E) Chemogenetic inhibition of NPFF-INs with CNO (3 mg/kg) does not change mechanical sensitivity when conducted either before (3-way repeated measures [RM] ANOVA: drug × virus × side, F1,20 = 0.7875, P = 0.3854) or after SNI (3-way RM ANOVA: drug × virus × side, F1,20 = 0.6462, P = 0.4309) (n = 6 mice/group). (F and G) Chemogenetic inhibition of NPFF-INs with CNO (3 mg/kg) does not change cold sensitivity pre-SNI (3-way RM ANOVA: drug × virus × side, F1,20 = 1.215, P = 0.2834) or post-SNI (3-way RM ANOVA: drug × virus × side, F1,20 = 0.1615, P = 0.6920) (n = 6 mice/group). Data shown as mean ± SEM.
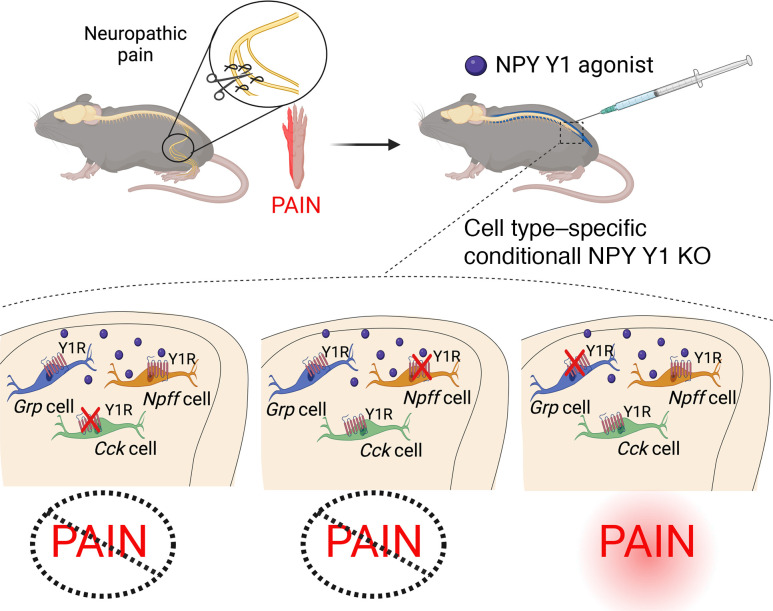
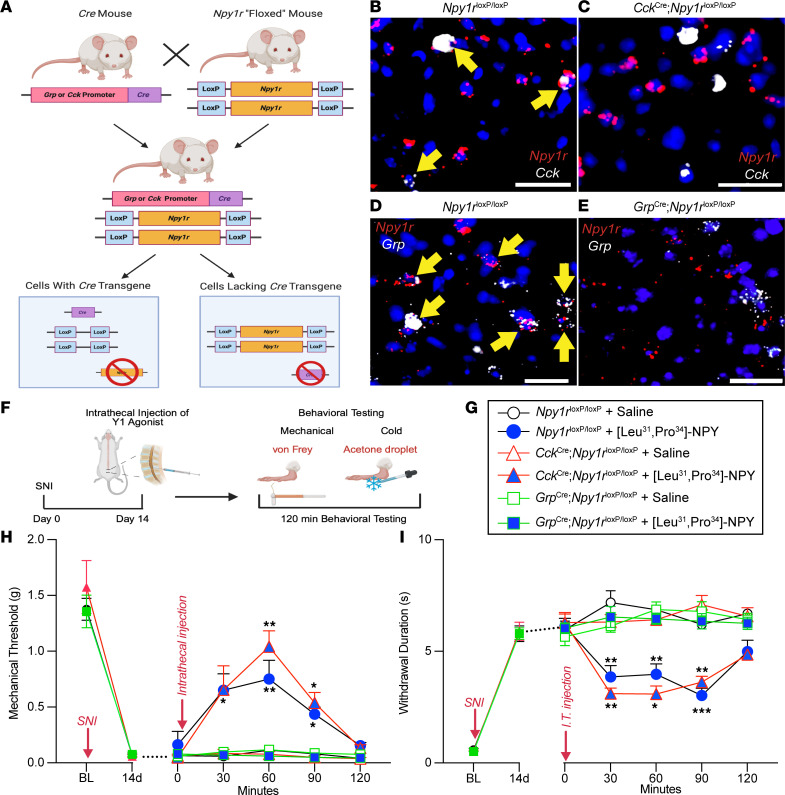
The Y1 agonist [Leu31, Pro34]-NPY eliminates nerve injury–induced mechanical and cold hypersensitivity in mice with conditional deletion of Cck but not Grp from Npy1r-INs.
In contrast to NPFF-INs, many CCK- and GRP-INs do not coexpress Npy1r. As a result, we could not inhibit the GrpCre or CckCre interneuron populations with a chemogenetic approach as we did with our NpffCre mice because this could also modulate neurons that do not express Npy1r (and there is no currently available Npy1rFlp mouse line to allow a dual recombinase chemogenetic approach). Instead, we pharmacologically inhibited Y1-INs with an intrathecal Y1 agonist in conditional genetic knockout lines. Specifically, we crossed Npy1rloxP/loxP mice (53) with either GrpCre (54) or CckCre (55) mice to selectively knock out Npy1r from GrpCre or CckCre neurons, respectively (Figure 6, A–E). Next, we performed SNI to produce mechanical and cold hypersensitivity (Figure 6, F–I). Two weeks later, we intrathecally administered the Y1-selective agonist, [Leu31, Pro34]-NPY, to inhibit spinal Y1-INs (24). [Leu31, Pro34]-NPY reduced SNI-induced allodynia in both control Npy1rloxP/loxP and Npy1rloxP/loxP CckCre mice but not in Npy1rloxP/loxP GrpCre mice (Figure 6, F–I). Similarly, intrathecal administration of [Leu31, Pro34]-NPY reduced SNI-induced allodynia in Npy1rloxP/loxP mice crossed with NpffCre mice (Supplemental Figure 2). Conditional genetic knockout of Npy1r was confirmed with FISH and quantified for all 3 mouse crosses (Supplemental Figure 3). These results indicate that NPY Y1 agonists inhibit behavioral signs of neuropathic pain by actions at spinal cord Npy1r-expressing interneurons that coexpress Grp but not Cck or Npff.
Figure 6. Grp/Npy1r-INs but not Cck/Npy1r-INs contribute to the inhibition of nerve injury–induced mechanical and cold allodynia by a Y1 agonist.
(A) Schematic representation of the conditional genetic knockout breeding protocol to delete Npy1r from CCK-Cre and GRP-Cre cells. (B–E) Confirmation of conditional deletion of Npy1r. FISH of sections of the lumbar spinal cord demonstrate that Npy1rloxP/loxP mice contain DH neurons that coexpress Npy1r and Cck as well as Npy1r and Grp. Conversely, Npy1rloxP/loxP CckCre mice lack expression of Npy1r in Cck-expressing neurons and Npy1rloxP/loxP GrpCre mice lack expression of Npy1r in Grp-expressing neurons. Yellow arrows indicate colocalization. Scale bars: 25 μm. (F) Experimental timeline for SNI, intrathecal pharmacology, and mechanical (von Frey) and cold (acetone droplet withdrawal) behavioral testing. (G) Genetic mouse lines and pharmacological drugs represented in H and I. (H) [Leu31, Pro34]-NPY abolishes SNI-induced mechanical allodynia in Npy1rloxP/loxP and Npy1rloxP/loxP CckCre mice but not in Npy1rloxP/loxP GrpCre mice (n = 8–9 mice/group). Three-way RM ANOVA: time × genotype × drug, F4,128 = 7.509, P < 0.0001; Tukey’s multiple-comparison tests. (I) [Leu31, Pro34]-NPY abolishes SNI-induced cold allodynia in Npy1rloxP/loxP and Npy1rloxP/loxP CckCre mice but not in Npy1rloxP/loxP GrpCre mice (n = 8–9 mice/group). Three-way RM ANOVA: time × genotype × drug, F4,128 = 6.322, P = 0.0001; Tukey’s multiple-comparison tests. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001. Data shown as mean ± SEM.
Discussion
In this study, we demonstrate that spinal Npy1r-expressing DH interneurons that coexpress Grp are evolutionarily conserved across rodent, nonhuman primate, and human and are necessary for the efficacy of spinally directed Y1 agonists to inhibit neuropathic pain. The Grp subpopulation of Npy1r-expressing DH INs represent an optimal and precise future pharmacotherapeutic target for the treatment of neuropathic pain.
Y1-INs segregate into 3 glutamatergic subpopulations.
Consistent with previous results (25, 26, 36, 46), we found that Y1-INs are almost entirely glutamatergic and segregate into 3 largely nonoverlapping excitatory subpopulations. These are demarcated by coexpression of Npy1r with Grp, Npff, or Cck, consistent with the 3 excitatory subpopulations (Glut2, Glut8, and Glut9) that Häring et al. identified using single-cell RNA sequencing (36). In that study, Cck-expressing neurons fell into 3 separate clusters: Glut1, Glut2, and Glut3. The laminar location of these Cck-expressing subpopulations varied with Glut2 neurons scattered in superficial laminae I-II (the same location we found our Cck/Npy1r-INs), Glut3 neurons forming a compact band in laminae IIi-III, and Glut1 Cck-expressing neurons (the largest population) restricted to the deep dorsal horn (laminae III-V) (36, 56). Thus, our Cck/Npy1r-INs are likely Glut2 neurons with reference to the Häring et al. data set. In the single-cell results, Npff was found exclusively in the Glut9 population; thus, our Npff/Npy1r-INs likely correspond to the Glut9 cluster in the Häring et al. data set (36, 38). In contrast, Grp expression was broadly distributed across the Glut5–12 populations in the Häring et al. data set. However, many of these GRP-INs (~30%) are in the Glut8 cluster (exhibiting Nmur2 and Reln expression but not Npff expression) (36, 40). Therefore, by process of elimination (lack of effect in Cck and Npff subpopulations), the Grp/Npy1r-INs that mediate neuropathic pain and its inhibition by Y1 agonists likely correspond to the Glut8 cluster.
Converging pieces of evidence implicate Glut8 neurons (exhibiting Nmur2 and Reln expression but not Npff expression) as both necessary and sufficient for the manifestation of mechanical and thermal hypersensitivity. First, the neuromedin U receptor 2 (encoded by Nmur2) is coupled to Gαq/11, and intrathecal administration of neuromedin U dose-dependently produces mechanical and heat hypersensitivity (57, 58). Second, Häring et al. found immediate early gene expression in Glut8 neurons in response to both noxious heat and cold stimuli (36). Third, Reln-expressing DH neurons also exhibit Fos expression in response to noxious thermal or mechanical stimulation (59). Fourth, recently uncovered ErbB4+ neurons that participate in spinal heat signaling are exclusively represented in a Nmur2-expressing excitatory neuron subpopulation (60). Last, SST-INs are necessary and sufficient for neuropathic pain (19, 20), and Sst expression is also detected in Glut8 neurons (albeit also in other excitatory populations) (36). Thus, we believe that the Reln/Nmur2/Sst/Grp/Npy1r population in all these studies converges on the Glut8 population and that it is a specific subpopulation that is necessary for neuropathic pain. Therefore, future studies could further investigate the contribution to neuropathic pain of Npff-Glut8 neurons (Reln+, Nmur2+). Future experiments may perhaps synthesize an Nmur2Cre mouse or use an intersectional approach that develops and utilizes a Npy1rFlp mouse crossed with a GrpCre mouse to perform more advanced analyses on the Glut8 neuron subpopulation to better define its role in neuropathic pain.
Grp/Npy1r-expressing DH interneurons mediate the antiallodynic actions of spinally directed NPY Y1 agonists.
Our results establish GRP-INs (specifically those that coexpress Npy1r) as a mediator of neuropathic allodynia. In contrast with our conclusions, GRP-INs were previously described to be exclusively involved in pruritis and not pain-like behavior (52, 61–63). However, previous Grp conclusions were made using a GENSAT BAC transgenic GrpCre mouse line, which only labeled about 25% of DH GRP-INs (40, 62) and prevented the functional interrogation of the entire GRP-IN population. By contrast, in this study we utilized a high-fidelity GrpCre mouse line, which labels approximately 90% of the DH GRP-INs (54). Additionally, for the first time to our knowledge we probed the role of GRP-INs in a chronic pain model (SNI) rather than acute pain modalities (i.e. heat, von Frey, pinprick) (54, 62). With these results, we conclude that GRP-INs are the target of intrathecal Y1 agonists to inhibit SNI-induced mechanical and cold hypersensitivity.
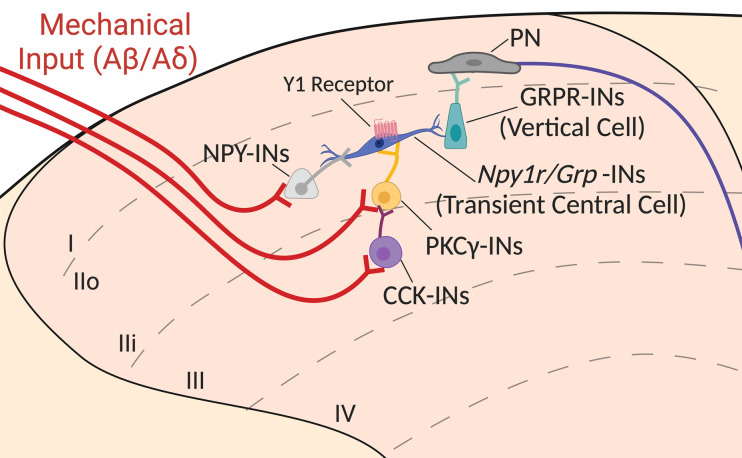
Mechanical allodynia is hypothesized to occur via a polysynaptic DH microcircuit that allows A-fibers to transmit innocuous mechanical input to “pain circuits” (15). Y1-INs and GRP-INs have both been characterized as “transient central cells” (neurons in lamina II outer with a central morphology that discharge action potentials transiently during a depolarizing step; ref. (64) in this circuit (24, 62, 65). We theorize that in the context of nerve injury, GRP-specific Y1-IN transient central cells drive both mechanical allodynia and pathological itch (66–68) via activation of GRPR-INs (Figure 7). Indeed, intrathecal administration of NPY inhibits not only the behavioral signs of neuropathic pain but also chemical and mechanical itch (26, 69, 70). Thus, our results support the idea that neuropathic pain and itch share a spinal circuitry that includes GRP/Y1-INs. Accordingly, a pharmacotherapeutic approach to inhibit GRP/Y1-INs could potentially reduce both chronic neuropathic pain-induced hypersensitivity as well as neuropathic itch (26).
Figure 7. Schematized model for NPY Y1 agonists to inhibit Grp/Npy1r-INs and dampen neuropathic pain, silencing a key component of the ascending circuit in the dorsal horn that mediates mechanical allodynia.
In the context of nerve injury, aberrant hyperexcitation of Npy1r/Grp-INs may drive allodynia. Exogenous administration of NPY or Y1 agonist binding to the Gi-coupled NPY Y1 receptor on Npy1r/Grp-INs results in cellular inhibition and the abolishment of peripheral nerve injury–induced mechanical allodynia. We posit that NPY Y1 agonists act by inhibiting a key neuron population implicated in the transduction of mechanical allodynia. Briefly, non-noxious mechanical stimuli activate Aß/Aδ myelinated afferents (shown in red) that project into the deeper laminae of the dorsal horn and synapse onto interneurons marked by the expression of CCK (purple) and PKCγ (yellow). Normally, feed-forward inhibition prevents the activation of these interneurons, and as a result light touch is perceived as nonpainful. For example, inhibitory NPY interneurons (light gray) may “gate” Npy1r/Grp-INs to prevent these neurons from being activated and driving pain-like behaviors. However, in the context of neuropathic pain, feed-forward inhibition is lost, and innocuous light touch inputs activate a theorized dorsally directed microcircuit to allow innocuous mechanical sensory information to be perceived as painful. In this theorized circuit, activated CCK and PKCγ interneurons excite transient central cells (theorized here as Npy1r/Grp-INs), which in turn synapse onto GRPR-INs (vertical cells), which then activate ascending projection neurons (PNs) that travel via the spinothalamic and spinoparabrachial tracts to be processed via higher order pain centers, such as the lateral parabrachial nucleus. Image is updated from our circuit diagrams previously published under CC BY license (24, 26).
Net DH excitation/inhibition balance or a subpopulation-specific effect.
In addition to our interpretation that intrathecal Y1 agonists act at GRP-INs to inhibit neuropathic pain, an alternative interpretation relates to the appropriate balance of excitation and inhibition (E/I) in the DH, which is critical for appropriate somatosensory processing (2). Peripheral nerve injury produces an enduring shift in the E/I balance of the DH toward an excitatory and pro-nociceptive state (13, 71–76); consequently, inhibition of pain-facilitatory excitatory Y1-INs could restore E/I balance and dampen pathogenic hyperexcitation. In this scenario, it is the total number of glutamatergic pro-nociceptive Y1-INs that are inhibited that is critical for the reduction of neuropathic pain. Therefore, loss of Npyr in the Grp/Npy1r-INs, the largest subpopulation of Y1-INs (~60%) in the superficial DH (Figure 2), might prevent intrathecal NPY from being able to inhibit enough pro-nociceptive Y1-INs to reduce neuropathic hypersensitivity. While this alternative interpretation may be valid, there is evidence to the contrary. For example, inhibition of the larger and predominately excitatory calretinin DH interneuron population (~30% of all superficial dorsal horn neurons are calretinin-immunoreactive, ref. 77; as opposed to ~25% expressing Npy1r, Figure 1) did not reduce peripheral nerve injury–induced allodynia (14). Therefore, we believe that neuronal subpopulation specificity, rather than total neuronal number, underlies the loss of Y1 agonist–induced antiallodynia following conditional knockout of Npy1r in GRP-INs.
Evolutionary conservation of Y1-IN subpopulations.
NPY binding sites were previously reported in the superficial laminae of the DH of mouse, rat, and monkey (29, 34, 78–82). Here, using FISH, we expanded these results to demonstrate Y1-IN subpopulations are conserved across the rodent, rhesus macaque, and human (Figure 3). This evolutionary conservation supports their translational relevance. One caveat is that we evaluated human cervical spinal cord as opposed to lumbar spinal cord in macaque and mouse, and we cannot rule out variance in RNA expression between cervical and lumbar spinal cord. However, a single-nucleosome RNA-sequencing database has recently emerged for the adult human lumbar spinal cord (82), and our FISH results obtained in cervical tissue recapitulate many of the single-cell results. Interestingly, in contrast to the mouse, RNA sequencing showed that NPY1R is broadly expressed in human dorsal horn across all glutamatergic subpopulations (Ex-Dorsal 1–12) (Supplemental Figure 4). However, like the mouse, specific subpopulations coexpress CCK (most densely Ex-Dorsal 2 and 4) or GRP (most densely Ex-Dorsal 11–12) (Supplemental Figure 4). One stark contrast with the mouse and macaque is that NPFF expression was not detected in the human single-nucleosome RNA-sequencing database (Supplemental Figure 4) (82). However, the lack of NPFF expression result further supports the integrity of our cervical FISH results (Figure 3H) and reveals the sparse to nondetectable expression of NPFF in the adult human spinal cord DH. Importantly, the GRP/NPY1R-IN population, the subpopulation that we believe is fundamental to the manifestation of neuropathic pain, is conserved in the macaque and human spinal cord.
NPY Y1/GRP interneurons — a new therapeutic target?
A rich 30-year history of preclinical research implicates NPY at the spinal cord as a potent inhibitor of acute and chronic pain (26), thereby prompting further investigation of Y1-selective therapeutics. Future investigations might test spinally directed NPY Y1 agonists in increasingly more translatable animal models (i.e., macaque, ref. 83) to perform pain behavioral testing as well as side effect profiling analyses. We are optimistic that Y1 agonism will safely and potently reduce the behavioral signs of neuropathic pain in larger mammalian species, and ultimately, humans with chronic pain. Further, our results suggest that GRP/NPY1R-INs are particularly well positioned in the spinal cord to modulate clinical neuropathic pain and thus represent a promising and refined therapeutic target for future drug development. In fact, the human RNA-sequencing data suggest that NPY1R expression is rather nonspecific across excitatory interneurons in humans, and a GRP/NPY1R-selective approach may therefore be optimal.
If GRP/NPY1R-INs are indeed a specific neuronal subpopulation within the human spinal cord that play a crucial role in pain transmission and perception, then the next challenge is to target them effectively and specifically with the delivery of therapeutic agents. A promising future approach is promoter-specific gene therapy (84). Promoters are DNA sequences that determine when and where a gene is expressed. Thus, viral vectors, nanoparticles, or CRISPR/Cas9 gene-editing techniques can be used to introduce GRP promoter-specific therapeutic genes into DH neurons, allowing functional modifications to reduce pain signals. The human single-nucleosome RNA sequencing suggests that NPY1R is broadly expressed across all excitatory neuronal subpopulations and therefore may not represent a precise therapeutic target. Rather, by using the promoter-specific gene therapy approach, we may specifically target the smaller GRP neuronal population and thereby minimize potential side effects of targeting all NPY1R neurons. For example, virally expressed genes could produce proteins that modulate pain signaling, reduce neuronal hyperexcitability, or enhance endogenous pain relief mechanisms. Promoter-specific gene therapy for pain is a promising avenue in the development of novel and effective treatments for chronic pain conditions. Long-term safety and efficacy will need to be thoroughly assessed through preclinical and clinical studies to determine its full potential and applicability in clinical settings.
Methods
Animals
Adult C57BL/6NCrl (Charles River, 027), Npy1rloxP/loxP (courtesy of Herbert Herzog, Garvan Institute, Sydney, New South Wales, Australia; ref. 53), GrpCre (courtesy of Zhou-Feng Chen; Department of Anesthesiology, Washington University School of Medicine, St. Louis, Missouri, USA ref. 85), CckCre (Jackson Laboratory, 012706), and NpffCre (see below; ref. 86) mice were group-housed; provided access to food and water ad libitum; and maintained on a 12-hour light/12-hour dark cycle (lights on at 7 am) in temperature- and humidity-controlled rooms. Male and female mice were used in all experiments. Although we were not powered to detect significant sex differences, no major/obvious trends in sex differences were observed; means from both sexes were pooled.
The NpffCre knockin mouse line (86) was generated by Taconic Biosciences GmbH (Leverkusen, Germany), using a conventional embryonic stem cell targeting strategy and homologous recombination. Briefly, the sequence for the T2A peptide and the open reading frame of improved Cre recombinase (iCre) were inserted between the last amino acid and the translation termination codon in exon 3 of the NPFF gene. A positive selection marker (puromycin resistance) flanked by FRT sites was removed by crossing NpffCre mice with germline Flpe mice. The presence of the T2A sequence should result in cotranslational cleavage between the NPFF and iCre proteins, resulting in coexpression of both proteins, under control of the Npff promoter.
Pharmacological testing
[Leu31, Pro34]-NPY (human, rat) (TOCRIS catalog 1176) was diluted in 0.9% sterile sodium chloride (Medline catalog 63323-186-10) and stored at –20°C. Intrathecal injections of [Leu31, Pro34]-NPY were performed in lightly restrained, unanesthetized mice. Briefly, a 30G needle attached to a Hamilton microsyringe was inserted between the L5/L6 vertebrae at the cauda equina, puncturing the dura (confirmed by presence of reflexive tail flick). We then injected a 5 μL volume of saline or [Leu31, Pro34]-NPY (10 μg/5 μL). Animals were injected twice using a crossover design with 3 to 7 days of separation between the 2 injections. For example, animals receiving saline for the first injection received [Leu31, Pro34]-NPY for the second, and animals receiving [Leu31, Pro34]-NPY for the first injection received saline for the second. In all cases, group means of saline and [Leu31, Pro34]-NPY did not differ on either injection day and so were combined for final analysis.
CNO (TOCRIS catalog 4936) was diluted in 0.9% sterile sodium chloride and stored at room temperature. Intraperitoneal injections of CNO (3 mg/kg) or saline were performed in lightly restrained, unanesthetized mice with a 27G permanently attached needle BD Tuberculin Syringe (catalog 305620). Animals were injected using a crossover design with at least 3 days of separation between saline and CNO injections. In all cases, group means of saline and CNO did not differ on either injection day and so were combined for final analysis.
Surgeries
SNI.
SNI was performed as previously described (23, 24). Briefly, mice were anesthetized with inhaled isoflurane (5% induction and 2% maintenance), and the left hind limb was shaved with trimmers and asepticized with 70% ethanol and a ChloraPrep swabstick (BD catalog 260100). A small incision was made in the skin of the hind left leg, and the underlying muscle was spread via blunt dissection to expose the underlying branches of the sciatic nerve. The peroneal and tibial nerves were then ligated with 6-0 silk sutures (Butler Schein Animal Health catalog NC0049524) and transected while carefully avoiding the sural nerve. The muscle tissue was then loosely sutured with 5-0 vicryl sutures (Med Vet International catalog 50-118-0847), and the skin was closed with 9 mm wound clips (Braintree Scientific catalog NC9281117). Topical triple antibiotic ointment (Neosporin Neomycin Sulfate/Bacitracin Zinc/Polymycin Ointment; Hanna Pharmaceutical Supply Co. catalog NC0100117) was applied to the wound. Wound clips were removed 7–10 days postsurgery, and behavioral experiments began 14 days after surgery.
Intraspinal AAV injections.
Mice were anesthetized with inhaled isoflurane (5% induction and 2% maintenance), and the skin of the back was shaved with trimmers and asepticized with 70% ethanol and a ChloraPrep swabstick. A midline incision was carefully made to allow visualization of the underlying L1 vertebrae. The L1 vertebrae was then removed by laminectomy, exposing the L4 segment of the spinal cord. A glass microelectrode was inserted into 3 separate locations along the rostral caudal axis of the L4 segment: in the middle and near the boundary with L3 and L5. At each injection site, the glass microelectrode was lowered to a depth of 250 μm below the dura using a stereotaxic frame (David Kopf Instruments Model 940). A total of 333.3 nL of virus (AAV8-hSyn-DIO-mCherry, Addgene catalog 50459-AAV8, or AAV8-hSyn-DIO-hM4Di-mCherry, Addgene catalog 44362-AAV8) was slowly injected into each of the 3 spots (5 nL/s) using a Nanoject III (Drummond catalog 3-000-207) with a 3-minute wait time after completion of each injection to permit adequate infusion. The lassimus dorsi was sutured with 5-0 nylon sutures to protect the exposed spinal cord, and the overlying skin was closed with 9 mm wound clips (Braintree Scientific catalog NC9281117). Topical triple antibiotic ointment (Neosporin Neomycin Sulfate/Bacitracin Zinc/Polymycin Ointment; Hanna Pharmaceutical Supply Co. catalog NC0100117) was applied to the wound. Ethiqa XR buprenorphine extended-release injectable suspension (Fidelis Pharmaceuticals catalog FP-001 072117, 3.25 mg/kg, subcutaneous injection) was utilized as a postoperative analgesic. Behavioral experiments began 21 days after surgery.
Behavioral testing
Mechanical withdrawal threshold.
Testing was performed as previously described (24, 87, 88). Mice were habituated to plexiglass chambers with opaque walls (15 × 4 × 4 cm) on a raised wire mesh platform for 30–60 minutes 1 day before and immediately prior to behavioral testing. Testing was performed using a calibrated set of logarithmically increasing von Frey monofilaments (Stoelting catalog 58011) that range in gram force from 0.007 to 6.0 g. Beginning with a 0.4 g filament, these were applied perpendicular to the lateral hind paw surface with sufficient force to cause a slight bending of the filament. A positive response was denoted as a rapid withdrawal of the paw within 4 seconds of application and was followed by application of the next lower filament. A negative response was followed by application of the next higher filament. An up-down method (89) was used to calculate the 50% withdrawal threshold for each mouse.
Cold withdrawal duration.
Immediately following von Frey testing, cold sensitivity testing was performed on mice in the same plexiglass chambers on a raised wire mesh platform. Using a syringe connected to PE-90 tubing, flared at the tip to a diameter of 3.5 mm, we applied a drop of acetone (VWR catalog BDH1101-1LP) to the lateral side of the hind plantar paw (sural nerve innervation territory). Surface tension maintained the volume of the drop to ~10 μL. The duration of time the animal lifted or shook its paw was recorded for 30 seconds. Three observations were averaged.
Hind paw brush for Fos.
To produce Fos activation, a light-touch stimulation protocol was initiated on the ipsilateral hind paw of SNI mice 14 days after nerve injury. Mice were anesthetized with isoflurane (5% induction, 2% maintenance), and the lateral surface of the left hind paw was gently stroked in the longitudinal plane with a cotton-tipped applicator for 3 seconds every 5 seconds, for 5 minutes. After an additional 60-minute awake and freely moving wait time in their home cage, mice received an overdose injection of sodium pentobarbital (Fatal-Plus, Vortech catalog 9373, 0.25 mL, intraperitoneal injection) and were transcardially perfused.
FISH (RNAscope)
Mice.
Mice were transcardially perfused with ice-cold 1× PBS followed by 10% buffered formalin. Spinal cords were extracted via blunt dissection, postfixed in 10% formalin (2–4 hours), and then placed in 30% sucrose at 4°C until the tissue sank (48–72 hours). L3 and L4 were sectioned (20 μm) on a vibrating microtome (Epredia), floated, mounted on Fisherbrand Superfrost plus microscope slides (Thermo Fisher Scientific), and air-dried overnight at room temperature. FISH pretreatment began with a 10-minute xylene bath, 4-minute 100% ethanol bath, and 2-minute RNAscope H2O2 treatment. Next, the FISH protocol for RNAscope Multiplex Fluorescent Detection Kit v2 Assay (Advanced Cell Diagnostics catalog 323100) was followed for hybridization to marker probes described in Table 1. Signal amplification was carried out using TSA Fluorescein (PerkinElmer catalog NEL701A001KT), Cyanine 3 (PerkinElmer catalog NEL704A001KT), and Cyanine 5 (PerkinElmer catalog NEL705A001KT) reagents at a 1:1,000 dilution. Slides were then coverslipped with VECTASHIELD HardSet Antifade Mounting Medium with DAPI (VECTOR Laboratories catalog H-1500-10) or processed for mCherry immunohistochemistry.
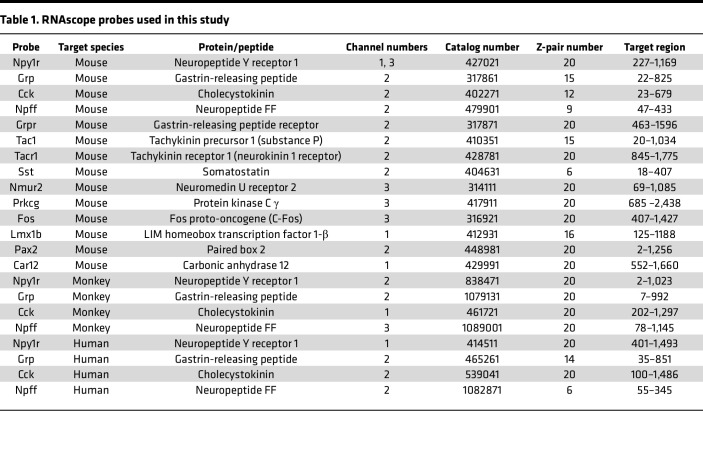
Table 1. RNAscope probes used in this study.
Rhesus macaques.
Two rhesus macaques (male, 4 years at time of death; and female, 4 years at time of death) were provided by David Lewis (University of Pittsburgh) and cared for under the guidelines of the NIH and as approved by the Institutional Animal Care and Use Committee of the University of Pittsburgh. No prior manipulations to the spinal cord were conducted in the macaques. At the time of tissue harvest, macaques were perfused with artificial cerebrospinal fluid, and L5–L7 spinal cord tissue was removed, placed in OCT, and immediately frozen on dry ice. Tissue sections were cut 20 μm thick using a cryostat and mounted onto Superfrost-charged slides, then stored at –80°C. After a 30-minute fixation step with cold 4% paraformaldehyde, the fresh-frozen FISH protocol for RNAscope Multiplex Fluorescent Detection Kit v2 Assay (Advanced Cell Diagnostics catalog 323100) was followed for hybridization to the marker probes described in Table 1. Signal amplification was carried out using TSA Fluorescein, Cyanine 3, and Cyanine 5 reagents (1:1500; Akoya Biosciences). All sections were costained for DAPI and coverslipped at the end of the assay.
Human donor spinal cord tissue.
Two human cervical spinal cord tissues were obtained freshly frozen from the NeuroBioBank, NIH (project no. 063772), and provided by Jill Glausier (University of Pittsburgh). All available evidence indicated that these individuals (male, 45 years at time of accidental death; female, 44 years at time of natural death) were not afflicted with any major psychiatric or neuropathological illnesses at the time of death. All procedures were approved by the Committee for the Oversight of Research and Clinical Training Involving Decedents at University of Pittsburgh, Pittsburgh, Pennsylvania. The fresh-frozen FISH protocol for RNAscope Fluorescent Multiplex Reagent Assay (Advanced Cell Diagnostics catalog 320850) was followed for hybridization to the marker probes described in Table 1. Briefly, 16 μm–thick, fresh-frozen human spinal cord sections were fixed in 4% paraformaldehyde, dehydrated, treated with protease for 15 minutes, and hybridized with gene- and species-specific probes.
Immunohistochemistry
Immediately following the RNAscope V2 protocol, mouse spinal cord sections were washed 3 times in PBS and then pretreated with blocking solution (3% normal goat serum and 0.3% Triton X-100 in PBS) for 1 hour. Sections were then incubated overnight on a slow rocker at 4°C in blocking solution containing the primary antibody (1:2,000, Anti-mCherry; Invitrogen catalog M11217). The sections were washed 3 times in 1× PBS and then incubated in secondary antibody (1:1,000, Alexa Fluor 568 Goat anti-Rat; Invitrogen catalog A11077) for 60 minutes. Finally, sections were washed in 3 times in 1× PBS and then 2 times in 0.01 M phosphate buffer without saline before mounting the tissue to Fisherbrand Superfrost Plus microscope slides (Thermo Fisher Scientific catalog 12-550-15) and coverslipping with VECTASHIELD HardSet Antifade Mounting Medium with DAPI (VECTOR Laboratories catalog H-1500-10).
Microscopy and quantification
All images were captured on a Nikon Eclipse Ti2 microscope using a 20× or 40× objective and analyzed using NIS-Elements Advanced Research software v5.02 (Nikon). Cells with at least 3 puncta associated with a DAPI+ nucleus were considered positive. In mouse, positive cells within the superficial 100 μm of the DH were quantified in 3–5 sections and averaged to yield 1 data point for each mouse. In macaque,s positive cells within the substantia gelatinosa (translucent area in the superficial DH) were quantified, and 1 spinal cord section represents 1 individual data point. Intense lipofuscin prevented quantification in the human spinal cord.
Masking procedures
Experimenter masking (blinding) was employed to promote research rigor. In all cases the experimenter was masked to drug treatments and transgenic mouse genotype. Intrathecal injections were performed by a laboratory colleague, thus providing complete anonymity of drug agent for each animal. In the chemogenetic experiments the experimenter was masked both at the time of surgery to virus (DREADD vs. control) and at the time of behavior to drug agent (CNO vs. saline). The code key was kept hidden in a notebook and not revealed until after the completion of each experiment.
Data presentation
Graphs and images were created in GraphPad Prism 9.0, meta-chart.com (Venn Diagram Maker Online), Adobe Illustrator 26.3, or BioRender.com.
Statistics
Data were graphed and analyzed using GraphPad Prism 9.0 using unpaired 2-tailed t tests for the data of Figure 4, B and C; 2-way ANOVA (mRNA expression × stimulation) followed by a Holm-Šídák test when appropriate for the data of Figure 4E; 3-way RM ANOVA (virus × side × drug as the repeated measure) for the data of Figure 5, D–G; or 3-way RM ANOVA (genotype × drug × time as the repeated measure) followed by Tukey’s test when appropriate for the data of Figure 6, H and I. Statistical significance was determined as P < 0.05. Data are presented as mean ± SEM.
Study approval
All procedures were consistent with the NIH’s Guide for the Care and Use of Laboratory Animals (National Academies Press, 2011), followed the guidelines for the treatment of animals of the International Association for the Study of Pain, and were approved by the Institutional Animal Care and Use Committee and the Committee for the Oversight of Research and Clinical Training Involving Decedents at University of Pittsburgh, Pittsburgh, Pennsylvania.
Data availability
All raw data are included in the Supporting Data Values file. Images are available from the corresponding author upon reasonable request.
Author contributions
TSN, BKT, and AJT designed research; TSN, HNA, PB, PP, EN, CMA, and DFSS performed research; TSN, HNA, PB, PP, BKT, and AJT analyzed data; TSN and BKT wrote the manuscript with contributions from HNA, AJT, and RPS; TSN, HNA, AJT, SER, and BKT received funding for the project; and BKT supervised the overall project.
Supplementary Material
Acknowledgments
This work was supported by NIH grants T32NS073548 (to TSN and HNA); F31NS117054 and F99NS124190 (to TSN); R01DA37621, R01NS45954, and R01NS62306 (to BKT); and R01NS096705 (to SER) and by the Ray and Elizabeth Bloch Charitable Foundation. The NpffCre mouse line was generated with a Wellcome Trust Grant: 102645/Z/13/Z (to AJT). Additionally, we thank Herbert Herzog for providing the Npy1rloxP/loxP mouse line and Zhou-Feng Chen for providing the GrpCre mouse line. We also thank Ronald Sivak (Department of Anesthesiology and Perioperative Medicine, University of Pittsburgh School of Medicine, Pittsburgh, Pennsylvania, USA) for genotyping the mouse lines.
Version 1. 10/12/2023
In-Press Preview
Version 2. 11/22/2023
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Copyright: © 2023, Nelson et al. This is an open access article published under the terms of the Creative Commons Attribution 4.0 International License.
Reference information: JCI Insight. 2023;8(22):e169554.https://doi.org/10.1172/jci.insight.169554.
Contributor Information
Tyler S. Nelson, Email: tn2341@nyu.edu.
Heather N. Allen, Email: ha2686@nyu.edu.
Paramita Basu, Email: pab171@pitt.edu.
Pranav Prasoon, Email: PRANAV25@pitt.edu.
Cynthia M. Arokiaraj, Email: CMA83@pitt.edu.
Diogo F.S. Santos, Email: diogof@pitt.edu.
Rebecca P. Seal, Email: rpseal@pitt.edu.
Sarah E. Ross, Email: saross@pitt.edu.
Andrew J. Todd, Email: Andrew.Todd@glasgow.ac.uk.
Bradley K. Taylor, Email: bkt@pitt.edu.
References
- 1.Merighi A. The histology, physiology, neurochemistry and circuitry of the substantia gelatinosa Rolandi (lamina II) in mammalian spinal cord. Prog Neurobiol. 2018;169:91–134. doi: 10.1016/j.pneurobio.2018.06.012. [DOI] [PubMed] [Google Scholar]
- 2.Todd AJ. Neuronal circuitry for pain processing in the dorsal horn. Nat Rev Neurosci. 2010;11(12):823–836. doi: 10.1038/nrn2947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braz J, et al. Transmitting pain and itch messages: a contemporary view of the spinal cord circuits that generate gate control. Neuron. 2014;82(3):522–536. doi: 10.1016/j.neuron.2014.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Todd AJ. Identifying functional populations among the interneurons in laminae I-III of the spinal dorsal horn. Mol Pain. 2017;13:1744806917693003. doi: 10.1177/1744806917693003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang X, et al. Excitatory superficial dorsal horn interneurons are functionally heterogeneous and required for the full behavioral expression of pain and itch. Neuron. 2013;78(2):312–324. doi: 10.1016/j.neuron.2013.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeilhofer HU, et al. Fast synaptic inhibition in spinal sensory processing and pain control. Physiol Rev. 2012;92(1):193–235. doi: 10.1152/physrev.00043.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finnerup NB, et al. Neuropathic pain: from mechanisms to treatment. Physiol Rev. 2021;101(1):259–301. doi: 10.1152/physrev.00045.2019. [DOI] [PubMed] [Google Scholar]
- 8.Jensen TS, Finnerup NB. Allodynia and hyperalgesia in neuropathic pain: clinical manifestations and mechanisms. Lancet Neurol. 2014;13(9):924–935. doi: 10.1016/S1474-4422(14)70102-4. [DOI] [PubMed] [Google Scholar]
- 9.Peirs C, et al. Recent advances in our understanding of the organization of dorsal horn neuron populations and their contribution to cutaneous mechanical allodynia. J Neural Transm (Vienna) 2020;127(4):505–525. doi: 10.1007/s00702-020-02159-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lolignier S, et al. Mechanical allodynia. Pflugers Arch. 2014;467(1):133–139. doi: 10.1007/s00424-014-1532-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malmberg AB, et al. Preserved acute pain and reduced neuropathic pain in mice lacking PKCgamma. Science. 1997;278(5336):279–283. doi: 10.1126/science.278.5336.279. [DOI] [PubMed] [Google Scholar]
- 12.Miracourt LS, et al. Glycine inhibitory dysfunction turns touch into pain through PKCgamma interneurons. PLoS One. 2007;2(11):e1116. doi: 10.1371/journal.pone.0001116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Petitjean H, et al. Dorsal horn parvalbumin neurons are gate-keepers of touch-evoked pain after nerve injury. Cell Rep. 2015;13(6):1246–1257. doi: 10.1016/j.celrep.2015.09.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Peirs C, et al. Mechanical allodynia circuitry in the dorsal horn is defined by the nature of the injury. Neuron. 2021;109(1):73–90. doi: 10.1016/j.neuron.2020.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu Y, et al. A feed-forward spinal cord glycinergic neural circuit gates mechanical allodynia. J Clin Invest. 2013;123(9):4050–4062. doi: 10.1172/JCI70026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Q, et al. Synaptic dynamics of the feed-forward inhibitory circuitry gating mechanical allodynia in mice. Anesthesiology. 2020;132(5):1212–1228. doi: 10.1097/ALN.0000000000003194. [DOI] [PubMed] [Google Scholar]
- 17.Neumann S, et al. Innocuous, not noxious, input activates PKCgamma interneurons of the spinal dorsal horn via myelinated afferent fibers. J Neurosci. 2008;28(32):7936–7944. doi: 10.1523/JNEUROSCI.1259-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nelson TS. Dorsal horn PKCγ interneurons mediate mechanical allodynia through 5-HT2AR-dependent structural reorganization. J Neurosci. 2019;39(32):6221–6223. doi: 10.1523/JNEUROSCI.0291-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Duan B, et al. Identification of spinal circuits transmitting and gating mechanical pain. Cell. 2014;159(6):1417–1432. doi: 10.1016/j.cell.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christensen AJ, et al. In vivo interrogation of spinal mechanosensory circuits. Cell Rep. 2016;17(6):1699–1710. doi: 10.1016/j.celrep.2016.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Y, et al. Touch and tactile neuropathic pain sensitivity are set by corticospinal projections. Nature. 2018;561(7724):547–550. doi: 10.1038/s41586-018-0515-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maiarù M, et al. Selective neuronal silencing using synthetic botulinum molecules alleviates chronic pain in mice. Sci Transl Med. 2018;10(450):eaar7384. doi: 10.1126/scitranslmed.aar7384. [DOI] [PubMed] [Google Scholar]
- 23.Qi Y, et al. Contribution of μ opioid receptor-expressing dorsal horn interneurons to neuropathic pain-like behavior in mice. Anesthesiology. doi: 10.1097/ALN.0000000000004735. [published online August 11, 2023]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nelson TS, et al. Spinal neuropeptide Y Y1 receptor-expressing neurons are a pharmacotherapeutic target for the alleviation of neuropathic pain. Proc Natl Acad Sci U S A. 2022;119(46):e2204515119. doi: 10.1073/pnas.2204515119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nelson TS, et al. Facilitation of neuropathic pain by the NPY Y1 receptor-expressing subpopulation of excitatory interneurons in the dorsal horn. Sci Rep. 2019;9(1):7248. doi: 10.1038/s41598-019-43493-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nelson TS, Taylor BK. Targeting spinal neuropeptide Y1 receptor-expressing interneurons to alleviate chronic pain and itch. Prog Neurobiol. 2021;196:101894. doi: 10.1016/j.pneurobio.2020.101894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Miyakawa A, et al. Action of neuropeptide Y on nociceptive transmission in substantia gelatinosa of the adult rat spinal dorsal horn. Neuroscience. 2005;134(2):595–604. doi: 10.1016/j.neuroscience.2005.04.045. [DOI] [PubMed] [Google Scholar]
- 28.Melnick IV. Cell type-specific postsynaptic effects of neuropeptide Y in substantia gelatinosa neurons of the rat spinal cord. Synapse. 2012;66(7):640–649. doi: 10.1002/syn.21550. [DOI] [PubMed] [Google Scholar]
- 29.Sinha GP, et al. Fast A-type currents shape a rapidly adapting form of delayed short latency firing of excitatory superficial dorsal horn neurons that express the neuropeptide Y Y1 receptor. J Physiol. 2021;599(10):2723–2750. doi: 10.1113/JP281033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moran TD, et al. Opioid-like actions of neuropeptide Y in rat substantia gelatinosa: Y1 suppression of inhibition and Y2 suppression of excitation. J Neurophysiol. 2004;92(6):3266–3275. doi: 10.1152/jn.00096.2004. [DOI] [PubMed] [Google Scholar]
- 31.Kuphal KE, et al. Y1 receptor knockout increases nociception and prevents the anti-allodynic actions of NPY. Nutrition. 2008;24(9):885–891. doi: 10.1016/j.nut.2008.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Intondi AB, et al. Intrathecal neuropeptide Y reduces behavioral and molecular markers of inflammatory or neuropathic pain. Pain. 2008;137(2):352–365. doi: 10.1016/j.pain.2007.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Malet M, et al. Spinal activation of the NPY Y1 receptor reduces mechanical and cold allodynia in rats with chronic constriction injury. Peptides. 2017;92:38–45. doi: 10.1016/j.peptides.2017.04.005. [DOI] [PubMed] [Google Scholar]
- 34.Brumovsky P, et al. The neuropeptide tyrosine Y1R is expressed in interneurons and projection neurons in the dorsal horn and area X of the rat spinal cord. Neuroscience. 2006;138(4):1361–1376. doi: 10.1016/j.neuroscience.2005.11.069. [DOI] [PubMed] [Google Scholar]
- 35.Brumovsky P, et al. Neuropeptide tyrosine and pain. Trends Pharmacol Sci. 2007;28(2):93–102. doi: 10.1016/j.tips.2006.12.003. [DOI] [PubMed] [Google Scholar]
- 36.Häring M, et al. Neuronal atlas of the dorsal horn defines its architecture and links sensory input to transcriptional cell types. Nat Neurosci. 2018;21(6):869–880. doi: 10.1038/s41593-018-0141-1. [DOI] [PubMed] [Google Scholar]
- 37.Gutierrez-Mecinas M, et al. A quantitative study of neurochemically defined excitatory interneuron populations in laminae I-III of the mouse spinal cord. Mol Pain. 2016;12:1744806916629065. doi: 10.1177/1744806916629065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gutierrez-Mecinas M, et al. Expression of neuropeptide FF defines a population of excitatory interneurons in the superficial dorsal horn of the mouse spinal cord that respond to noxious and pruritic stimuli. Neuroscience. 2019;416:281–293. doi: 10.1016/j.neuroscience.2019.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Polgár E, et al. Grpr expression defines a population of superficial dorsal horn vertical cells that have a role in both itch and pain. Pain. 2023;164(1):149–170. doi: 10.1097/j.pain.0000000000002677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bell AM, et al. Expression of green fluorescent protein defines a specific population of lamina II excitatory interneurons in the GRP:eGFP mouse. Sci Rep. 2020;10(1):13176. doi: 10.1038/s41598-020-69711-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng L, et al. Lbx1 and Tlx3 are opposing switches in determining GABAergic versus glutamatergic transmitter phenotypes. Nat Neurosci. 2005;8(11):1510–1515. doi: 10.1038/nn1569. [DOI] [PubMed] [Google Scholar]
- 42.Hernandez-Miranda LR, et al. The dorsal spinal cord and hindbrain: From developmental mechanisms to functional circuits. Dev Biol. 2017;432(1):34–42. doi: 10.1016/j.ydbio.2016.10.008. [DOI] [PubMed] [Google Scholar]
- 43.Del Barrio MG, et al. A transcription factor code defines nine sensory interneuron subtypes in the mechanosensory area of the spinal cord. PLoS One. 2013;8(11):e77928. doi: 10.1371/journal.pone.0077928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Szabo NE, et al. Hoxb8 intersection defines a role for Lmx1b in excitatory dorsal horn neuron development, spinofugal connectivity, and nociception. J Neurosci. 2015;35(13):5233–5246. doi: 10.1523/JNEUROSCI.4690-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Russ DE, et al. A harmonized atlas of mouse spinal cord cell types and their spatial organization. Nat Commun. 2021;12(1):5722. doi: 10.1038/s41467-021-25125-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sathyamurthy A, et al. Transcriptional repressor HIC1 contributes to suppressive function of human induced regulatory T cells. Cell Rep. 2018;22(8):2094–2106. doi: 10.1016/j.celrep.2018.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peirs C, et al. Dorsal horn circuits for persistent mechanical pain. Neuron. 2015;87(4):797–812. doi: 10.1016/j.neuron.2015.07.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Torres R, et al. Mice genetically deficient in neuromedin U receptor 2, but not neuromedin U receptor 1, have impaired nociceptive responses. Pain. 2007;130(3):267–278. doi: 10.1016/j.pain.2007.01.036. [DOI] [PubMed] [Google Scholar]
- 49.Chamessian A, et al. Transcriptional profiling of somatostatin interneurons in the spinal dorsal horn. Sci Rep. 2018;8(1):6809. doi: 10.1038/s41598-018-25110-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Decosterd I, Woolf CJ. Spared nerve injury: an animal model of persistent peripheral neuropathic pain. Pain. 2000;87(2):149–158. doi: 10.1016/S0304-3959(00)00276-1. [DOI] [PubMed] [Google Scholar]
- 51.Jendryka M, et al. Pharmacokinetic and pharmacodynamic actions of clozapine-N-oxide, clozapine, and compound 21 in DREADD-based chemogenetics in mice. Sci Rep. 2019;9(1):4522. doi: 10.1038/s41598-019-41088-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kiguchi N, et al. Chemogenetic activation of central gastrin-releasing peptide-expressing neurons elicits itch-related scratching behavior in male and female mice. Pharmacol Res Perspect. 2021;9(3):e00790. doi: 10.1002/prp2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Howell OW, et al. Neuropeptide Y is neuroproliferative for post-natal hippocampal precursor cells. J Neurochem. 2003;86(3):646–659. doi: 10.1046/j.1471-4159.2003.01895.x. [DOI] [PubMed] [Google Scholar]
- 54.Barry DM, et al. Exploration of sensory and spinal neurons expressing gastrin-releasing peptide in itch and pain related behaviors. Nat Commun. 2020;11(1):1397. doi: 10.1038/s41467-020-15230-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Taniguchi H, et al. A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron. 2011;71(6):995–1013. doi: 10.1016/j.neuron.2011.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gutierrez-Mecinas M, et al. Expression of cholecystokinin by neurons in mouse spinal dorsal horn. J Comp Neurol. 2019;527(11):1857–1871. doi: 10.1002/cne.24657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu XH, et al. Pro-nociceptive effects of neuromedin U in rat. Neuroscience. 2003;120(2):467–474. doi: 10.1016/S0306-4522(03)00300-2. [DOI] [PubMed] [Google Scholar]
- 58.Cao CQ, et al. A pro-nociceptive role of neuromedin U in adult mice. Pain. 2003;104(3):609–616. doi: 10.1016/S0304-3959(03)00118-0. [DOI] [PubMed] [Google Scholar]
- 59.Yvone GM, et al. Disabled-1 dorsal horn spinal cord neurons co-express Lmx1b and function in nociceptive circuits. Eur J Neurosci. 2017;45(5):733–747. doi: 10.1111/ejn.13520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang H, et al. A novel spinal neuron connection for heat sensation. Neuron. 2022;110(14):2315–2333. doi: 10.1016/j.neuron.2022.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chen Z-F. A neuropeptide code for itch. Nat Rev Neurosci. 2021;22(12):758–776. doi: 10.1038/s41583-021-00526-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Albisetti GW, et al. Dorsal horn gastrin-releasing peptide expressing neurons transmit spinal itch but not pain signals. J Neurosci. 2019;39(12):2238–2250. doi: 10.1523/JNEUROSCI.2559-18.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pagani M, et al. How gastrin-releasing peptide opens the spinal gate for itch. Neuron. 2019;103(1):102–117. doi: 10.1016/j.neuron.2019.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grudt TJ, Perl ER. Correlations between neuronal morphology and electro-physiological features in the rodent superficial dorsal horn. J Physiol. 2002;540(pt 1):189–207. doi: 10.1113/jphysiol.2001.012890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dickie AC, et al. Morphological and functional properties distinguish the substance P and gastrin-releasing peptide subsets of excitatory interneuron in the spinal cord dorsal horn. Pain. 2019;160(2):442–462. doi: 10.1097/j.pain.0000000000001406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Steinhoff M, et al. Clinical presentation, management, and pathophysiology of neuropathic itch. Lancet Neurol. 2018;17(8):709–720. doi: 10.1016/S1474-4422(18)30217-5. [DOI] [PubMed] [Google Scholar]
- 67.Meixiong J, et al. Neuropathic itch. Cells. 2020;9(10):2263. doi: 10.3390/cells9102263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Misery L, et al. Neuropathic pruritus. Nat Rev Neurol. 2014;10(7):408–416. doi: 10.1038/nrneurol.2014.99. [DOI] [PubMed] [Google Scholar]
- 69.Jakobsson JET, et al. Neuropeptide Y in itch regulation. Neuropeptides. 2019;78:101976. doi: 10.1016/j.npep.2019.101976. [DOI] [PubMed] [Google Scholar]
- 70.Gao T, et al. The neuropeptide Y system regulates both mechanical and histaminergic itch. J Invest Dermatol. 2018;138(11):2405–2411. doi: 10.1016/j.jid.2018.05.008. [DOI] [PubMed] [Google Scholar]
- 71.Zeilhofer HU, et al. Chronic pain states: pharmacological strategies to restore diminished inhibitory spinal pain control. Annu Rev Pharmacol Toxicol. 2012;52:111–133. doi: 10.1146/annurev-pharmtox-010611-134636. [DOI] [PubMed] [Google Scholar]
- 72.Lee KY, et al. Excitatory neurons are more disinhibited than inhibitory neurons by chloride dysregulation in the spinal dorsal horn. Elife. 2019;8:e49753. doi: 10.7554/eLife.49753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gong N, et al. Functional reorganization of local circuit connectivity in superficial spinal dorsal horn with neuropathic pain states. eNeuro. 2019;6(5):ENEURO.0272–19.2019. doi: 10.1523/ENEURO.0272-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Inquimbert P, et al. Peripheral nerve injury produces a sustained shift in the balance between glutamate release and uptake in the dorsal horn of the spinal cord. Pain. 2012;153(12):2422–2431. doi: 10.1016/j.pain.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Leitner J, et al. Impaired excitatory drive to spinal GABAergic neurons of neuropathic mice. PLoS One. 2013;8(8):e73370. doi: 10.1371/journal.pone.0073370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Alles SRA, Smith PA. Etiology and pharmacology of neuropathic pain. Pharmacol Rev. 2018;70(2):315–347. doi: 10.1124/pr.117.014399. [DOI] [PubMed] [Google Scholar]
- 77.Smith KM, et al. Distinct forms of synaptic inhibition and neuromodulation regulate calretinin-positive neuron excitability in the spinal cord dorsal horn. Neuroscience. 2016;326:10–21. doi: 10.1016/j.neuroscience.2016.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Zhang X, et al. Neuropeptide Y and galanin binding sites in rat and monkey lumbar dorsal root ganglia and spinal cord and effect of peripheral axotomy. Eur J Neurosci. 1995;7(3):367–380. doi: 10.1111/j.1460-9568.1995.tb00332.x. [DOI] [PubMed] [Google Scholar]
- 79.Zhang X, et al. Localization of neuropeptide Y Y1 receptors in the rat nervous system with special reference to somatic receptors on small dorsal root ganglion neurons. Proc Natl Acad Sci U S A. 1994;91(24):11738–11742. doi: 10.1073/pnas.91.24.11738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ji R, et al. Expression of neuropeptide Y and neuropeptide Y (Y1) receptor mRNA in rat spinal cord and dorsal root ganglia following peripheral tissue inflammation. J Neurosci. 1994;14(11):6423–6434. doi: 10.1523/JNEUROSCI.14-11-06423.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kopp J, et al. Expression of the neuropeptide Y Y1 receptor in the CNS of rat and of wild-type and Y1 receptor knock-out mice. Focus on immunohistochemical localization. Neuroscience. 2010;111(3):443–532. doi: 10.1016/S0306-4522(01)00463-8. [DOI] [PubMed] [Google Scholar]
- 82.Kar S, Quirion R. Quantitative autoradiographic localization of [125I]neuropeptide Y receptor binding sites in rat spinal cord and the effects of neonatal capsaicin, dorsal rhizotomy and peripheral axotomy. Brain Res. 1992;574(1–2):333–337. doi: 10.1016/0006-8993(92)90836-X. [DOI] [PubMed] [Google Scholar]
- 83.Hama AT, et al. Beyond rodent models of pain: non-human primate models for evaluating novel analgesic therapeutics and elaborating pain mechanisms. CNS Neurol Disord Drug Targets. 2013;12(8):1257–1270. doi: 10.2174/18715273113129990111. [DOI] [PubMed] [Google Scholar]
- 84.Chen C, et al. Promoter-operating targeted expression of gene therapy in cancer: current stage and prospect. Mol Ther Nucleic Acids. 2018;11:508–514. doi: 10.1016/j.omtn.2018.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kim AY, et al. Pirt, a phosphoinositide-binding protein, functions as a regulatory subunit of TRPV1. Cell. 2008;133(3):475–485. doi: 10.1016/j.cell.2008.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Quillet R, et al. Characterisation of NPFF-expressing neurons in the superficial dorsal horn of the mouse spinal cord. Sci Rep. 2023;13(1):5891. doi: 10.1038/s41598-023-32720-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nelson TS, et al. Endogenous μ-opioid-neuropeptide Y Y1 receptor synergy silences chronic postoperative pain in mice. PNAS Nexus. 2023;2(8):pgad261. doi: 10.1093/pnasnexus/pgad261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fu W, et al. An NPY Y1 receptor antagonist unmasks latent sensitization and reveals the contribution of protein kinase A and Epac to chronic inflammatory pain. Pain. 2019;160(8):1754–1765. doi: 10.1097/j.pain.0000000000001557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Chaplan SR, et al. Quantitative assessment of tactile allodynia in the rat paw. J Neurosci Methods. 1994;53(1):55–63. doi: 10.1016/0165-0270(94)90144-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data are included in the Supporting Data Values file. Images are available from the corresponding author upon reasonable request.