Abstract
The bZIP transcription factor ABSCISIC ACID INSENSITIVE5 (ABI5) is a master regulator of seed germination and post-germinative growth in response to abscisic acid (ABA), but the detailed molecular mechanism by which it represses plant growth remains unclear. In this study, we used proximity labeling to map the neighboring proteome of ABI5 and identified FCS-LIKE ZINC FINGER PROTEIN 13 (FLZ13) as a novel ABI5 interaction partner. Phenotypic analysis of flz13 mutants and FLZ13-overexpressing lines demonstrated that FLZ13 acts as a positive regulator of ABA signaling. Transcriptomic analysis revealed that both FLZ13 and ABI5 downregulate the expression of ABA-repressed and growth-related genes involved in chlorophyll biosynthesis, photosynthesis, and cell wall organization, thereby repressing seed germination and seedling establishment in response to ABA. Further genetic analysis showed that FLZ13 and ABI5 function together to regulate seed germination. Collectively, our findings reveal a previously uncharacterized transcriptional regulatory mechanism by which ABA mediates inhibition of seed germination and seedling establishment.
Key words: ABA, ABI5, FLZ, gene expression, seed germination
FLZ13 physically interacts with ABI5 to downregulate the expression of ABA-repressed and growth-related genes, thereby inhibiting seed germination and seedling establishment in response to ABA.
Introduction
Abscisic acid (ABA), a sesquiterpene compound discovered in the 1960s (Eagles and Wareing, 1963; Ohkuma et al., 1963; Cornforth et al., 1965; Addicott et al., 1968), is important for plant stress tolerance, growth, and development (Leung and Giraudat, 1998; Holdsworth et al., 2008; Cutler et al., 2010; Chen et al., 2020). ABA is perceived by the soluble ABA receptor PYRABACTIN RESISTANCE1 (PYR1)/PYR1-LIKE (PYL)/REGULATORY COMPONENT OF ABA RECEPTOR (RCAR) (Ma et al., 2009; Nishimura et al., 2009; Park et al., 2009; Santiago et al., 2009). Upon ABA binding, these receptors can form a complex with TYPE 2C PROTEIN PHOSPHATASES (PP2Cs), thereby relieving PP2C inhibition of SUCROSE NONFERMENTING-1-RELATED PROTEIN KINASE 2s (SnRK2s) from subfamily III, including SnRK2.2, SnRK2.3, and SnRK2.6 (Ma et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009; Park et al., 2009; Cutler et al., 2010; Chen et al., 2020). In addition, B2- and B3-type Raf protein kinases directly phosphorylate and activate subclass III SnRK2s (Saruhashi et al., 2015; Nguyen et al., 2019; Katsuta et al., 2020; Takahashi et al., 2020; Lin et al., 2021). Activated SnRK2s phosphorylate and activate downstream transcription factors such as ABA-responsive element (ABRE)-binding protein/ABRE-binding factors (AREB/ABFs), which subsequently modulate the expression of ABA-responsive genes (Kobayashi et al., 2005; Furihata et al., 2006; Fujii et al., 2007; Fujii and Zhu, 2009; Nakashima et al., 2009).
ABA INSENSITIVE 5 (ABI5), a bZIP transcription factor that is preferentially expressed in dry seeds and strongly induced by exogenous ABA, plays a critical role in ABA-mediated seed germination and post-germinative growth in Arabidopsis (Finkelstein, 1994; Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Lopez-Molina et al., 2001, 2002; Brocard et al., 2002; Finkelstein et al., 2005; Yu et al., 2015; Skubacz et al., 2016). ABI5 loss-of-function mutants are hyposensitive to ABA, whereas plants overexpressing ABI5 display hypersensitivity to ABA (Finkelstein, 1994; Finkelstein and Lynch, 2000; Lopez-Molina and Chua, 2000; Zhou et al., 2015). As a transcription factor, ABI5 functions mainly by modulating the expression of its target genes (Skubacz et al., 2016). Previous studies have identified thousands of ABI5-targeted genes (Lee et al., 2012; Nakabayashi et al., 2005; Nakashima et al., 2009; Reeves et al., 2011; O'Malley et al., 2016); however, the detailed functions of these genes in ABA-regulated seed germination remain elusive. For example, EARLY METHIONINE-LABELED 6 (Em6) is a well-known ABA-induced target gene of ABI5 (Carles et al., 2002), but the em6-1 mutant does not display an obvious defect in ABA-repressed seed germination (Manfre et al., 2006). Recently, Huang et al. reported that PHOSPHATE1 (PHO1), an important regulator of phosphorus transport in Arabidopsis (Hamburger et al., 2002), is an ABA-repressed gene that is targeted by ABI5 to control seed germination (Huang et al., 2017). Although loss-of-function mutations in PHO1 completely abolish the ABA-hyposensitive phenotype of the abi5-8 mutant (Zhou et al., 2015), the function of PHO1 in ABA-inhibited seed germination remains unknown (Huang et al., 2017). Thus, the mechanisms underlying ABI5-mediated ABA responses during seed germination and seedling establishment require further characterization.
ABI5 is often associated with other transcriptional regulators, such as ABI3, DELLA, BRI1-EMS-SUPPRESSOR1 (BES1), JASMONATE ZIM-DOMAIN (JAZ) proteins, VQ18/26, and INDUCER OF CBF EXPRESSION1 (ICE1), to regulate ABA responses (Lim et al., 2013; Pan et al., 2018; Hu et al., 2019; Ju et al., 2019; Zhao et al., 2019). Most of the aforementioned interactors function as negative regulators to suppress the transcriptional regulatory activity of ABI5, whereas the positive regulators of ABI5 are less well understood. Several studies have demonstrated the formation of an ABI3–ABI5 complex to activate downstream genes containing ABREs (Nakamura et al., 2001; Gampala et al., 2002; Finkelstein et al., 2005), and other studies have suggested that ABI5 can act as a transcription factor without ABI3 (Lopez-Molina et al., 2002). Thus, ABI5 can rescue the abi3-1 mutant and act downstream of ABI3 to induce growth arrest (Lopez-Molina et al., 2002). Recently, the circadian clock proteins PSEUDO-RESPONSE REGULATOR 5 (PRR5) and PRR7 were found to interact with and stimulate ABI5 to positively modulate ABA signaling during seed germination (Yang et al., 2021). Despite these findings, details of the transcriptional regulatory mechanisms of ABI5 and its cofactors remain to be fully characterized.
Previous studies have reported that endospermic ABA represses expression of cutin biosynthetic genes in the embryo through ABI5 action, which favors ABA-mediated inhibition of the embryo-to-seedling transition (De Giorgi et al., 2015). Our recent study revealed that the plant-specific endosomal sorting complex required for transport (ESCRT) component FYVE DOMAIN PROTEIN REQUIRED FOR ENDOSOMAL SORTING 1 (FREE1) physically interacts with ABI5 and interferes with its DNA-binding ability to negatively regulate seedling establishment in the presence of ABA (Li et al., 2019, 2022a). To gain further insight into the mechanism of ABI5-engaged transcriptional regulation, we used TurboID, an enzyme with high activity in protein proximity labeling (Mair et al., 2019), to map the neighboring proteome of ABI5 and identified a hitherto unknown ABI5-interacting protein, FCS-LIKE ZINC FINGER PROTEIN 13 (FLZ13). Further studies showed that FLZ13 functions together with ABI5 to determine the transcription of ABA-repressed and growth-related genes, thus positively modulating ABA responses during seed germination and post-germinative growth.
Results
Proximity labeling proteomics identifies FLZ13 as a novel ABI5-interacting protein
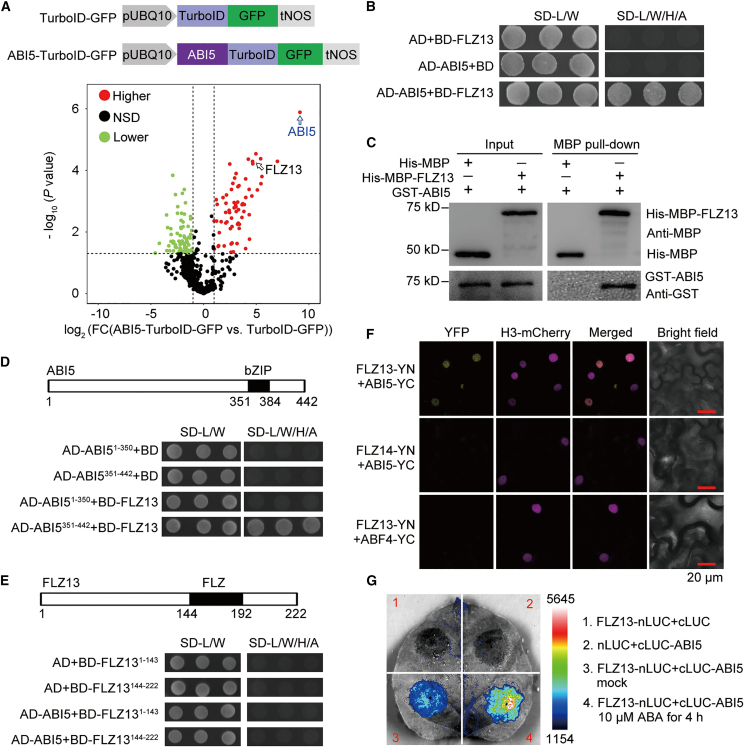
To map the neighboring proteome of ABI5, we generated transgenic plants expressing either the ABI5-TurboID-GFP fusion or TurboID-GFP as a control in the Col-0 background (Figure 1A). The ABI5-TurboID-GFP fusion was functional in planta, because ABI5-TurboID-GFP was correctly localized in the nucleus and its overexpressing seedlings were hypersensitive to 0.5 μM ABA treatment compared with the TurboID-GFP plants (Supplemental Figure 1). Five-day-old seedlings were incubated with 100 μM ABA for 4 h, then treated with 50 μM biotin for another 1 h before being harvested to isolate total proteins for affinity purification with streptavidin-coated beads, followed by label-free quantitative mass spectrometry. Using P value <0.05 and fold change >2 as cutoffs, we obtained 67 ABI5-specific prey (Figure 1A; Supplemental Table 1). Among them, ABI5 BINDING PROTEIN1 (AFP1) and FREE1 were previously reported as ABI5-interacting proteins (Lopez-Molina et al., 2003; Garcia et al., 2008; Li et al., 2019), indicating that our TurboID-based proximity labeling was effective in isolating ABI5 interactors. To gain an overview of these candidates, 48 nuclear-localized proteins out of the 67 highly enriched proteins were used to construct a protein–protein interaction (PPI) network to show the known interlinks among them (Supplemental Table 2). As shown in Supplemental Figure 2A, 44 of 48 nuclear proteins were included in this network, indicating that they were indeed associated in plant cells. To enrich the ABI5-interacting network, a broader network was generated using the 48 nuclear proteins identified in this study with 37 known ABI5-interacting proteins (Supplemental Table 3). Eighty-two out of 83 unique proteins could be connected in this network (Supplemental Figure 2B), indicating that ABI5 potentially associated with diverse proteins to generate multiple functional complexes and that TurboID-based proximity labeling is a powerful approach for identifying the protein interactome in plants.
Figure 1.
Identification of FLZ13 as a novel interactor of ABI5.
(A) Volcano plot showing protein abundances in the ABI5-TurboID-GFP vs. TurboID-GFP pair. The constructs encoding the ABI5-TurboID-GFP and TurboID-GFP fusion proteins are shown. Integrated label-free quantitation (LFQ) peptide intensity data from three biological replicates of each sample were normalized and subjected to ratiometric analysis and plotting using the Perseus program. In total, 611 proteins identified in both the ABI5-TurboID-GFP and TurboID-GFP samples were plotted. Preys with fold change >2 (log2(FC) > 1) and P < 0.05 (−log10[P value] > 1.301) are indicated by colored dots. Green and red dots represent significantly decreased and increased protein levels, respectively. NSD stands for no significant difference.
(B) Y2H assay showing the interaction between ABI5 and FLZ13. Protein interactions were determined by growth of yeast cells co-transformed with various combinations of plasmids on synthetic dropout medium lacking Leu and Trp (SD-L/W) and lacking Leu, Trp, His, and Ade (SD-L/W/H/A), as indicated.
(C)In vitro pull-down assay showing physical binding of ABI5 and FLZ13. Equal amounts of GST-ABI5 were incubated with His-MBP and His-MBP-FLZ13 bound to MBP magnetic beads and then eluted and analyzed by immunoblotting using anti-GST and anti-MBP antibodies.
(D and E) Mapping of the interaction region of ABI5 and FLZ13 by Y2H. Diagrams of the ABI5 (D) and FLZ13 (E) protein structures are shown. Protein interactions were determined by growth of yeast cells co-transformed with various combinations of plasmids on synthetic dropout medium lacking Leu and Trp (SD-L/W) and lacking Leu, Trp, His, and adenine (SD-L/W/H/A), as indicated.
(F) BiFC assay revealing the association between ABI5 and FLZ13 in the nuclei of N. benthamiana leaves. FLZ13 fused to the N-terminal half of YFP (FLZ13-YN) was co-infiltrated with ABI5 fused to the C-terminal half of YFP (ABI5-YC). The FLZ14-YN/ABI5-YC and FLZ13-YN/ABF4-YC pairs were used as negative controls. H3, histone 3.
(G) LCI assay showing the interaction between ABI5 and FLZ13 in N. benthamiana leaves. Agrobacteria harboring FLZ13-nLUC (N-terminal part of luciferase) were co-infiltrated with Agrobacteria harboring cLUC-ABI5 into N. benthamiana leaves. FLZ13-nLUC/cLUC and cLUC-ABI5/nLUC pairs were used as negative controls. After 36 h of co-infiltration, leaves were treated with mock and 10 μM ABA for 4 h, followed by luminescence imaging.
Among the ABI5-neighboring proteins identified in this study, FLZ13 is the most interesting because its gene expression is induced by ABA (Supplemental Table 2) and its molecular function has not been characterized. FLZs are a group of plant-specific regulatory proteins containing a common FCS-LIKE ZINC FINGER domain (FLZ; also known as Domain of Unknown Function 581) (Jamsheer and Laxmi, 2015) whose biological functions are just beginning to be explored. Therefore, we chose FLZ13 as an example and characterized its function in the ABI5-mediated signaling pathway. Yeast two-hybrid (Y2H) analysis showed that FLZ13 could specifically interact with ABI5 but not with other transcription factors in ABA signaling, including ABI3, ABI4, ABF2, and ABF4 (Figure 1B and SupplementalFigure 3). The direct interaction between FLZ13 and ABI5 was further corroborated by an in vitro pull-down assay in which GST-ABI5 was specifically precipitated by His-MBP-FLZ13 but not by His-MBP alone (Figure 1C). Additional Y2H assays with truncated proteins showed that the C-terminal part of ABI5 (351–442 amino acids) harboring a bZIP domain, but not the N-terminal part of ABI5 (1–350 amino acids), was able to bind FLZ13 (Figure 1D). Furthermore, truncated FLZ13 lacking either the N-terminal or C-terminal half failed to interact with ABI5 (Figure 1E). These Y2H results suggest that the C-terminal bZIP domain of ABI5 mediates its interaction with full-length FLZ13. To confirm the interaction between ABI5 and FLZ13 in planta, we performed bimolecular fluorescence complementation (BiFC) and luciferase complementation imaging (LCI) assays. The BiFC results revealed that ABI5 interacted with FLZ13 in the nucleus (Figure 1F). Furthermore, co-expression of FLZ13-nLUC with cLUC-ABI5 in tobacco leaves resulted in obvious LUC signals (Figure 1G and Supplemental Figure 4) that were significantly increased by ABA treatment, indicating that ABA may promote the formation of the ABI5–FLZ13 complex (Figure 1G and Supplemental Figure 4). Collectively, these results clearly demonstrate that FLZ13 physically interacts with ABI5, suggesting that FLZ13 might function as a working partner of ABI5 to modulate ABA signaling.
FLZ13 is responsive to ABA in germinating seeds and positively regulates ABA response
Given that FLZ13 is a novel interactor of ABI5, we examined the transcription and protein expression of FLZ13 during seed germination with or without exogenous ABA treatment. Consistent with previous reports (Lopez-Molina et al., 2001; Brocard et al., 2002), the transcription of ABI5 decreased substantially following seed germination but increased upon 0.5 μM ABA treatment (Supplemental Figures 5A and 5B). However, FLZ13 displayed a slightly increased expression pattern during seed germination and ABA treatment (Supplemental Figures 5A and 5B). To determine the protein abundance of ABI5 and FLZ13 in germinating seeds, we generated transgenic Arabidopsis lines expressing a pFLZ13::FLZ13-GFP fusion in the Col-0 background. As presented in Supplemental Figure 5C, the ABI5 protein showed remarkably decreased expression following seed germination, whereas the FLZ13-GFP protein displayed a gradual accumulation. When germinating seeds were exposed to 0.5 μM ABA, the protein levels of FLZ13-GFP did not change significantly compared with the control group (0 μM ABA), whereas ABI5 levels clearly increased (Supplemental Figure 5C). These results indicate the co-existence of FLZ13 and ABI5 in the presence of ABA, meeting the requirement for their interaction in planta.
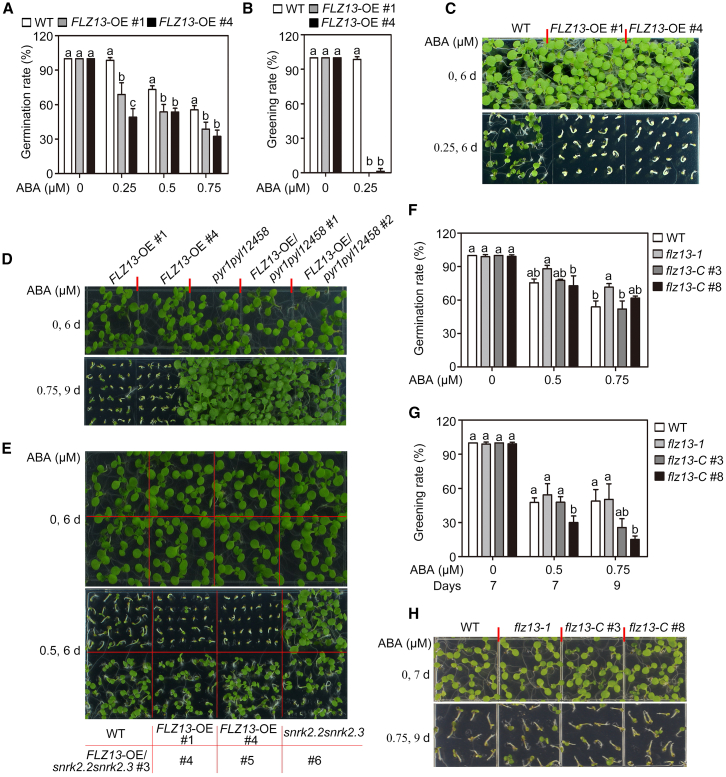
The biological function of FLZ13 in Arabidopsis has not yet been documented. As FLZ13 physically interacts with ABI5 (Figure 1) and responds to ABA in germinating seeds (Supplemental Figure 5), we hypothesized that FLZ13 might be involved in ABI5-mediated ABA signaling during seed germination. To test this possibility, we first generated FLZ13 overexpression (OE) transgenic plants in the Col-0 background. Two representative homozygous lines (pUBQ10::FLZ13-GFP #1 and pUBQ10::FLZ13-GFP #4, termed FLZ13-OE #1 and OE #4) with higher FLZ13 gene expression levels under normal growth conditions were chosen for further study (Supplemental Figure 6). As shown in Figure 2A–2C, seed germination and seedling growth of wild-type (WT) and FLZ13-OE lines did not show obvious differences in medium without exogenous ABA, whereas seeds of FLZ13-OE lines had much lower germination and greening rates than Col-0 in the presence of ABA, suggesting that overexpression of FLZ13 conferred ABA hypersensitivity on seed germination and seedling establishment. We also overexpressed FLZ13 in pyr1pyl1pyl2pyl4pyl5pyl8 (pyr1pyl12458) and snrk2.2snrk2.3 backgrounds for genetic analysis of FLZ13 and other components involved in ABA signaling (Fujii et al., 2007; Gonzalez-Guzman et al., 2012). The FLZ13-OE/pyr1pyl12458 and FLZ13-OE/snrk2.2snrk2.3 lines almost phenocopied the pyr1pyl12458 and snrk2.2snrk2.3 mutants, respectively (Figure 2D and 2E), suggesting that a functional ABA signaling pathway is required to show the effects of FLZ13 overexpression on ABA hypersensitivity.
Figure 2.
FLZ13 positively regulates ABA response during seed germination.
(A) Germination rates of the WT and FLZ13-OE lines. Seed germination rates were recorded 4 days after plating on ½ MS medium supplemented with the indicated concentrations of ABA. Data are presented as mean ± SD (n = 3 biological replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
(B) Cotyledon greening rates of the WT and FLZ13-OE lines. Cotyledon greening rates were scored 6 days after plating on ½ MS medium supplemented with 0 or 0.25 μM ABA. Data are presented as mean ± SD (n = 3 biological replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
(C) Photographs of seedlings of WT and two FLZ13-OE lines germinated on ½ MS medium containing 0 or 0.25 μM ABA for 6 days.(D) Photographs of seedlings of FLZ13-OE, pyr1pyl12458 (pyr1pyl1pyl2pyl4pyl5pyl8), and FLZ13-OE/pyr1pyl12458 lines after 0 μM ABA treatment for 6 days or 0.75 μM ABA treatment for 9 days.(E) Photographs of seedlings of WT, FLZ13-OE, snrk2.2snrk2.3, and FLZ13-OE/snrk2.2snrk2.3 lines after 0 or 0.5 μM ABA treatment for 6 days.(F) Germination rates of the WT, flz13-1, and two gFLZ13/flz13-1 complementation lines (flz13-C #3 and flz13-C #8). Seed germination rates were recorded 4 days after plating on ½ MS medium supplemented with various concentrations of ABA, as indicated. Data are presented as mean ± SD (n = 3 biological replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
(G) Cotyledon greening rates of WT, flz13-1, and two gFLZ13/flz13-1 complementation lines (flz13-C #3 and flz13-C #8) treated with various ABA concentrations as recorded at the indicated time points. Data are presented as mean ± SD (n = 3 biological replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
(H) Photographs of seedlings of WT, flz13-1, and two gFLZ13/flz13-1 complementation lines (flz13-C #3 and flz13-C #8) after treatment with 0 μM ABA for 7 days or 0.75 μM ABA for 9 days.
To further confirm the role of FLZ13 in ABA signaling, we obtained an FLZ13 transfer DNA insertion mutant (flz13-1, SALK_142112C) harboring a transfer DNA insertion 100 base pairs upstream of the 5′ untranslated region (UTR), which resulted in an obvious reduction (∼20% of WT expression) in the transcript level of FLZ13 (Supplemental Figure 6). An ABA sensitivity test showed that the flz13-1 mutant was slightly more resistant to ABA than Col-0 in terms of seed germination rate (Figures 2F and 3A). Next, we introduced a construct containing the entire genomic sequence of FLZ13 into the flz13-1 mutant (Supplemental Figure 6), resulting in 1.9-fold (flz13-C #3) and 2.38-fold (flz13-C #8) expression of FLZ13. Phenotype analysis showed that the two independent complementation lines germinated like WT plants in response to 0.5 and 0.75 μM ABA treatment (Figure 2F) but exhibited lower greening rates than the WT and flz13-1 (Figure 2G and 2H), suggesting that the reduced ABA sensitivity of the flz13-1 mutant was indeed due to knockdown of FLZ13. To obtain additional mutant alleles, CRISPR–Cas9 technology was used to edit FLZ13 in Col-0 WT plants. We obtained one edited line, flz13-2, with a deletion of 16 nucleotides (164–179) predicted to result in a truncated FLZ13 protein (Supplemental Figure 7A). The flz13-2 mutant showed reduced sensitivity to ABA, as revealed by comparison of the seed germination rate and cotyledon greening rate in the presence of 1.0 μM ABA (Supplemental Figure 7B and 7C). We further tested the responses of the flz13-1 and flz13-2 mutants to a relatively high concentration (2.0 μM) of ABA, and the results showed that they were less sensitive to ABA treatment than Col-0 plants, with higher cotyledon greening rates (Supplemental Figure 8). Taken together, these data suggest that FLZ13 positively regulates ABA signaling to repress seed germination and early seedling growth in Arabidopsis.
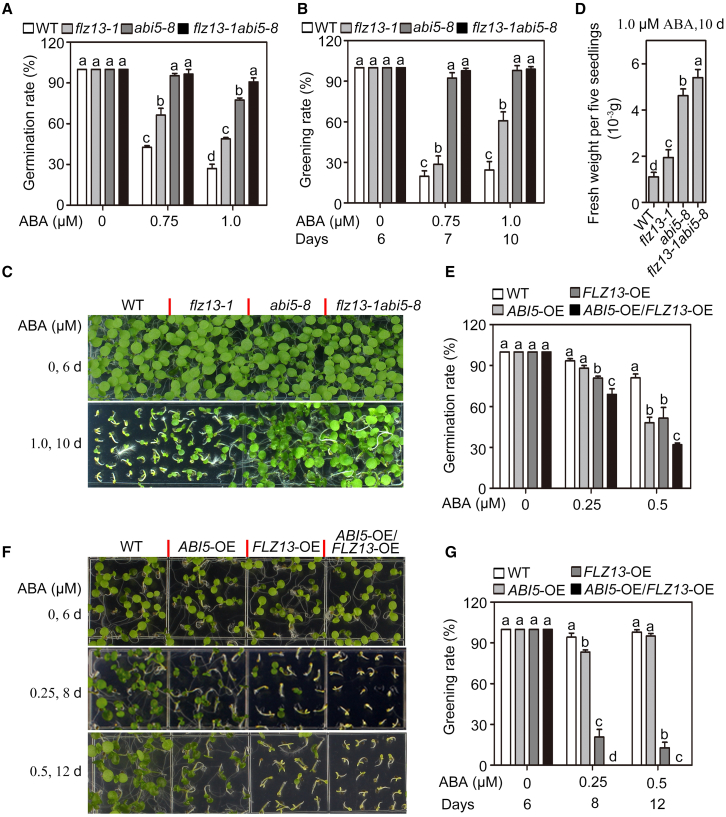
Figure 3.
Genetic interaction between FLZ13 and ABI5 in ABA signaling during seed germination.
(A) Germination rates of the WT and flz13-1, abi5-8, and flz13-1abi5-8 mutants treated with the indicated concentrations of ABA. Germination rates were recorded 4 days after plating on ½ MS medium. Data are presented as mean ± SD (n = 3 biological replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
(B) Cotyledon greening rates of WT, flz13-1, abi5-8, and flz13-1abi5-8 plants treated with various concentrations of ABA at different time points as indicated. Data are presented as mean ± SD (n = 3 biological replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
(C) Photographs of seedlings of WT, flz13-1, abi5-8, and flz13-1abi5-8 treated with various concentrations of ABA at different time points, as indicated.
(D) Fresh weight of five seedlings of WT, flz13-1, abi5-8, and flz13-1abi5-8 treated with 1.0 μM ABA for 10 days. Data are presented as mean ± SD (n = 5 biological replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
(E) Germination rates of WT, ABI5-OE, FLZ13-OE, and ABI5-OE/FLZ13-OE plants treated with the indicated concentrations of ABA. Germination rates were recorded 4 days after plating on ½ MS medium. Data are presented as mean ± SD (n = 3 biological replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
(F) Photographs of WT, ABI5-OE, FLZ13-OE, and ABI5-OE/FLZ13-OE seedlings germinated on media containing various concentrations of ABA at different time points as indicated.
(G) Cotyledon greening rates of WT, ABI5-OE, FLZ13-OE, and ABI5-OE/FLZ13-OE plants treated with various concentrations of ABA at different time points as indicated. Data are presented as mean ± SD (n = 3 biological replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
FLZ13 and ABI5 function additively to modulate ABA signaling
To determine the functional relationship between FLZ13 and ABI5 in ABA signaling, we crossed flz13-1 with abi5-8 to generate an flz13-1abi5-8 double mutant for phenotypic analysis. Consistent with a previous study (Zhou et al., 2015), abi5-8 seeds germinated and grew much faster than WT seeds on ABA-containing medium (Figure 3A–3D), and flz13-1 seeds also showed less sensitivity to ABA than WT seeds (Figure 3A–3D). Notably, the flz13-1abi5-8 double mutant displayed less sensitivity to 1.0 or 2.0 μM ABA treatments compared with the abi5-8 and flz13-1 single mutants (Figure 3A–3D and SupplementalFigure 8), suggesting that FLZ13 and ABI5 may function additively to regulate ABA signaling during seed germination. We also crossed our previously established pUBQ10::ABI5-GFP #1 line (ABI5-OE #1, Li et al., 2019) with FLZ13-OE #1 to generate plants that simultaneously overexpressed ABI5 and FLZ13 (ABI5-OE/FLZ13-OE) to test ABA sensitivity. As expected, the progenies of ABI5-OE/FLZ13-OE displayed significantly lower rates of seed germination and cotyledon greening than those of ABI5-OE and FLZ13-OE in media with various concentrations of ABA (Figure 3E–3G), further confirming the additive effect of ABI5 and FLZ13 on plant ABA response. Collectively, these data demonstrate that FLZ13 and ABI5 function additively to repress seed germination upon ABA treatment.
FLZ13 and ABI5 regulate common target genes in response to ABA
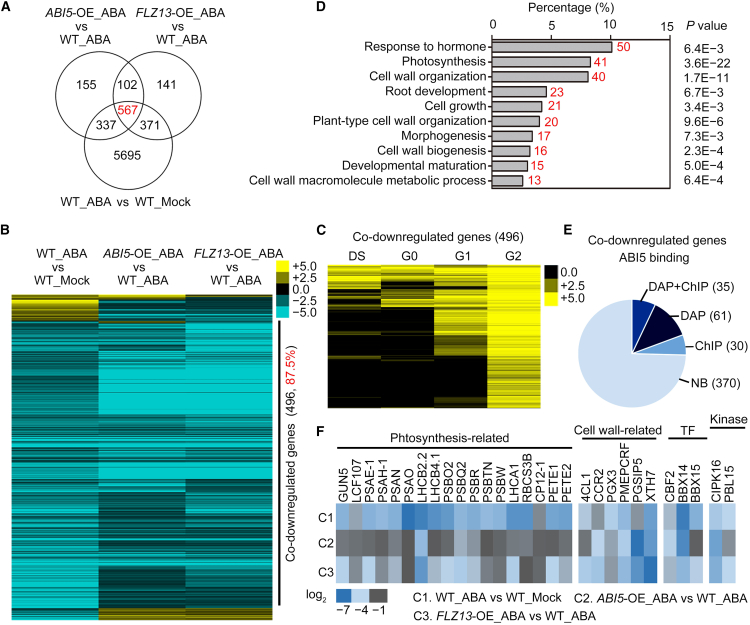
Observations of the protein and genetic interactions between FLZ13 and ABI5 motivated us to identify genes co-regulated by FLZ13/ABI5 in response to ABA during seed germination. RNA sequencing (RNA-seq) analyses were performed using germinating seeds of WT, ABI5-OE #1, and FLZ13-OE #1 treated with 0 or 0.5 μM ABA for 3 days. Without ABA treatment, all the seeds of the different genotypes germinated, and the cotyledons were green. With 0.5 μM ABA treatment, approximately 50% of WT seeds germinated, whereas the germination rates of ABI5-OE #1 and FLZ13-OE #1 seeds were lower than 10%. In WT seeds, 6970 differentially expressed genes (DEGs) (fold change ≥2, false discovery rate [FDR] <0.01) were identified (Figure 4A; Supplemental Table 4), including 77 genes previously found to be induced by ABA (Supplemental Table 5), indicating that this batch of RNA-seq data were reliable. Compared with the WT, 1161 and 1181 DEGs were identified in ABI5-OE #1 and FLZ13-OE #1 seeds with 0.5 μM ABA treatment (Figure 4A; Supplemental Tables 6 and 7).
Figure 4.
Identification of ABA/ABI5/FLZ13 co-regulated genes.
(A) Venn diagram showing the overlap of ABA/ABI5/FLZ13-regulated genes.
(B) Heatmap showing the expression of ABA/ABI5/FLZ13 co-regulated genes. The expression data were clustered using Cluster 3.0 software and edited in TreeView.
(C) Heatmap showing the expression of ABA/ABI5/FLZ13 co-repressed genes during seed germination. The expression data were obtained from a publicly available database (http://bioinfo.sibs.ac.cn/plant-regulomics/; Ran et al., 2020), clustered with Cluster 3.0 software, and edited in TreeView. DS, dry seed; G0, G1, and G2 indicate germinating seeds at 0, 1, and 2 days, respectively.
(D) Representative enriched BP terms of the ABA/ABI5/FLZ13 co-repressed genes. GO enrichment analysis was performed using an online program (https://david.ncifcrf.gov; Huang et al., 2009). Red numbers indicate gene numbers.
(E) Pie chart showing the number of ABA/ABI5/FLZ13 co-repressed genes bound by ABI5 in ChIP-seq and/or DAP-seq assays (O'Malley et al., 2016). NB, not bound.
(F) Heatmap showing the expression of well-annotated ABI5 target genes that are co-repressed by ABA/ABI5/FLZ13.
Figure 7.
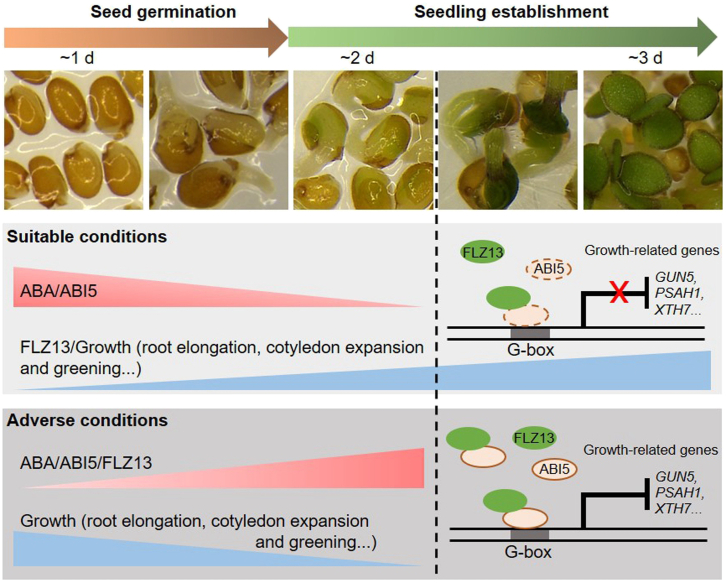
A working model for the ABI5–FLZ13 module in seed germination and seedling establishment.
After seed germination under suitable conditions, the level of ABA and the activity of ABI5 decrease, diminishing the suppression of downstream growth-related genes to promote normal germination and seedling establishment. If germinating seeds are exposed to adverse conditions, the increased ABA activates ABI5, and the ABI5–FLZ13 module represses transcription of growth-related genes to arrest seedling establishment.
To identify ABA-responsive genes that were also co-regulated by ABI5 and FLZ13, we selected genes that met the following three criteria: (1) genes were differentially expressed after ABA treatment in the WT; (2) their expression differed between WT and ABI5-OE #1 after ABA treatment; and (3) their expression differed between WT and FLZ13-OE #1 after ABA treatment. Finally, 567 genes were identified as ABA-responsive and ABI5–FLZ13 co-regulated (Figure 4A; Supplemental Table 8). A heatmap showed that only four genes were upregulated, whereas most genes (496, 87.5%) were simultaneously suppressed by ABA, ABI5, and FLZ13 (Figure 4B), and they all showed induced expression patterns during seed germination (Figure 4C). Gene Ontology (GO) term enrichment analysis revealed that these 496 DEGs were mainly enriched in categories related to growth, including “photosynthesis” (41 genes, P = 3.6E−22) and “cell wall organization” (40 genes, P = 1.7E−11) (Figure 4D; Supplemental Table 9). Furthermore, among the 496 genes, 126 were targeted by ABI5, as verified by chromatin immunoprecipitation sequencing (ChIP-seq) and/or DNA affinity purification sequencing (DAP-seq) assays (O'Malley et al., 2016) (Figure 4E; Supplemental Table 10). The set of 35 high-confidence ABI5-targeted genes, as verified by both ChIP-seq and DAP-seq assays, contained genes encoding 18 photosynthesis-related proteins, six cell wall–modification related proteins, three transcription factors, and two kinases (Figure 4F; Supplemental Table 11). Collectively, these results indicate that FLZ13 and ABI5 display overlapping functions in determining plant responses to ABA by co-targeting a subset of ABA-responsive genes, especially ABA-repressed and growth-related genes.
FLZ13 activation of ABA signaling requires a functional ABI5
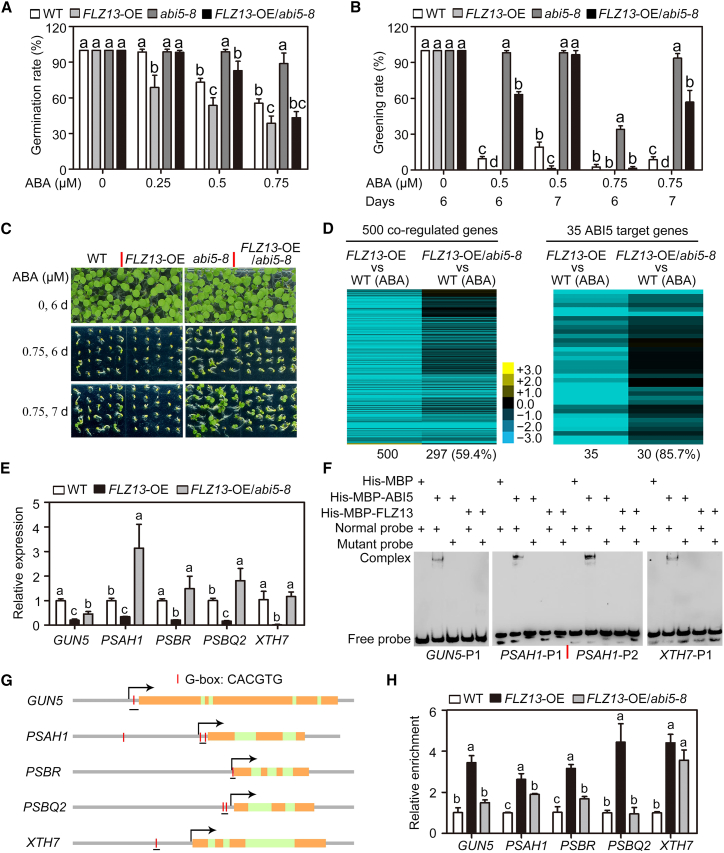
To study the genetic epistasis between FLZ13 and ABI5, we explored whether the action of FLZ13 in mediating ABA signaling requires a functional ABI5 by generating plants that overexpressed FLZ13 in the abi5-8 mutant background. Progenies of FLZ13-OE/abi5-8 were hyposensitive to 0.5 μM ABA treatment during seed germination, with higher seed germination percentages and cotyledon greening rates than WT and FLZ13-OE plants (Figure 5A–5C). However, compared with the abi5-8 mutant, FLZ13-OE/abi5-8 was slightly more sensitive to 0.75 μM ABA, indicating that ABA hypersensitivity conferred by FLZ13 overexpression was largely but not fully dependent on ABI5 (Figure 5A–5C).
Figure 5.
FLZ13 regulates ABA response during seed germination in an ABI5-dependent manner.
(A) Germination rates of WT, FLZ13-OE, abi5-8, and FLZ13-OE/abi5-8 treated with the indicated concentrations of ABA. Germination rates were recorded 4 days after plating on ½ MS medium. Data are presented as mean ± SD (n = 3 biological replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
(B) Cotyledon greening rates of WT, FLZ13-OE, abi5-8, and FLZ13-OE/abi5-8 treated with various concentrations of ABA at different time points, as indicated. Data are presented as mean ± SD (n = 3 biological replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
(C) Photographs of WT, FLZ13-OE, abi5-8, and FLZ13-OE/abi5-8 seedlings germinated on medium containing various concentrations of ABA for the indicated time periods.
(D) Heatmap showing the expression of genes synchronously co-regulated by ABA/ABI5/FLZ13 in FLZ13-OE and FLZ13-OE/abi5-8 lines in the presence of ABA. Three-day-old germinating seeds with 0.5 μM ABA treatment were collected for the RNA-seq assay.
(E) Quantitative RT–PCR assay showing expression of the selected genes in WT, FLZ13-OE, and FLZ13-OE/abi5-8 upon ABA treatment. Three-day-old germinating seeds with 0.5 μM ABA treatment were collected for the qRT–PCR assay. PP2A was used as an internal control. Data are presented as mean ± SD (n = 3 technical replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05). This experiment was repeated twice with similar results.
(F) EMSA showing the binding ability of ABI5 and FLZ13 to the ABI5 target genes GUN5, PSAH1, and XTH7. The DNA probes used in this experiment are shown in Supplemental Figure 9. In the mutant probes, the G-box sequence (CACGTG) was replaced with CTTTTG. The probe sequences are listed in Supplemental Table 14.
(G) Structure of selected ABI5 target genes. The positions of the G-box and fragments used for the ChIP–PCR assay are marked.
(H) ChIP–qPCR assay showing the relative enrichment of FLZ13 at the selected gene locus. Data are presented as mean ± SD (n = 3 technical replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05). This experiment was repeated twice with similar results.
To assess the effect of ABI5 on the expression of FLZ13-regulated genes, we carried out RNA-seq analysis using 3-day-old germinating FLZ13-OE/abi5-8 seeds treated with 0.5 μM ABA (Supplemental Table 12). A heatmap of 500 ABA/ABI5/FLZ13 synchronously co-regulated genes showed altered expression patterns. Two-hundred and ninety-seven out of these 500 genes (59.4%) exhibited significant alterations in FLZ13-OE/abi5-8 (Figure 5D, left panel), supporting the notion that FLZ13-mediated transcriptional regulation largely depends on the function of ABI5. For example, 30 of the 35 ABI5 target genes showed differential expression between FLZ13-OE/abi5-8 and FLZ13-OE plants after ABA treatment (Figure 5D, right panel). We also performed qRT–PCR to confirm the transcript levels of five ABI5 target genes (GUN5, PSAH1, PSBR, PSBQ2, and XTH7) (Supplemental Figure 9). The results revealed that mutation of ABI5 largely compromised FLZ13-repressed expression of these genes (Figure 5E). The above ABA sensitivity test and gene expression analysis clearly demonstrated that the function of FLZ13 in ABA signaling depends largely on a functional ABI5.
To further explore how ABI5 affects the action of FLZ13, we used an electrophoretic mobility shift assay (EMSA) to examine the DNA-binding ability of ABI5 and FLZ13 to their co-regulated genes. As shown in Figure 5F, ABI5 directly bound to the promoter regions of GUN5, PSAH1, and XTH7 via a conserved G-box cis element, whereas FLZ13 failed to bind to these regions. To check these bindings in vivo, we performed chromatin immunoprecipitation (ChIP) using ABA-treated germinating seeds of FLZ13-OE and FLZ13-OE/abi5-8 plants. ChIP–qPCR analysis showed that FLZ13 was enriched at ABI5-bound genomic loci, including GUN5, PSAH1, PBSR, PBSQ2, and XTH7, in FLZ13-OE seeds (Figure 5G and 5H). However, enrichment of FLZ13 on their promoters was generally lower in FLZ13-OE/abi5-8 than in FLZ13-OE (Figure 5G and 5H). These findings suggest that FLZ13 is recruited to the promoters of GUN5, PSAH1, PBSR, PBSQ2, and XTH7 at least in part through its interaction with ABI5.
Knockdown of FLZ13 compromises the function of ABI5
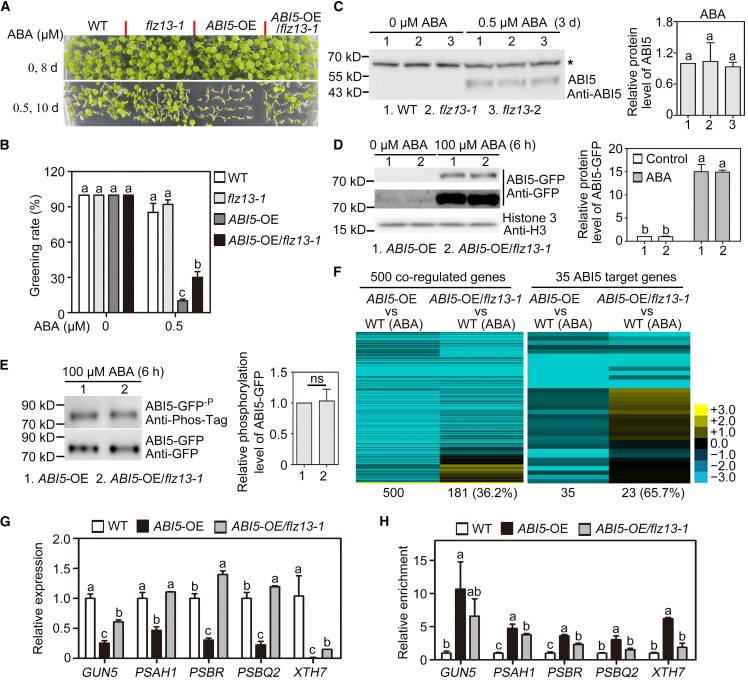
To elucidate the role of FLZ13 in ABI5 activity, we generated an ABI5-OE/flz13-1 plant by crossing and performed an ABA sensitivity test. As shown in Figure 6A and 6B, knockdown of FLZ13 significantly increased the cotyledon greening rate of ABI5-OE plants in the presence of 0.5 μM ABA, suggesting that FLZ13 contributes to the ABI5-modulated ABA response. To explore how FLZ13 functions in the ABI5-mediated ABA response, we analyzed whether FLZ13 affects ABI5 protein stability and phosphorylation, two decisive factors that contribute to ABI5 activity during ABA signaling (Yu et al., 2015). We first examined endogenous ABI5 protein levels in 3-day-old germinating seeds of the WT, flz13-1, and flz13-2 lines with 0 or 0.5 μM ABA treatment. ABI5 was too low to be detected in all the genotypes without ABA treatment but could be detected in these seeds with 0.5 μM ABA treatment, although no obvious differences in ABI5 protein levels were observed among the genotypes (Figure 6C). We then compared ABI5-GFP protein expression in ABI5-OE and ABI5-OE/flz13-1 plants. Immunoblotting results demonstrated that the abundance of ABI5-GFP protein was similar in flz13-1 and WT backgrounds under both normal growth conditions and ABA treatment (Figure 6D), further suggesting that FLZ13 does not regulate the stability of ABI5. Moreover, the phosphorylation levels of immunoprecipitated ABI5-GFP from ABA-treated samples were similar in flz13-1 and WT backgrounds (Figure 6E), suggesting that FLZ13 did not affect the phosphorylation of ABI5 in response to ABA.
Figure 6.
The function of ABI5 during seed germination is partially compromised in FLZ13 mutants.
(A) Photographs of WT, flz13-1, ABI5-OE, and ABI5-OE/flz13-1 seedlings germinated on media containing various concentrations of ABA for the indicated time periods.
(B) Cotyledon greening rates of WT, flz13-1, ABI5-OE, and ABI5-OE/flz13-1 plants upon treatment with various concentrations of ABA at the indicated time points. Data are presented as mean ± SD (n = 3 biological replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
(C) Immunoblotting analysis of ABI5 protein in the WT and flz13 mutants. Three-day-old germinating seeds sown on ½ MS plates with 0 or 0.5 μM ABA treatment were harvested for protein extraction and western blot analysis using anti-ABI5 antibodies. The relative protein levels of ABI5 in ABA-treated samples are shown on the left. Data are presented as mean ± SD (n = 4 biological replicates). An asterisk (∗) indicates non-specific bands that were used as loading controls. The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
(D) Immunoblotting analysis of ABI5-GFP protein in the WT and flz13-1 backgrounds. Whole seedlings of 7-day-old ABI5-OE and ABI5-OE/flz13-1 were treated with 0 (Control) or 100 μM ABA for 6 h before protein extraction for western blot analysis using anti-GFP and anti-H3 antibodies. Short (upper panel) and long (middle panel) exposures of blots for ABI5-GFP are shown. The protein levels of ABI5-GFP relative to that of histone 3 are shown on the left. Data are presented as mean ± SD (n = 3 biological replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05).
(E) Phosphorylation of ABI5-GFP protein in the WT and flz13-1 background upon ABA treatment. Whole seedlings of 7-day-old ABI5-OE and ABI5-OE/flz13-1 plants were treated with 100 μM ABA for 6 h before protein extraction for immunoprecipitation using GFP-Trap beads. The samples were then subjected to western blot analysis using anti-GFP and anti-Phos-tag antibodies. Relative phosphorylation levels of ABI5-GFP are shown on the left. Data are presented as mean ± SD (n = 3 biological replicates). Statistical analysis was performed using Student’s t-test. ns, no significant difference.
(F) Heatmap showing the expression of genes synchronously co-regulated by ABA/ABI5/FLZ13 in ABI5-OE and ABI5-OE/flz13-1 lines in the presence of ABA. Three-day-old germinating seeds with 0.5 μM ABA treatment were collected for the RNA-seq assay.
(G) Quantitative RT–PCR assay showing the expression of selected genes in WT, ABI5-OE, and ABI5-OE/flz13-1 upon ABA treatment. Three-day-old germinating seeds with 0.5 μM ABA treatment were collected for the qRT–PCR assay. PP2A was used as an internal control. Data are presented as mean ± SD (n = 3 technical replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05). This experiment was repeated twice with similar results.
(H) ChIP–qPCR assay showing the relative enrichment of ABI5 at the selected gene locus. Data are presented as mean ± SD (n = 3 technical replicates). The different letters above each bar indicate statistically significant differences determined by one-way ANOVA followed by Tukey’s multiple comparison test (P < 0.05). This experiment was repeated twice with similar results.
We next analyzed the expression of 500 genes synchronously co-regulated by ABA/ABI5/FLZ13 in 3-day-old germinating seeds of ABI5-OE/flz13-1 upon 0.5 μM ABA treatment using an RNA-seq assay (Supplemental Table 13). A heatmap showed significantly altered expression levels of 181 genes (36.2%) (Figure 6F, left panel), indicating that ABI5-mediated transcriptional regulation of these genes was affected by FLZ13. For instance, 23 of 35 ABI5 target genes showed differential expression between ABI5-OE/flz13-1 and ABI5-OE plants treated with ABA (Figure 6F, right panel). We also performed qRT–PCR to confirm the transcript levels of GUN5, PSAH1, PBSR, PBSQ2, and XTH7 and found that mutations in FLZ13 largely compromised the ABI5-repressed expression of these genes (Figure 6G). Based on these results, we speculated that FLZ13 may affect the DNA-binding ability of ABI5 to its downstream genes. To test this possibility, we performed ChIP–qPCR analysis using ABA-treated germinating seeds of ABI5-OE and ABI5-OE/flz13-1. Enrichment of ABI5 on the promoters of GUN5, PSAH1, PBSR, PBSQ2, and XTH7 was generally lower in ABI5-OE/flz13-1 than in ABI5-OE (Figure 6H), indicating that FLZ13 knockdown decreased the binding of ABI5 to its target genes in planta.
Discussion
ABA modulates plant growth or stress responses by triggering massive transcriptional reprogramming that depends mainly on several ABA-responsive transcription factors. In particular, ABI5 functions as the master regulator during ABA-inhibited seed germination and seedling establishment (Chen et al., 2020). However, how ABI5 modulates ABA-repressed seed germination and seedling establishment remains largely unknown because only a few target genes of ABI5 are directly associated with these processes (Lee and Luan, 2012; Skubacz et al., 2016). Furthermore, the precise regulatory mechanism of the ABI5-engaged transcriptional regulation machinery also remains to be fully clarified.
In this study, we used TurboID-mediated proximity labeling to obtain the ABI5 interactome and identified FLZ13 as a new protein that interacts with ABI5 (Figure 1). FLZs are plant-specific regulatory proteins, and the Arabidopsis genome contains 19 FLZ proteins (Jamsheer and Laxmi, 2015). The first FLZ protein, MEDIATOR OF ABA-REGULATED DORMANCY 1 (MARD1/FLZ19), was identified from senescence-related enhancer trapping and has been implicated in ABA-mediated seed germination in Arabidopsis (He et al., 2001; He and Gan, 2004). Expression of MARD1 was induced by ABA treatment, and the seeds of mard1 were relatively resistant to external ABA during germination (He and Gan, 2004). Recently, FLZ6, FLZ8, and FLZ10 were also reported to participate in plant ABA responses by interfering with SUCROSE NON-FERMENTING1 (SNF1)-RELATED KINASE 1 (SnRK1) and TARGET OF RAPAMYCIN (TOR) signaling (Jamsheer et al., 2018, 2022). In an ABA-inhibited root elongation assay, flz6.1 and flz10.1 showed slightly higher sensitivity to external ABA, whereas flz8.1 showed lower sensitivity (Jamsheer et al., 2018, 2022). Here, we offer several lines of evidence to show that FLZ13 positively regulates the ABA response during the embryo-to-seedling transition. First, FLZ13 directly interacts with ABI5, a core transcription factor in ABA signaling (Skubacz et al., 2016), and this interaction is responsive to ABA treatment (Figure 1 and SupplementalFigure 4). Second, progenies of the flz13 mutant were less sensitive to ABA, whereas those of FLZ13-overexpressing plants were much more sensitive to ABA than WT plants during seed germination and seedling establishment (Figures 2 and 3 and SupplementalFigures 7 and 8). Third, FLZ13 and ABI5 co-regulate a large subset of genes in response to ABA (Figure 4). Finally, FLZ13 and ABI5 function interdependently to mediate the ABA response during seed germination and seedling establishment (Figures 5 and 6). These results strongly suggest that FLZ13 works together with ABI5 to positively regulate ABA signaling during seed germination. We also verified that ABA receptors and subclass III SnRK2s are required for FLZ13 to positively regulate ABA signaling (Figure 2D and 2E). However, it is worth noting that the phenotypes of flz13 mutants were much weaker than those of abi5-8 in response to ABA treatment (Figure 3A and 3B and SupplementalFigure 8). In the Arabidopsis genome, FLZ13 has two close homologs (FLZ12 and FLZ14) (Jamsheer and Laxmi, 2015), which raises the possibility that FLZ13 may function redundantly with FLZ12 and FLZ14 to regulate ABA responses, although the detailed biological roles of FLZ12 and FLZ14 remain to be determined.
The flz13-1abi5-8 double mutant exhibited higher germination and greening percentages than flz13-1 and abi5-8 single mutants in the presence of ABA, and FLZ13-OE/ABI5-OE showed higher sensitivity to ABA than FLZ13-OE and ABI5-OE (Figure 3), indicating that ABI5 and FLZ13 have additive effects on the promotion of ABA response during seed germination. As shown in Figure 4A, transcriptome analyses identified 567, 337, and 371 ABA-responsive genes that were co-regulated or specifically regulated by ABI5 and FLZ13, respectively, implying that FLZ13 and ABI5 can also function with other modulators to regulate different gene sets to determine ABA signaling. For example, PRR5 and PRR7 can interact with ABI5 to enhance its function in ABA signaling and inhibit seed germination in the presence of ABA (Yang et al., 2021). Although we failed to observe an interaction between FLZ13 and other critical transcription factors of ABA signaling such as ABI3, ABI4, ABF2, and ABF4 (Supplemental Figure 3) in the Y2H assay, we cannot exclude the possibility that FLZ13 associates with other modulators to affect their activity in ABA signaling. For instance, a recent study showed that ZmFLZ25 interacts with several ABA receptors to confer hypersensitivity to ABA during seed germination (Chen et al., 2021).
Interestingly, the ABA sensitivity of the ABI5-OE/flz13-1 progenies was between that of flz13-1 and ABI5-OE plants (Figure 6A and 6B), indicating that the action of ABI5 during ABA signaling is partially dependent on FLZ13. FLZ13 did not affect the protein stability or phosphorylation of ABI5 in the presence of ABA (Figure 6C–6E), but it was partially required for the DNA binding of ABI5 to downstream target genes (Figure 6H). The bZIP domain associated with ABI5 dimerization and DNA binding (Finkelstein and Lynch, 2000) was involved in the interaction with FLZ13 (Figure 1D), and FLZ13 is an FCS-like zinc-finger protein that can be recruited to GUN5, PBSR, PSAH1, PBSQ2, and XTH7 promoters through interaction with ABI5 (Figure 5H). It is therefore possible that the FLZ13–ABI5 complex functions similarly to the ABI5 dimer and shows increased binding activity to promoters of target genes. Future studies need to be performed to explore the detailed biochemical mechanisms by which FLZ13 synergizes with ABI5 to modulate downstream genes.
RNA-seq analysis identified 567 genes co-regulated by ABA, ABI5, and FLZ13 (Figure 4A; Supplemental Table 8). Interestingly, most of the FLZ13 and ABI5 co-regulated genes were repressed by ABA (Figure 4B), suggesting that ABI5 and FLZ13 play important roles in ABA-mediated transcriptional repression. Interestingly, these co-repressed genes were mainly enriched in categories related to growth, including “photosynthesis” (41 genes, P = 3.6E−22) and “cell wall organization” (40 genes, P = 1.7E−11) (Figure 4D; Supplemental Table 9). However, ABI5 is a transcription factor with strong transcriptional activation activity, and how the ABI5–FLZ13 complex represses target gene expression remains unclear. It is possible that the ABI5–FLZ13 complex recruits transcriptional corepressors to target gene loci and modulates their expression. For example, the mediator (a bridge between transcription factors and RNA polymerase II during transcription) subunit MED16 interacts with ABI5 to repress the expression of ABI5 target genes (Guo et al., 2021). In our ABI5-interactome data, several transcriptional corepressors, such as LEUNIG, SUESS, and TETRATRICOPEPTIDE REPEAT 2 (TPR2) (Franks et al., 2002; Long et al., 2006), showed significantly higher enrichment in the ABI5-TurboID-GFP vs. TurboID-GFP pairs (Supplemental Table 1). They were positioned at the central nodes of the PPI networks (Supplemental Figure 2), indicating their important roles in ABI5-mediated transcriptional repression. Further investigation of the underlying molecular mechanisms will provide new insights into the genetic basis of ABI5–FLZ13-inhibited seed germination and post-germination growth.
In summary, our study sheds new light on how ABI5-mediated ABA signaling represses seed germination and seedling establishment (Figure 7). After seed germination under suitable conditions, ABA levels and ABI5 activity decrease, resulting in diminished suppression of downstream growth-related genes to ensure normal germination and seedling establishment. If adverse conditions are imposed on germinating seeds, elevated ABA activates ABI5, and the ABI5–FLZ13 module represses transcription of growth-related genes to arrest seedling establishment until suitable conditions appear. These findings might be helpful in resetting the balance between growth and stress resistance based on the ABA pathway, thus enabling the engineering of stress-resistant and high-yielding crops in the future.
Methods
Plant materials and growth conditions
All Arabidopsis (Arabidopsis thaliana) lines used in this study were in the Col-0 background. The abi5-8 mutant and ABI5 overexpression plants (pUBQ10::ABI5-GFP, ABI5-OE #1) were used as described previously (Zhou et al., 2015; Li et al., 2019). The flz13-1 mutant (SALK_142112C) was obtained from the ABRC (https://abrc.osu.edu/). The FLZ13-Cas9 line (flz13-2) was generated using the CRISPR–Cas9 system with two guide RNA sequences (Xing et al., 2014). To generate FLZ13-OE transgenic lines, full-length FLZ13 was cloned in-frame into the binary vector pCAMBIA1300 fused with GFP at the C terminus under the control of the Arabidopsis UBQ10 promoter. The resulting construct was introduced into Col-0, pyr1pyl1pyl2pyl4pyl5pyl8 (pyr1pyl12458) (Gonzalez-Guzman et al., 2012), and snrk2.2snrk2.3 (Fujii et al., 2007) by Agrobacterium-mediated transformation using the floral dip method (Clough and Bent, 1998). To generate pFLZ13::FLZ13-GFP plants, the entire genomic sequence of FLZ13 containing a ∼2.0-kb promoter sequence was PCR amplified and inserted into the binary vector pCAMBIA1300 and fused in-frame with GFP at the C terminus. The resulting construct was introduced into Col-0 via Agrobacterium-mediated transformation using the floral dip method. To generate gFLZ13/flz13-1 (flz13-C) lines, the entire genomic sequence of FLZ13 containing a ∼2.0-kb promoter sequence and a ∼1.0-kb terminator sequence was PCR amplified and inserted into the binary vector pCAMBIA1300. The resulting construct was introduced into flz13-1 via Agrobacterium-mediated transformation using the floral dip method. Full-length TurboID was cloned into pCAMBIA1300 and fused in-frame with GFP at the C terminus under the control of the Arabidopsis UBQ10 promoter to generate the TurboID-GFP construct. To generate ABI5-TurboID-GFP, full-length ABI5 was inserted into TurboID-GFP to fuse it with TurboID-GFP at the C terminus. The resulting constructs were introduced into Col-0 via Agrobacterium-mediated transformation using the floral dip method. T0 seeds were screened in ½ Murashige and Skoog (MS) medium containing hygromycin (25 mg/ml). Homozygous plants from the T3 generation were used in subsequent studies. The flz13-1abi5-8, FLZ13-OE/abi5-8, and ABI5-OE/flz13-1 plants were generated by crossing and genotyping. The primers used are listed in Supplemental Table 14.
Seeds were surface sterilized with 30% (v/v) bleach for 5 min, washed with double-distilled H2O three times (3 min each time), kept at 4°C in the dark for 2–3 days, and then sown on ½ MS plates supplemented with 0.8% (w/v) agar and 1% sucrose. After 6 days, the well-established seedlings were transferred to soil and grown in a growth chamber at 22°C under long-day conditions (LD, 16 h light/8 h dark). Nicotiana benthamiana plants were grown in the same chambers under LD conditions for BiFC and LCI assays.
ABA sensitivity test
Germination and greening of seeds from different genotypes were determined as previously described (Li et al., 2019). In brief, seeds were surface sterilized and then cold stratified at 4°C in the dark for 2–3 days before being sown on ½ MS medium plates with or without various concentrations of ABA (Sangon Biotech, E813BA0006). The plates were placed in a growth chamber at 22°C under LD conditions for germination. After 3–4 days, germination was determined from the appearance of the embryonic axis (appearance of radicle protrusion) as observed under a microscope. After 6–12 days, pictures were taken to calculate the greening rate (appearance of green cotyledons on seedlings) using Adobe Photoshop software. These experiments were performed at least three times with similar results, and around 50–100 seeds of each genotype were used for each biological replicate.
Proximity labeling, affinity purification, and mass spectrometry
Five-day-old TurboID-GFP and ABI5-TurboID-GFP transgenic seedlings grown on plates with ½ MS medium were transferred to ½ MS liquid medium supplemented with 100 μM ABA for 4 h. This was followed by incubation with 50 μM biotin for 1 h. Then, the seedlings were rinsed with ice-cold water three times for 5 min each to stop the labeling reaction. The seedlings were blotted dry with paper towels and separated into aliquots of approximately 0.6 g fresh weight for the three biological replicates, then stored at −80°C until use.
Total proteins were extracted from the samples with 6 ml of buffer (50 mM Tris [pH 7.5], 150 mM NaCl, 0.1% SDS, 1 mM EDTA, 1% Triton X-100, 0.5% Na-deoxycholate, 1 mM DTT, 1× complete protease inhibitor cocktail, and 1 mM PMSF). Before affinity purification with streptavidin-coated magnetic beads (MedChemExpress, HY-K0208), the protein extract was desalted to remove free biotin using Zeba Spin Desalting Columns (Thermo Fisher, 89893) according to the manufacturer’s instructions. Affinity purification was performed at 4°C for approximately 16 h. After affinity purification, the beads were washed five times with the extraction buffer. The samples were then digested on beads with trypsin and analyzed by liquid chromatography–tandem mass spectrometry following established protocols (Branon et al., 2018; Mair et al., 2019).
Proteomic analysis and PPI network construction
Detailed proteomic analysis was performed as described previously (Mair et al., 2019). Filtering and statistical analyses were performed using Perseus (Tyanova et al., 2016). The output file from MaxQuant was imported into Perseus, using label-free quantitation (LFQ) intensities as the main category. The data matrix was filtered to remove proteins marked as “only identified by site,” “reverse,” and “potential contaminants.” LFQ values were log2 transformed, and proteins that were identified in only one of three replicates of a single genotype were removed. The criteria for identification of ABI5-specific substrates were fold change (ABI5-TurboID-GFP vs. TurboID-GFP) >2 and P value <0.05. Finally, a volplot was generated using the OmicShare tools platform (http://www.omicshare.com/tools), and PPI networks were constructed online (https://metascape.org/; Zhou et al., 2019).
Protein interaction analysis
-
(1)
Yeast two-hybrid (Y2H) assay. Full-length and truncated versions of FLZ13 or other tested proteins were cloned into pGBKT7 to generate bait vectors with the Gal4 DNA-binding domain. The primers used are listed in Supplemental Table 14. Full-length and truncated versions of ABI5 were cloned into pGADT7 to generate prey vectors with the Gal4 activation domain, as described previously (Li et al., 2019). The Y2H assay was performed as described previously (Yang et al., 2018). The bait and prey vectors were co-transformed into the yeast strain AH109, and physical interactions were determined by the growth of co-transfected yeast cells on synthetic dropout medium (SD)-Leu-Trp or SD-Leu-Trp-His-adenine plates for 3–4 days after plating.
-
(2)
BiFC. Full-length coding sequences of FLZ13 and ABI5 were cloned into the pCAMBIA1300-YN or pCAMBIA1300-YC vector to generate FLZ13-YN and ABI5-YC, respectively. The primers used are listed in Supplemental Table 14. The resulting constructs were transformed into Agrobacterium tumefaciens strain GV3101. A. tumefaciens containing FLZ13-YN, A. tumefaciens containing ABI5-YC, and A. tumefaciens containing Histone3-mCherry were co-injected into fully expanded leaves of 4-week-old N. benthamian plants at a ratio of 2:2:1. After infiltration for 3 days, the infiltrated regions were observed and imaged using a confocal microscope.
-
(3)
LCI assays. Full-length coding sequences of FLZ13 and ABI5 were cloned into the pCAMBIA1300-nLUC and pCAMBIA1300-cLUC vectors to generate FLZ13-nLUC and cLUC-ABI5, respectively. The primers used are listed in Supplemental Table 14. The resulting constructs were transformed into A. tumefaciens strain GV3101. A. tumefaciens containing FLZ13-nLUC, A. tumefaciens containing cLUC-ABI5, and A. tumefaciens containing P19 were co-injected into fully expanded leaves of 4-week-old N. benthamiana plants at a ratio of 2:2:1. After infiltration for 40 hours, the infiltrated regions were injected with 1 mM luciferin before capturing LUC activity with a Tanon 5200 imaging system. For ABA treatment, leaves were pre-treated with 10 μM ABA for 4 h.
-
(4)
Pull-down assay. Full-length FLZ13 and ABI5 were cloned into pMAL-c2× and pGEX4T-3 plasmids, respectively. The primers used are listed in Supplementary Table 14. The recombinant vectors were transformed into Escherichia coli BL21 (DE3) to express His-MBP-FLZ13 and GST-ABI5 proteins at 25°C for 6 h with 0.4 mM IPTG induction. The supernatant containing His-MBP or His-MBP-FLZ13 proteins was mixed with 50 μl of MBP-magic beads (Sangon) and incubated for 2 h at 4°C. The beads were then washed six times with 1 ml of PBS and incubated with the supernatant containing GST-ABI5 protein for 4 h at 4°C. After six washes with 1 ml of PBS, the beads were boiled in 30 μl of 1× SDS–PAGE loading buffer. Finally, the presence of GST- and MBP-tagged proteins was detected by western blotting using anti-GST and anti-MBP antibodies.
Gene expression analysis by qRT–PCR
To analyze FLZ13 and ABI5 expression during seed germination and its response to ABA, 50 mg of Col-0 seeds were surface sterilized and then cold stratified at 4°C in the dark for 3 days before being sown on ½ MS medium plates with or without 0.5 μM ABA. The plates were placed in a growth chamber at 22°C under LD conditions for germination, and the seeds were harvested at the indicated time points for total RNA isolation using a HiPure Plant RNA Mini Kit (Magen). Total RNA (1 μg) was reverse transcribed using HiScript II Q Select RT SuperMix (Vazyme), and the resulting first-strand cDNA was used as a template for quantitative real-time PCR performed on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad) with iTaq Universal SYBR Green Supermix (Bio-Rad). The expression of PP2A or Actin2 was used as an internal control, and relative expression was calculated using the 2−ΔΔCt method. The primer pairs used for the qRT–PCR assay are listed in Supplemental Table 14.
RNA-seq and data analyses
To perform RNA-seq analysis, 200 mg of seeds were surface sterilized and then cold stratified at 4°C in the dark for 3 days before being sown on ½ MS medium plates with or without 0.5 μM ABA. The plates were placed in a growth chamber at 22°C under LD conditions for germination. After 3 days, the seeds were harvested for total RNA isolation using a HiPure Plant RNA Mini Kit (Magen). RNA-seq was performed by Biomarker Technologies. Genes with a log2 (fold change) ≥1 and an average reads per kilobase of transcript per million reads mapped (RPKM) value from three biological repeats >1.0 in at least in one pair were identified as reliable DEGs. Multiple testing was corrected via FDR estimation, and an FDR <0.01 was considered to indicate differential expression. The transcriptome data have been deposited in the Genome Sequence Archive at the National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences (https://ngdc.cncb.ac.cn/gsa, CRA011240) and the NCBI GEO repository (http://www.ncbi.nlm.nih.gov/geo, PRJNA885244). GO enrichment analysis was performed using DAVID (https://david.ncifcrf.gov) (Huang et al., 2009). Heatmaps were generated using Cluster 3 software and visualized using TreeView.
ChIP–qPCR assay
ChIP assays were performed as described previously (Gendrel et al., 2005; Yang et al., 2020). In brief, 3-day-old germinating seeds of WT, FLZ13-OE, FLZ13-OE/abi5-8, ABI5-OE, and ABI5-OE/flz13-1 treated with 0.5 μM ABA were cross-linked with 1% formaldehyde. The chromatin was then extracted and sheared to approximately 500 base pairs by sonication before being immunoprecipitated with a GFP-Trap (ChromoTek). After cross-linking was reversed, the amount of each precipitated DNA fragment was determined by quantitative PCR. The relative quantity was calculated using the 2−ΔΔCt method and presented as the ratio of immunoprecipitation (IP) to input. Primer pairs used in the ChIP–qPCR assay are listed in Supplemental Table 14.
EMSA
EMSA was performed as described previously (Li et al., 2022b). In brief, full-length ABI5 and FLZ13 were separately fused with a His-MBP tag at the C terminus to generate His-MBP-ABI5 and His-MBP-FLZ13 vectors. The resulting vectors were transformed into E. coli BL21 (DE3) strains to express the recombinant proteins at 25°C for 6 h with 0.4 mM isopropyl β-D-1-thiogalactopyranoside (IPTG) induction. The expressed proteins were purified from the soluble fractions using His-Magic beads (Sangon Biotech). Biotin-labeled DNA probes were purchased from Sangon Biotech (Shanghai, China) and are listed in Supplemental Table 14. The EMSA was performed using a non-radioactive electrophoretic mobility shift assay kit (Viagene Biotech) following the manufacturer’s instructions.
Protein detection and phosphorylation analysis
To analyze the protein levels of FLZ13 and ABI5 during seed germination and their response to ABA, 100 seeds of pFLZ13::FLZ13-GFP #9 were surface sterilized and then cold stratified at 4°C in the dark for 3 days before being sown on ½ MS medium plates with or without 0.5 μM ABA. The plates were placed in a growth chamber at 22°C under LD conditions for germination, and the seeds were harvested at the indicated time points. To check the protein levels of ABI5, 100 seeds of WT, flz13-1, and flz13-2 were surface sterilized and then cold stratified at 4°C in the dark for 3 days before being sown on ½ MS medium plates with or without 0.5 μM ABA. The plates were placed in a growth chamber at 22°C under LD conditions for germination, and the seeds were harvested after incubation for 3 days. To check the protein levels of ABI5-GFP, 7-day-old ABI5-OE and ABI5-OE/flz13-1 were exposed to 100 μM ABA for 6 h before being harvested. Total protein was extracted from the harvested samples using 20 mM Tris–HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, and 1× protease inhibitor cocktail (Roche Diagnostics). The primary antibodies used in this study were anti-ABI5 (Stone et al., 2006; 1:3000 dilution), anti-GFP (HT801-02; 1:2000 dilution), anti-MBP (TransGen, HT701-02; 1:3000 dilution), anti-GST (TransGen, HT601-02; 1:3000 dilution), and anti-H3 (Abcam, ab1791; 1:5000 dilution).
For phosphorylation analysis of ABI5-GFP, 7-day-old ABI5-OE and ABI5-OE/flz13-1 plants were exposed to 100 μM ABA for 6 h before being harvested for protein isolation. The ABI5-GFP proteins were purified by GFP-Trap (ChromoTek) and then subjected to phosphorylation analysis using a Phos-tag Biotin BTL-111 kit as described previously (Li et al., 2019).
Data processing and statistical analysis
Histograms with average values and standard deviations were constructed using GraphPad Prism software, and the original data are presented in Supplemental Table 15. Unprocessed images of the western blots are shown in Supplemental Figure 10. ImageJ was used to quantify blot signals. Statistical differences were calculated using one-way analysis of variance (ANOVA) in SPSS software or two-tailed Student’s t-test in Microsoft Excel. Different letters indicate means that are significantly different according to Tukey’s multiple comparison test (P < 0.05). ∗P < 0.05.
Funding
This work was supported by grants from the Open Competition Program of Top Ten Critical Priorities of Agricultural Science and Technology Innovation for the 14th Five-Year Plan of Guangdong Province (2022SDZG05) and the National Natural Science Foundation of China (32270291, 32061160467, 31870171) to C.G.; the Youth Innovation Promotion Association, Chinese Academy of Sciences (2023364), the Guangdong Basic and Applied Basic Research Foundation (2022A1515012319), and the Guangzhou Basic and Applied Basic Research Foundation (2023A04J0094) to C.Y.; the Sub-Project of Chinese Academy of Sciences Pilot Project (XDA24030502) and the Guangdong Provincial Special Fund for Modern Agriculture Industry Technology Innovation Teams (2020KJ148) to Y.W.; and the National Natural Science Foundation of China (32170362), the Guangdong Natural Science Funds for Distinguished Young Scholars (2022B1515020026), the Youth Innovation Promotion Association, Chinese Academy of Sciences (Y2021094), and the Fund of South China Botanical Garden, Chinese Academy of Sciences (QNXM-02) to M.L.
Author contributions
C.Y., C.G., Y.W., and M.L. conceived of the project and designed the experiments. C.Y., X.L., S.C., C.L., L.Y., K.L., J.L., X.Z., and H.L. performed the experiments. C.Y., C.G., Y.L., S.Z., X.Z., P.R., Y.W., and M.L. analyzed the results. C.Y., C.G., Y.L., X.Z., P.R., Y.W., and M.L. wrote and edited the manuscript. All authors have read and approved the manuscript.
Acknowledgments
We thank Faqiang Li (South China Agricultural University) and Richard Vierstra (University of Wisconsin, Madison) for providing ABI5 antibody. No conflict of interest is declared.
Published: June 9, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Contributor Information
Chao Yang, Email: chaoyang@scbg.ac.cn.
Ming Luo, Email: luoming@scbg.ac.cn.
Ying Wang, Email: yingwang@scib.ac.cn.
Caiji Gao, Email: gaocaiji@m.scnu.edu.cn.
Supplemental information
References
- Addicott F.T., Lyon J.L., Ohkuma K., Thiessen W.E., Carns H.R., Smith O.E., Cornforth J.W., Milborrow B.V., Ryback G., Wareing P.F. Abscisic acid: a new name for abscisin II (dormin) Science. 1968;159:1493. doi: 10.1126/science.159.3822.1493.b. [DOI] [PubMed] [Google Scholar]
- Branon T.C., Bosch J.A., Sanchez A.D., Udeshi N.D., Svinkina T., Carr S.A., Feldman J.L., Perrimon N., Ting A.Y. Efficient proximity labeling in living cells and organisms with TurboID. Nat. Biotechnol. 2018;36:880–887. doi: 10.1038/nbt.4201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocard I.M., Lynch T.J., Finkelstein R.R. Regulation and role of the Arabidopsis abscisic acid-insensitive 5 gene in abscisic acid, sugar, and stress response. Plant Physiol. 2002;129:1533–1543. doi: 10.1104/pp.005793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carles C., Bies-Etheve N., Aspart L., Léon-Kloosterziel K.M., Koornneef M., Echeverria M., Delseny M. Regulation of Arabidopsis thaliana Em genes: role of ABI5. Plant J. 2002;30:373–383. doi: 10.1046/j.1365-313x.2002.01295.x. [DOI] [PubMed] [Google Scholar]
- Chen K., Li G.J., Bressan R.A., Song C.P., Zhu J.K., Zhao Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020;62:25–54. doi: 10.1111/jipb.12899. [DOI] [PubMed] [Google Scholar]
- Chen S., Li X., Yang C., Yan W., Liu C., Tang X., Gao C. Genome-wide identification and characterization of FCS-like zinc finger (FLZ) family genes in maize (Zea mays) and functional analysis of ZmFLZ25 in plant abscisic acid response. Int. J. Mol. Sci. 2021;22:3529. doi: 10.3390/ijms22073529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Cornforth J.W., Milborrow B.V., Ryback G., Wareing P.F. Chemistry and physiology of ‘Dormins’ in sycamore: identity of sycamore ‘dormin’ with abscisin II. Nature. 1965;205:1269–1270. doi: 10.1038/2051269b0. [DOI] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R., Abrams S.R. Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- De Giorgi J., Piskurewicz U., Loubery S., Utz-Pugin A., Bailly C., Mène-Saffrané L., Lopez-Molina L. An endosperm-associated cuticle is required for Arabidopsis seed viability, dormancy and early control of germination. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eagles C.F., Wareing P.F. Dormancy regulators in woody plants: experimental induction of dormancy in betula pubescens. Nature. 1963;199:874–875. doi: 10.1038/199874a0. [DOI] [Google Scholar]
- Finkelstein R., Gampala S.S.L., Lynch T.J., Thomas T.L., Rock C.D. Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI)5 and ABRE-BINDING FACTOR (ABF)3. Plant Mol. Biol. 2005;59:253–267. doi: 10.1007/s11103-005-8767-2. [DOI] [PubMed] [Google Scholar]
- Finkelstein R.R. Mutations at two new Arabidopsis ABA response loci are similar to the abi3 mutations. Plant J. 1994;5:765–771. doi: 10.1046/j.1365-313X.1994.5060765.x. [DOI] [Google Scholar]
- Finkelstein R.R., Lynch T.J. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell. 2000;12:599–609. doi: 10.1105/tpc.12.4.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Zhu J.K. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. USA. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Verslues P.E., Zhu J.K. Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell. 2007;19:485–494. doi: 10.1105/tpc.106.048538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T., Maruyama K., Fujita Y., Umezawa T., Yoshida R., Shinozaki K., Yamaguchi-Shinozaki K. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. USA. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franks R.G., Wang C., Levin J.Z., Liu Z. SEUSS, a member of a novel family of plant regulatory proteins, represses floral homeotic gene expression with LEUNIG. Development. 2002;129:253–263. doi: 10.1242/dev.129.1.253. [DOI] [PubMed] [Google Scholar]
- Gampala S.S.L., Finkelstein R.R., Sun S.S.M., Rock C.D. ABI5 interacts with abscisic acid signaling effectors in rice protoplasts. J. Biol. Chem. 2002;277:1689–1694. doi: 10.1074/jbc.M109980200. [DOI] [PubMed] [Google Scholar]
- Garcia M.E., Lynch T., Peeters J., Snowden C., Finkelstein R. A small plant-specific protein family of ABI five binding proteins (AFPs) regulates stress response in germinating Arabidopsis seeds and seedlings. Plant Mol. Biol. 2008;67:643–658. doi: 10.1007/s11103-008-9344-2. [DOI] [PubMed] [Google Scholar]
- Gendrel A.V., Lippman Z., Martienssen R., Colot V. Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods. 2005;2:213–218. doi: 10.1038/nmeth0305-213. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Guzman M., Pizzio G.A., Antoni R., Vera-Sirera F., Merilo E., Bassel G.W., Fernández M.A., Holdsworth M.J., Perez-Amador M.A., Kollist H., Rodriguez P.L. Arabidopsis PYR/PYL/RCAR receptors play a major role in quantitative regulation of stomatal aperture and transcriptional response to abscisic acid. Plant Cell. 2012;24:2483–2496. doi: 10.1105/tpc.112.098574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo P., Chong L., Wu F., Hsu C.C., Li C., Zhu J.K., Zhu Y. Mediator tail module subunits MED16 and MED25 differentially regulate abscisic acid signaling in Arabidopsis. J. Integr. Plant Biol. 2021;63:802–815. doi: 10.1111/jipb.13062. [DOI] [PubMed] [Google Scholar]
- Hamburger D., Rezzonico E., MacDonald-Comber Petétot J., Somerville C., Poirier Y. Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell. 2002;14:889–902. doi: 10.1105/tpc.000745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Tang W., Swain J.D., Green A.L., Jack T.P., Gan S. Networking senescence-regulating pathways by using Arabidopsis enhancer trap lines. Plant Physiol. 2001;126:707–716. doi: 10.1104/pp.126.2.707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y., Gan S. A novel zinc-finger protein with a proline-rich domain mediates ABA-regulated seed dormancy in Arabidopsis. Plant Mol. Biol. 2004;54:1–9. doi: 10.1023/B:PLAN.0000028730.10834.e3. [DOI] [PubMed] [Google Scholar]
- Holdsworth M.J., Bentsink L., Soppe W.J.J. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008;179:33–54. doi: 10.1111/j.1469-8137.2008.02437.x. [DOI] [PubMed] [Google Scholar]
- Hu Y., Han X., Yang M., Zhang M., Pan J., Yu D. The transcription factor INDUCER OF CBF EXPRESSION1 interacts with ABSCISIC ACID INSENSITIVE5 and DELLA proteins to fine-tune abscisic acid signaling during seed germination in Arabidopsis. Plant Cell. 2019;31:1520–1538. doi: 10.1105/tpc.18.00825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang Y., Sun M.M., Ye Q., Wu X.Q., Wu W.H., Chen Y.F. Abscisic acid modulates seed germination via ABA INSENSITIVE5-mediated PHOSPHATE1. Plant Physiol. 2017;175:1661–1668. doi: 10.1104/pp.17.00164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamsheer K M., Laxmi A. Expression of Arabidopsis FCS-Like Zinc finger genes is differentially regulated by sugars, cellular energy level, and abiotic stress. Front. Plant Sci. 2015;6:746. doi: 10.3389/fpls.2015.00746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamsheer K M., Sharma M., Singh D., Mannully C.T., Jindal S., Shukla B.N., Laxmi A. FCS-like zinc finger 6 and 10 repress SnRK1 signalling in Arabidopsis. Plant J. 2018;94:232–245. doi: 10.1111/tpj.13854. [DOI] [PubMed] [Google Scholar]
- Jamsheer K M., Jindal S., Sharma M., Awasthi P., S S., Sharma M., Mannully C.T., Laxmi A. A negative feedback loop of TOR signaling balances growth and stress-response trade-offs in plants. Cell Rep. 2022;39 doi: 10.1016/j.celrep.2022.110631. [DOI] [PubMed] [Google Scholar]
- Ju L., Jing Y., Shi P., Liu J., Chen J., Yan J., Chu J., Chen K.M., Sun J. JAZ proteins modulate seed germination through interaction with ABI5 in bread wheat and Arabidopsis. New Phytol. 2019;223:246–260. doi: 10.1111/nph.15757. [DOI] [PubMed] [Google Scholar]
- Katsuta S., Masuda G., Bak H., Shinozawa A., Kamiyama Y., Umezawa T., Takezawa D., Yotsui I., Taji T., Sakata Y. Arabidopsis Raf-like kinases act as positive regulators of subclass III SnRK2 in osmostress signaling. Plant J. 2020;103:634–644. doi: 10.1111/tpj.14756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Murata M., Minami H., Yamamoto S., Kagaya Y., Hobo T., Yamamoto A., Hattori T. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005;44:939–949. doi: 10.1111/j.1365-313X.2005.02583.x. [DOI] [PubMed] [Google Scholar]
- Lee K.P., Piskurewicz U., Turečková V., Carat S., Chappuis R., Strnad M., Fankhauser C., Lopez-Molina L. Spatially and genetically distinct control of seed germination by phytochromes A and B. Genes Dev. 2012;26:1984–1996. doi: 10.1101/gad.194266.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.C., Luan S. ABA signal transduction at the crossroad of biotic and abiotic stress responses. Plant Cell Environ. 2012;35:53–60. doi: 10.1111/j.1365-3040.2011.02426.x. [DOI] [PubMed] [Google Scholar]
- Leung J., Giraudat J. Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;49:199–222. doi: 10.1146/annurev.arplant.49.1.199. [DOI] [PubMed] [Google Scholar]
- Li H., Wei J., Liao Y., Cheng X., Yang S., Zhuang X., Zhang Z., Shen W., Gao C. MLKs kinases phosphorylate the ESCRT component FREE1 to suppress abscisic acid sensitivity of seedling establishment. Plant Cell Environ. 2022;45:2004–2018. doi: 10.1111/pce.14336. [DOI] [PubMed] [Google Scholar]
- Li H., Li Y., Zhao Q., Li T., Wei J., Li B., Shen W., Yang C., Zeng Y., Rodriguez P.L., et al. The plant ESCRT component FREE1 shuttles to the nucleus to attenuate abscisic acid signalling. Nat. Plants. 2019;5:512–524. doi: 10.1038/s41477-019-0400-5. [DOI] [PubMed] [Google Scholar]
- Li X., Huang Y., Han Y., Yang Q., Zheng Y., Li W., Shen W., Yang C., Gao C. Arabidopsis flowering integrator SOC1 transcriptionally regulates autophagy in response to long-term carbon starvation. J. Exp. Bot. 2022;73:6589–6599. doi: 10.1093/jxb/erac298. [DOI] [PubMed] [Google Scholar]
- Lim S., Park J., Lee N., Jeong J., Toh S., Watanabe A., Kim J., Kang H., Kim D.H., Kawakami N., Choi G. ABA-insensitive3, ABA-insensitive5, and DELLAs Interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell. 2013;25:4863–4878. doi: 10.1105/tpc.113.118604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Li Y., Wang Y., Liu X., Ma L., Zhang Z., Mu C., Zhang Y., Peng L., Xie S., et al. Initiation and amplification of SnRK2 activation in abscisic acid signaling. Nat. Commun. 2021;12:2456. doi: 10.1038/s41467-021-22812-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long J.A., Ohno C., Smith Z.R., Meyerowitz E.M. TOPLESS regulates apical embryonic fate in Arabidopsis. Science. 2006;312:1520–1523. doi: 10.1126/science.1123841. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Chua N.H. A null mutation in a bZIP factor confers ABA-insensitivity in Arabidopsis thaliana. Plant Cell Physiol. 2000;41:541–547. doi: 10.1093/pcp/41.5.541. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Chua N.H. A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2001;98:4782–4787. doi: 10.1073/pnas.081594298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., McLachlin D.T., Chait B.T., Chua N.H. ABI5 acts downstream of ABI3 to execute an ABA-dependent growth arrest during germination. Plant J. 2002;32:317–328. doi: 10.1046/j.1365-313x.2002.01430.x. [DOI] [PubMed] [Google Scholar]
- Lopez-Molina L., Mongrand S., Kinoshita N., Chua N.H. AFP is a novel negative regulator of ABA signaling that promotes ABI5 protein degradation. Genes Dev. 2003;17:410–418. doi: 10.1101/gad.1055803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., Grill E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- Manfre A.J., Lanni L.M., Marcotte W.R., Jr. The Arabidopsis group 1 LATE EMBRYOGENESIS ABUNDANT protein ATEM6 is required for normal seed development. Plant Physiol. 2006;140:140–149. doi: 10.1104/pp.105.072967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mair A., Xu S.L., Branon T.C., Ting A.Y., Bergmann D.C. Proximity labeling of protein complexes and cell-type-specific organellar proteomes in Arabidopsis enabled by TurboID. Elife. 2019;8 doi: 10.7554/eLife.47864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K.I., Miyakawa T., Sawano Y., Kubota K., Kang H.J., Asano A., Miyauchi Y., Takahashi M., Zhi Y., Fujita Y., et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- Nakabayashi K., Okamoto M., Koshiba T., Kamiya Y., Nambara E. Genome-wide profiling of stored mRNA in Arabidopsis thaliana seed germination: epigenetic and genetic regulation of transcription in seed. Plant J. 2005;41:697–709. doi: 10.1111/j.1365-313X.2005.02337.x. [DOI] [PubMed] [Google Scholar]
- Nakamura S., Lynch T.J., Finkelstein R.R. Physical interactions between ABA response loci of Arabidopsis. Plant J. 2001;26:627–635. doi: 10.1046/j.1365-313x.2001.01069.x. [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., Maruyama K., Yoshida T., Ishiyama K., Kobayashi M., et al. Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 2009;50:1345–1363. doi: 10.1093/pcp/pcp083. [DOI] [PubMed] [Google Scholar]
- Nguyen Q.T.C., Lee S.J., Choi S.W., Na Y.J., Song M.R., Hoang Q.T.N., Sim S.Y., Kim M.S., Kim J.I., Soh M.S., Kim S.Y. Arabidopsis Raf-like kinase Raf10 is a regulatory component of core ABA signaling. Mol. Cells. 2019;42:646–660. doi: 10.14348/molcells.2019.0173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N., Hitomi K., Arvai A.S., Rambo R.P., Hitomi C., Cutler S.R., Schroeder J.I., Getzoff E.D. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Malley R.C., Huang S.S.C., Song L., Lewsey M.G., Bartlett A., Nery J.R., Galli M., Gallavotti A., Ecker J.R. Cistrome and epicistrome features shape the regulatory DNA landscape. Cell. 2016;165:1280–1292. doi: 10.1016/j.cell.2016.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohkuma K., Lyon J.L., Addicott F.T., Smith O.E. Abscisin II, an abscission-accelerating substance from Young cotton fruit. Science. 1963;142:1592–1593. doi: 10.1126/science.142.3599.1592. [DOI] [PubMed] [Google Scholar]
- Pan J., Wang H., Hu Y., Yu D. Arabidopsis VQ18 and VQ26 proteins interact with ABI5 transcription factor to negatively modulate ABA response during seed germination. Plant J. 2018;95:529–544. doi: 10.1111/tpj.13969. [DOI] [PubMed] [Google Scholar]
- Park S.Y., Fung P., Nishimura N., Jensen D.R., Fujii H., Zhao Y., Lumba S., Santiago J., Rodrigues A., Chow T.F.F., et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran X., Zhao F., Wang Y., Liu J., Zhuang Y., Ye L., Qi M., Cheng J., Zhang Y. Plant Regulomics: a data-driven interface for retrieving upstream regulators from plant multi-omics data. Plant J. 2020;101:237–248. doi: 10.1111/tpj.14526. [DOI] [PubMed] [Google Scholar]
- Reeves W.M., Lynch T.J., Mobin R., Finkelstein R.R. Direct targets of the transcription factors ABA-Insensitive(ABI)4 and ABI5 reveal synergistic action by ABI4 and several bZIP ABA response factors. Plant Mol. Biol. 2011;75:347–363. doi: 10.1007/s11103-011-9733-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J., Dupeux F., Round A., Antoni R., Park S.Y., Jamin M., Cutler S.R., Rodriguez P.L., Márquez J.A. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–668. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- Saruhashi M., Kumar Ghosh T., Arai K., Ishizaki Y., Hagiwara K., Komatsu K., Shiwa Y., Izumikawa K., Yoshikawa H., Umezawa T., et al. Plant Raf-like kinase integrates abscisic acid and hyperosmotic stress signaling upstream of SNF1-related protein kinase2. Proc. Natl. Acad. Sci. USA. 2015;112:E6388–E6396. doi: 10.1073/pnas.1511238112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skubacz A., Daszkowska-Golec A., Szarejko I. The role and regulation of ABI5 (ABA-Insensitive 5) in plant development, abiotic stress responses and phytohormone crosstalk. Front. Plant Sci. 2016;7:1884. doi: 10.3389/fpls.2016.01884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S.L., Williams L.A., Farmer L.M., Vierstra R.D., Callis J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell. 2006;18:3415–3428. doi: 10.1105/tpc.106.046532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Zhang J., Hsu P.K., Ceciliato P.H.O., Zhang L., Dubeaux G., Munemasa S., Ge C., Zhao Y., Hauser F., Schroeder J.I. MAP3 Kinase-dependent SnRK2-kinase activation is required for abscisic acid signal transduction and rapid osmotic stress response. Nat. Commun. 2020;11:12. doi: 10.1038/s41467-019-13875-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyanova S., Temu T., Sinitcyn P., Carlson A., Hein M.Y., Geiger T., Mann M., Cox J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods. 2016;13:731–740. doi: 10.1038/nmeth.3901. [DOI] [PubMed] [Google Scholar]
- Xing H.L., Dong L., Wang Z.P., Zhang H.Y., Han C.Y., Liu B., Wang X.C., Chen Q.J. A CRISPR/Cas9 toolkit for multiplex genome editing in plants. BMC Plant Biol. 2014;14:327. doi: 10.1186/s12870-014-0327-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Ma Y., He Y., Tian Z., Li J. OsOFP19 modulates plant architecture by integrating the cell division pattern and brassinosteroid signaling. Plant J. 2018;93:489–501. doi: 10.1111/tpj.13793. [DOI] [PubMed] [Google Scholar]
- Yang C., Shen W., Yang L., Sun Y., Li X., Lai M., Wei J., Wang C., Xu Y., Li F., et al. HY5-HDA9 module transcriptionally regulates plant autophagy in response to light-to-dark conversion and nitrogen starvation. Mol. Plant. 2020;13:515–531. doi: 10.1016/j.molp.2020.02.011. [DOI] [PubMed] [Google Scholar]
- Yang M., Han X., Yang J., Jiang Y., Hu Y. The Arabidopsis circadian clock protein PRR5 interacts with and stimulates ABI5 to modulate abscisic acid signaling during seed germination. Plant Cell. 2021;33:3022–3041. doi: 10.1093/plcell/koab168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Wu Y., Xie Q. Precise protein post-translational modifications modulate ABI5 activity. Trends Plant Sci. 2015;20:569–575. doi: 10.1016/j.tplants.2015.05.004. [DOI] [PubMed] [Google Scholar]
- Zhao X., Dou L., Gong Z., Wang X., Mao T. BES1 hinders ABSCISIC ACID INSENSITIVE5 and promotes seed germination in Arabidopsis. New Phytol. 2019;221:908–918. doi: 10.1111/nph.15437. [DOI] [PubMed] [Google Scholar]
- Zhou X., Hao H., Zhang Y., Bai Y., Zhu W., Qin Y., Yuan F., Zhao F., Wang M., Hu J., et al. SOS2-LIKE PROTEIN KINASE5, an SNF1-RELATED PROTEIN KINASE3-type protein kinase, is important for abscisic acid responses in Arabidopsis through phosphorylation of ABSCISIC ACID-INSENSITIVE5. Plant Physiol. 2015;168:659–676. doi: 10.1104/pp.114.255455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O., Benner C., Chanda S.K. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 2019;10:1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.