Abstract
The sessile lifestyle of plants requires an immediate response to environmental stressors that affect photosynthesis, growth, and crop yield. Here, we showed that three abiotic perturbations—heat, cold, and high light—triggered considerable changes in the expression signatures of 42 epitranscriptomic factors (writers, erasers, and readers) with putative chloroplast-associated functions that formed clusters of commonly expressed genes in Arabidopsis. The expression changes under all conditions were reversible upon deacclimation, identifying epitranscriptomic players as modulators in acclimation processes. Chloroplast dysfunctions, particularly those induced by the oxidative stress-inducing norflurazon in a largely GENOME UNCOUPLED-independent manner, triggered retrograde signals to remodel chloroplast-associated epitranscriptomic expression patterns. N6-methyladenosine (m6A) is known as the most prevalent RNA modification and impacts numerous developmental and physiological functions in living organisms. During cold treatment, expression of components of the primary nuclear m6A methyltransferase complex was upregulated, accompanied by a significant increase in cellular m6A mRNA marks. In the cold, the presence of FIP37, a core component of the writer complex, played an important role in positive regulation of thylakoid structure, photosynthetic functions, and accumulation of photosystem I, the Cytb6f complex, cyclic electron transport proteins, and Curvature Thylakoid1 but not that of photosystem II components and the chloroplast ATP synthase. Downregulation of FIP37 affected abundance, polysomal loading, and translation of cytosolic transcripts related to photosynthesis in the cold, suggesting m6A-dependent translational regulation of chloroplast functions. In summary, we identified multifaceted roles of the cellular m6A RNA methylome in coping with cold; these were predominantly associated with chloroplasts and served to stabilize photosynthesis.
Key words: m6A, RNA methylation, cold acclimation, stress response, Arabidopsis thaliana, photosynthesis, chloroplast
Environment- and chloroplast dysfunction-dependent remodeling of gene expression of 42 epitranscriptomic factors associated with chloroplasts revealed their involvement in acclimation processes. The most prevalent m6A marks and the expression of m6A writers increased during cold treatment, positively regulating photosynthetic functions and thylakoid structure through regulation of cytoplasmic translation. These findings highlight the important role of m6A RNA modification in stabilizing photosynthesis during cold stress in chloroplasts.
Introduction
Natural environmental fluctuations can cause unfavorable temperatures, light levels, and water accessibility that negatively affect plants on multiple levels, such as growth, development, and photosynthesis. In addition to regulation of gene expression, mechanisms of posttranscriptional regulation at the RNA and protein levels represent important layers in the control of plant responses to adverse environmental conditions (Chinnusamy et al., 2007; Nakaminami and Seki, 2018). Therefore, a complex response to stressors must also be tightly regulated at multiple levels. Notably, many of these reactions involve chloroplasts, which constantly act as environmental sensors and targets of challenging and ever-changing conditions (Crosatti et al., 2013; Crawford et al., 2018; Manavski et al., 2018; Kleine et al., 2021; Schwenkert et al., 2021). Nonetheless, the molecular mechanisms used to overcome these challenges are far from being fully understood.
Posttranscriptional RNA modifications are widespread in living organisms, and over 170 types of chemical modification have been identified to date (Boccaletto et al., 2021). Regulatory proteins that install (writers), remove (erasers), and interpret (readers) such marks in a site-specific manner are involved in these epitranscriptomic modifications (Manavski et al., 2021a). By affecting RNA stability, splicing, transport, assembly, interactions, and translation, these modifications modulate the fate of numerous coding and non-coding RNAs (Shi et al., 2020). N6-methyladenosine (m6A) is considered the most common internal modification in eukaryotic mRNAs. Importantly, m6A methylation marks are known to be pervasive in nucleus-derived mRNAs related to chloroplast categories and are also highly enriched in chloroplast mRNAs (Luo et al., 2014; Wang et al., 2017; Manavski et al., 2021a; Qin et al., 2022).
In plants, the major and conserved nuclear m6A writer complex contains the two annotated methyltransferases MTA and MTB, the methylation factor VIRILIZER (VIR), the conserved E3 ubiquitin ligase-like HAKAI, and the central component FIP37 (Shen et al., 2016; Růžička et al., 2017; Shen, 2023). With the exception of HAKAI, other null alleles of this writer complex are arrested at the globular stage of embryonic development (Růžička et al., 2017).
Besides developmental processes, m6A is also linked to biotic and abiotic stress responses in living organisms (Liang et al., 2020: Manavski et al., 2021a; Yu et al., 2021). For example, under salt stress, m6A marks protect several transcripts related to salt and osmotic stress response from degradation in Arabidopsis (Anderson et al., 2018). Accordingly, m6A writer mutants also displayed salt-sensitive phenotypes (Hu et al., 2021). Under heat, responsive transcripts are relocated to stress granules by the m6A reader ECT2, potentially playing a role in RNA stability and translation (Scutenaire et al., 2018). VIR has been shown to impact various plant developmental programs and has a positive effect on photosynthesis under high light (Růžička et al., 2017; Zhang et al., 2022). Despite rapid progress in plant research on m6A-methylation marks, their importance for coping with environmental changes has only recently been explored (Manavski et al., 2021a; Zhang et al., 2022).
Here, we show that genes encoding 42 mostly uncharacterized epitranscriptomic factors with putative chloroplast function and/or localization were reversibly differentially expressed in response to perturbations by heat, high light, and cold and that they responded to the functional state of the chloroplast largely independently of Genome Uncoupled (GUN)1 and GUN5. Moreover, we show that both expression of components of the major m6A writer complex and cellular m6A mRNA methylation marks increased under cold acclimation and that FIP37, as a major component of the writer complex, was important for maintaining efficient photosynthesis and other chloroplast functions. Thus, we identified the cellular m6A RNA methylome as a central and multifaceted hub important for stabilizing functional photosynthesis in the cold and revealed the chloroplast as the main player in addressing cold response.
Results
Expression of genes encoding chloroplast-associated epitranscriptomic factors is responsive to cold, heat, high light, and the functional state of the chloroplast
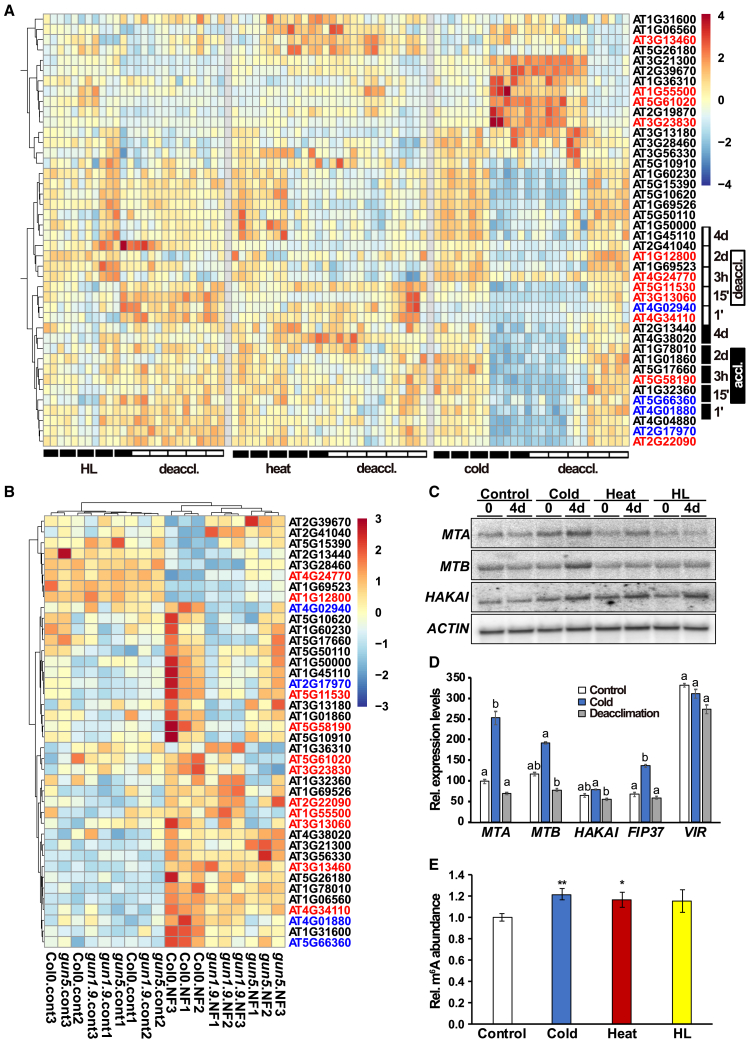
Previous transcriptome-wide maps have revealed the involvement of m6A in developmental processes and certain plant stress responses (Shen et al., 2016; Anderson et al., 2018; Hu et al., 2021; Qin et al., 2022). Because the chloroplast appears to play a crucial role as a sensor of environmental changes (Kleine et al., 2021), we selected 42 annotated and largely uncharacterized epitranscriptomic players (writers, erasers, and readers) from Arabidopsis databases that were either potentially associated with chloroplast functions and/or located within this organelle (Supplemental Table 1). We then investigated the expression behavior of these factors upon 4-day cold, heat, and high light treatments, conditions under which we recently observed global transcriptomic changes (Garcia-Molina et al., 2020). Normalized transcript values were used to create a heatmap with hierarchical clustering to capture overall expression patterns. Remarkably, heatmaps showed that all 42 selected genes displayed changes in transcript abundance in at least one of the three treatments (Figure 1A). Depending on the condition, transcripts formed clusters of co-expressed genes that were either up- or downregulated, suggesting coordinated control during acclimation and an important link between epitranscriptomic marks and chloroplast perception of external stimuli. Importantly, the expression levels of all players returned to levels of the previous standard conditions within 4 days of deacclimation, suggesting that the factors are indeed directly involved in the acclimation response (Figure 1A).
Figure 1.
Changes in transcript levels of writers, erasers, and readers during acclimation to high light, heat, and cold followed by deacclimation.
(A and B) Heatmaps with hierarchical clustering of transcript values for chloroplast-associated epitranscriptomic players (writers, erasers, and readers in black, blue, and red, respectively) were elaborated according to the Ward d2 method using (A) z-means of plants (de)acclimated to high light, heat, and cold treatments for 4 days as reported recently (Garcia-Molina et al., 2020) or (B)Z scores of normalized 5-day-old WT (Col0), genome uncoupled (gun)1-9, and gun5 mutant seedlings under control or norflurazon treatment as reported by Koussevitzky et al. (2007).
(C) RNA gel blot analysis of selected genes of the m6A writer complex in the WT before and after 4 days of cold, heat, and high light treatment using 8 μg leaf RNA. An actin probe was used as the loading control.
(D) Reversibility of upregulation of genes encoding m6A writer components during deacclimation, as revealed by RNA sequencing analysis. Bar plots depict means ± standard deviation for normalized counts of selected transcripts in pretreated, cold-treated, and deacclimated plants at day 4. Statistical significance was tested by one-way ANOVA with post hoc Tukey’s honestly significant difference (HSD) test (p ≤ 0.05).
(E) Quantification of m6A marks in poly(A)-enriched RNA samples using LC–MS. m6A abundance was normalized to that of the control for each sample (n = 5), and statistical significance among treatments was determined using one-way ANOVA and Tukey’s HSD post hoc test (∗p ≤ 0.05; ∗∗p ≤ 0.01). Normalization to the control was performed for each sample.
It is tempting to assume that the chloroplast itself plays a central role in regulating m6A methylation because it is generally thought to sense environmental changes that can trigger retrograde signals (Kleine et al., 2021). To investigate the relevance of chloroplast signaling for the cellular m6A-methylation machinery, transcript levels of 40 epitranscriptomic factors under standard conditions were compared with those of norflurazon (NF)-treated wild-type (WT), gun1-9, and gun5 mutant seedlings (Koussevitzky et al., 2007) (Figure 1B). The heatmap depicted two main trends corresponding to core sets of transcripts that tended to be either down- (upper 8 transcripts) or upregulated (remaining transcripts) upon NF treatment. Interestingly, transcript levels of the selected genes displayed similar values under standard conditions for all three genotypes, indicating that most genes responded to the functional state of the chloroplast largely independently of GUN1 and GUN5 proteins. The degree of induction or repression upon NF treatment changed only partially in both gun mutants compared with the WT (Figure 1B). Thus, expression of the epitranscriptomic factors is also responsive to the functional state of the chloroplast, but gun1-9 and gun5 do not markedly affect this communication for most genes.
Given that m6A is the most frequent RNA modification, we examined gene expression of three representative members of the main nuclear m6A writer complex, MTA, MTB, and HAKAI, using RNA gel blot analysis under control conditions and the three treatments (Figure 1C). Interestingly, expression of MTA, MTB, and HAKAI was upregulated after 4 days of cold treatment. HAKAI also showed increased expression under high light treatment, and both HAKAI and MTA showed increased expression in response to heat. Thus, our results suggest an individual response of the components of the m6A writer complex to all three environmental conditions and, in particular, a concerted activation of the three components under cold. To corroborate these data under cold conditions and verify the ability to deacclimate, we retrieved expression data for the five components of the writer complex from our previous transcriptomic study (Garcia-Molina et al., 2020) (Figure 1D). Indeed, expression of MTA, MTB, HAKAI, and FIP37 was clearly increased after cold treatment, whereas that of VIR showed little change. Again, expression fell to levels of pretreated plants within 4 days after transfer to standard conditions, indicating deacclimation of the cold response. These data confirm the important role of the writer complex as a modulator of the acclimation response, with individual contributions of its components.
m6A methylation of mRNAs in Arabidopsis increases in the cold
Because our results demonstrated that expression of genes encoding components of the major nuclear m6A writer complex and other chloroplast-associated epitranscriptomic players is increased upon cold treatment, it is reasonable to assume that m6A marks of cellular mRNAs also increase during cold acclimation. Absolute quantification of m6A requires normalization to the amount of injected RNA, and highly abundant tRNAs and rRNAs were therefore carefully removed prior to our liquid chromatography with mass spectrometry (LC–MS) analysis. tRNAs were removed through size-exclusion chromatography, whereas long RNA fractions were rRNA depleted through poly(A) enrichment. Quality assurance of poly(A) fractions was performed by automated gel electrophoresis (Supplemental Figure 1). Notably, the m6A content of poly(A) RNAs was significantly enriched by approximately 20% in cold- and heat-treated samples compared with control plants (Figure 1E). The observed increase in m6A under high light was not statistically relevant. It should be noted that the increase in m6A represents the sum of up- and downregulated m6A methylations and that different sites and transcripts are likely to be targeted for m6A methylation under the three chosen conditions. In this regard, recent studies have performed GO analysis of cold-treated WT and m6A mutant plants and provided the sequence context of m6A marks (Govindan et al., 2022; Wang et al., 2023). This is all consistent with the fact that the m6A-methylation pattern in heat and cold is associated with the activation of heat- and cold-sensitive genes, respectively, suggesting that different transcripts and sites are labeled with m6A under different conditions (Figure 1A) (Govindan et al., 2022; Wang et al., 2022). In summary, upregulation of m6A mRNA labeling in cold is congruent with increased cold-dependent expression of genes encoding components of the nuclear m6A writer complex (Figures 1C–1E).
Writer mutants are sensitive to cold
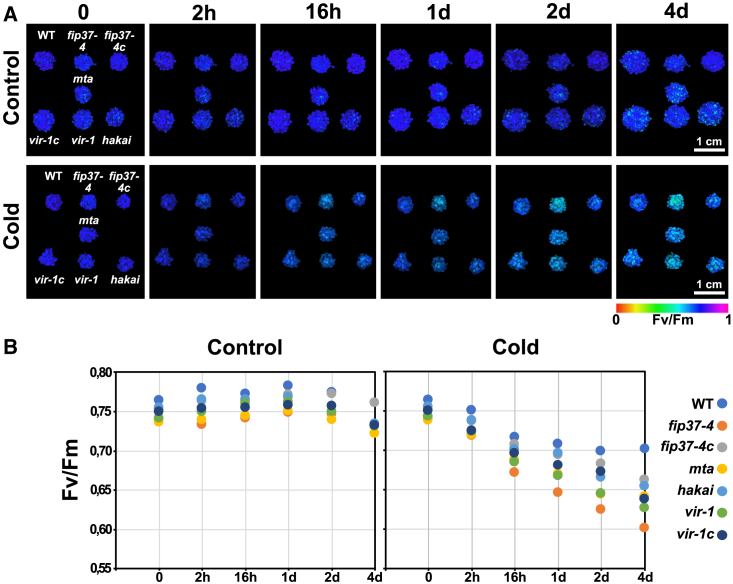
Photosynthesis is the main biological process for capturing light energy to form carbohydrates and is strongly affected by exposure to cold (Theocharis et al., 2012; Kleine et al., 2021). Because the m6A content of mRNAs was considerably enriched in Arabidopsis under cold, we next investigated how previously described mutants of individual subunits of the major writer complex and corresponding complemented lines behaved under this condition (Shen et al., 2016; Růžička et al., 2017; Qin et al., 2022; Shen, 2023; Wong et al., 2023). Hypomorphic alleles of FIP37, VIR, and MTA showed a decrease in m6A levels to about 10%–15%, and the hakai knockout decreased to about 65% (Růžička et al., 2017; Parker et al., 2020). We initially measured photosystem II (PSII) integrity (Fv/Fm) of the four fip37-4, mta, vir-1, and hakai mutant lines of the m6A writer complex during cold acclimation. None of the mutant lines showed a marked decrease in the maximum quantum yield of PSII, measured as Fv/Fm under standard conditions, indicating that photosynthesis was not primarily impaired in the mutants (Figure 2A). However, Fv/Fm decreased much faster in these lines than in the WT just a few hours after cold exposure (Figure 2A). After 16 h of cold acclimation, Fv/Fm stagnated at about 0.7 in the WT, whereas it still showed a continuous decline within the first 4 days in the mutants (Figure 2B). The complemented vir1 and fip37-4 mutant lines (referred to here as vir-1c and fip37-4c) performed much better than the single mutants but failed to fully recover the Fv/Fm level, indicating incomplete complementation, probably due to the presence of C-terminal hemagglutinin tags in both lines, which could slightly affect the stability of the writer complex with functional interdependent subunits (Shen, 2023). Because the strongest effect of cold on Fv/Fm was observed for fip37-4 (Figure 2), we took a closer look at FIP37 to obtain a comprehensive picture of the writer complex and its specific role in cold acclimation.
Figure 2.
Photosynthetic performance of m6A writer complex mutants is negatively affected by cold treatment.
(A) Imaging of Fv/Fm values for the WT, knockdowns (fip37-4, mta, vir-1, hakai), and corresponding complemented lines (fip37-4c, vir-1c) grown for 10 days on medium. Scale bar: 1 cm.
(B) Fv/Fm data were collected at several time points of cold treatment as indicated. The corresponding numeric Fv/Fm values reflect the cold sensitivity of the writer mutants.
If FIP37 were indeed involved in cold acclimation, we would expect the effects of cold on photosynthesis to be reversible in the corresponding mutant. Indeed, after cold treatment, fip37-4 plants were completely deacclimated upon exposure to standard conditions for another 4 days (Supplemental Figure 2). Taken together, these results suggest that the activity of the writer complex plays a distinct role in facilitating efficient photosynthesis under cold acclimation. Importantly, the fact that Fv/Fm values returned to levels of pretreated leaves within 4 days demonstrates that FIP37, and presumably the entire m6A writer complex, is a true acclimation factor or modulator (Kleine et al., 2021).
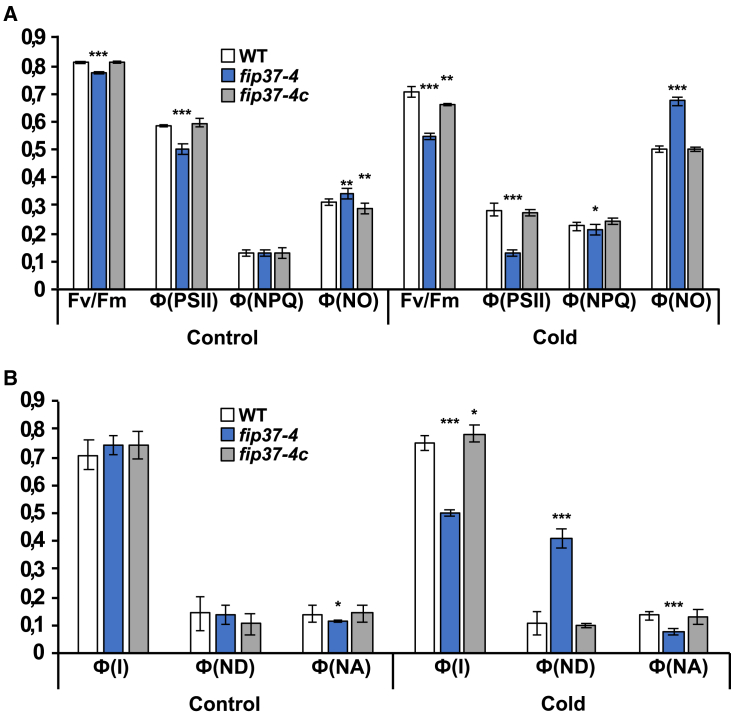
Photosynthetic efficiency is severely decreased in fip37-4 in the cold
We next investigated the effect of cold on defined photosynthetic parameters in the fip37-4 mutant to identify the primary deficiencies. Chlorophyll content, Fv/Fm, the efficient quantum yield of PSII (Ф(II)), non-photochemical quenching (Ф(NPQ)), and the non-regulated, basal non-photochemical energy loss in PSII (Φ(NO)) were not markedly changed under standard growth conditions in the mutant (Figure 3A and Supplemental Figures 2 and 3). In the cold, chlorophyll fluorescence of fip37-4 remained at higher levels during induction and decreased only slowly compared with that of the WT (Supplemental Figure 3A). This behavior is indicative of mutants with a defective electron transport chain or PSI (Meurer et al., 1996). After cold exposure, Fv/Fm and Ф(II) values were severely decreased in fip37-4 compared with WT (0.54 vs. 0.70 and 0.12 vs. 0.28, respectively), whereas Ф(NPQ) remained largely unchanged in all samples (Figure 3A). Ф(NPQ), however, increased slowly compared with the values of WT and fip37-4c, indicating that cold-treated fip37-4 was unable to properly regulate photosynthesis after the sudden onset of light (Supplemental Figure 3B). As expected, Ф(NO) levels increased in all samples in the cold, with the highest value in the mutant (0.49 vs. 0.67), indicating elevated oxidative pressure (Figure 3A). We concluded that cold treatment has a drastic and rapid effect on photosynthetic electron transport, causing increased oxidative damage in the mutant. In agreement with the PSII measurements, we found no changes in the activity of PSI under standard conditions. All three parameters, photosystem I yield (Ф(I)) and the donor and acceptor side limitations, Ф(ND) and Ф(NA), were apparently unaffected in the mutant (Figure 3B). In the cold, however, the ∼25% decrease in Ф(I) was due primarily to a four-fold increase in limitation on the donor side but not on the acceptor side, again suggesting that electron flow toward PSI is greatly reduced in cold-treated fip37-4 mutants (Figure 3B).
Figure 3.
Cold-induced photosynthetic deficiency in fip37-4.
(A) The photosynthetic parameters Fv/Fm, Φ(II), Φ(NPQ), and Φ(NO) were calculated from plants grown in soil for 14 days under standard conditions and then subjected to 4 days of cold treatment.
(B) PSI parameters Φ(I), Φ(ND), and Φ(NA) were calculated from plants grown as described in (A).
Statistically significant differences compared with the control condition were assessed using Student’s t-tests in (A) and (B) (∗p ≤ 0.05; ∗∗p ≤ 0.01; ∗∗∗p ≤ 0.001).
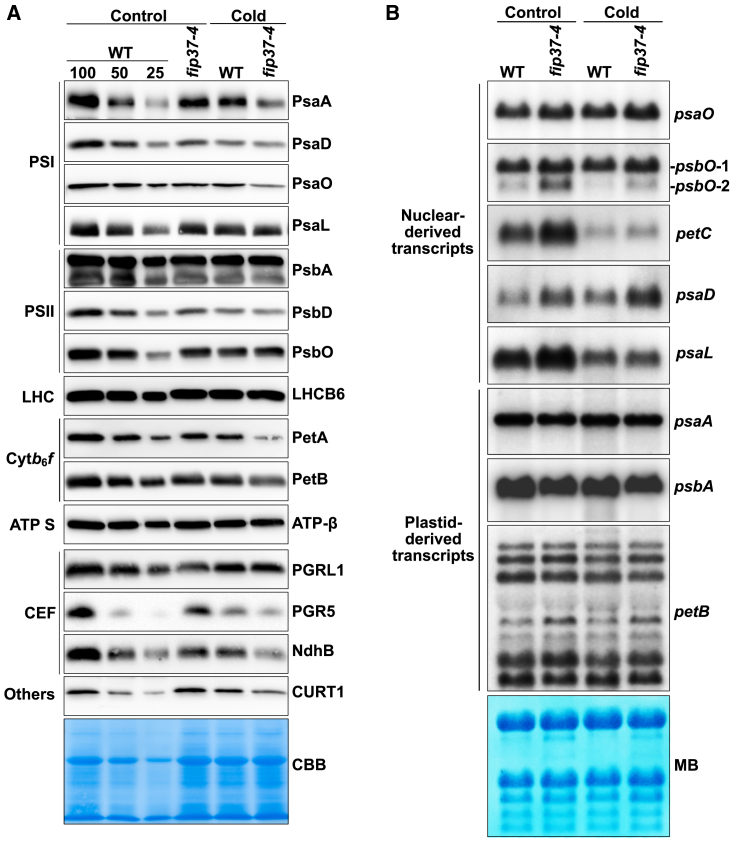
FIP37 deficiency affects accumulation of chlorophyll and specific photosynthetic complexes in the cold
We next wanted to determine whether cold treatment also affects chloroplast pigment and protein content. Chlorophyll levels in fip37-4 were reduced only in the cold to 86.68% ± 5.67% compared with those in the WT. Representative subunits of major thylakoid membrane complexes were examined by immunoblot analysis. Under standard conditions, there was a marginal decrease in all analyzed subunits of PSI (PsaA, PsaD, PsaO, PsaO), PSII (PsbA, PsbD, PsbO, LhcB6), the Cytb6f complex (Cytf, Cytb6), ATP synthase (AtpB), and the cyclic electron transport (CET) (PGR5, PGRL1, NdhB) in fip37-4 compared with the WT (Figure 4A). After a 4-day cold treatment, levels of proteins were generally slightly decreased in the WT. However, the decrease in protein levels of PSI (PsaA, PsaO), the Cytb6f complex, and the CET (PGRL1, NdhB) was much more pronounced in fip37-4 (Figure 4A). Levels of PSII and ATP synthase proteins and of PGR5 were comparable in cold-treated WT and mutant plants, indicating a specific deficiency in thylakoid membrane complexes. Furthermore, levels of CurT1, which is required for curvature of the thylakoid membrane (Armbruster et al., 2013), were also decreased after exposure to 4°C.
Figure 4.
Accumulation of photosynthetic proteins and RNAs in FIP37-deficient plants.
(A) Steady-state levels of representative subunits of photosynthetic complexes. Immunoblot analyses were performed with plants grown for 10 days under standard conditions and an additional 4 days under cold conditions. Coomassie brilliant blue staining served as a loading control. ATP S, ATP synthase.
(B) RNA gel blot analysis of plastid- and nuclear-derived transcripts encoding chloroplast proteins in WT and fip37-4 under control and cold conditions. 3 μg total RNA was loaded. Methylene blue was used as a loading control.
Expression of photosynthetic transcripts is changed in cold-treated fip37-4
We next asked whether protein levels are determined by mRNA levels and examined the expression of representative plastid and nuclear genes from PSI (PsaA, PsaD, PsaL, PsaO), the Cytb6f complex (PetB, PetC), and PSII (PsbA, PsbO1, PsbO2). Whereas levels of petC and psaL transcripts were severely downregulated in the cold in both WT and fip37-4, expression of all other genes was comparable under standard and cold conditions. Under control and cold conditions, levels of nuclear- (psaO, psbO1, psaD) and plastid-derived (psaA, psbA, petB) transcripts were slightly higher and lower, respectively, in fip37-4 than in the WT (Figure 4B). Strikingly, expression of PsbO2 and PetC was considerably upregulated in fip37-4 under standard conditions but returned to lower levels after cold treatment. However, levels of cytoplasmic transcripts still remained higher in fip37-4 than in the WT, presumably to compensate for protein deficiencies.
As has often been observed in mutants not directly affected in chloroplast RNA metabolism, slight changes in processed petB transcripts occurred between WT and fip37-4 (Figure 4B). Thus, we conclude that the observed differences in gene expression do not correlate with the decreases in the set of proteins, emphasizing that posttranscriptional changes are responsible for the observed deficiencies in protein levels.
Cytoplasmic translation is reduced in cold-treated fip37-4
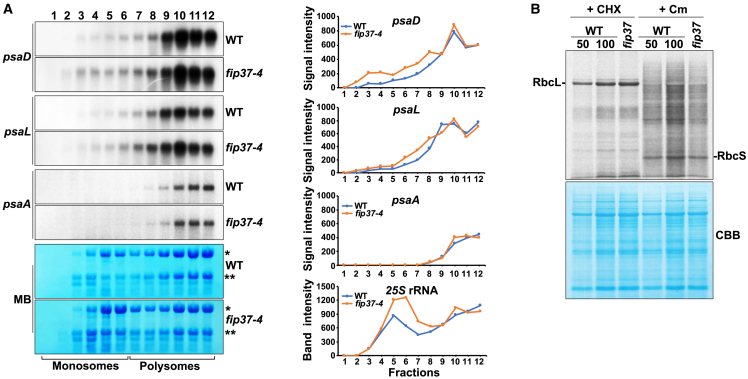
In contrast to those associated with most other GO terms, the cold-enriched m6A peaks of cytoplasmic transcripts associated with photosynthesis and chloroplast organization showed preferentially lower ribosome loading (Govindan et al., 2022), presumably explaining the decreased levels of most investigated nuclear-encoded proteins in WT plants in the cold (Figure 4). We therefore examined ribosomal occupancy of photosynthetic transcripts with altered levels in fip37-4 in the cold (Figure 5A). No obvious differences between WT and fip37-4 were observed for general ribosomal loading of chloroplast transcripts, including psaA. However, methylene blue staining of the polysome fractions showed that, in contrast to WT, a substantial proportion of cytoplasmic 25S and 18S ribosomes was found in monosomes in fip37-4 in the cold. To examine whether polysome loading of nuclear-derived photosynthetic transcripts was affected, we probed fractions with psaD and psaL. In WT and fip37-4, both transcripts were found mainly in high-molecular-weight polysomes in the cold; however, in fip37-4, some of these transcripts were also present in monosomes (Figure 5A). This is also consistent with reduced PsaD and PsaL protein levels in cold-treated fip37-4 plants and suggests a mechanism by which translation of even upregulated nucleus-derived transcripts with photosynthetic functions is downregulated owing to the deficiency of most m6A marks.
Figure 5.
Polysome loading and in vivo radiolabeling of cytoplasmic and chloroplast proteins.
(A) Separation of intact polysomes fractionated by ultracentrifugation. Fractions were stained with methylene blue. Monosomal and polysomal fractions are indicated. Isolated RNA was denatured and subjected to gel blot analysis using psaA, psaD, and psaL probes. The signal intensity of the fractions is represented graphically on the right side. MB, methylene blue.
(B) Cytoplasmic and chloroplast proteins were freshly radiolabeled in the presence of chloramphenicol or cycloheximide, respectively. In vivo labeled proteins were subjected to SDS–PAGE. RbcL and RbcS bands are indicated in the chloroplast and cytosolic labels, respectively. Asterisks show the cytoplasmic 25S (∗) and the 18S (∗∗) rRNAs. Coomassie brilliant blue staining was used as a loading control. CHX, cycloheximide; Cm, chloramphenicol.
To examine translation further, we performed in vivo labeling experiments using cycloheximide and chloramphenicol as inhibitors of cytoplasmic and chloroplast translation, respectively (Figure 5B). The data clearly indicated that general chloroplast translation remained unchanged in the fip37-4 mutant, whereas overall cytoplasmic translation was downregulated by approximately 50% when comparing protein loading. This finding confirms that reduced cytoplasmic translation of RNAs encoding chloroplast proteins is likely to be the cause of the observed deficits in plastid proteins in fip37-4. In addition, unchanged protein biosynthesis in chloroplasts suggests that decreased plastid-encoded proteins are unstable owing to the reduced amount of specific nuclear-encoded chloroplast proteins constituting the same photosynthetic complexes.
Assembly studies of photosynthetic complexes
In addition to steady-state levels of proteins, the assembly of photosynthetic thylakoid membrane complexes was analyzed by blue native PAGE using extracts from fip37-4 and WT plants exposed to cold (Supplemental Figure 4). The overall assembly of complexes did not appear to be affected. Immunological analysis of the second dimension showed that levels of all detectable PSII complexes and D1 appeared comparable in fip37-4 and the WT. However, when using equal chlorophyll loading, the mutant exhibited decreased levels of the PSI proteins PsaA and PsaG, as well as reduced levels of Cytb6 in the more prominent dimeric Cytb6f complexes compared with monomers (Supplemental Figure 4).
Role of FIP37 in thylakoid organization in the cold
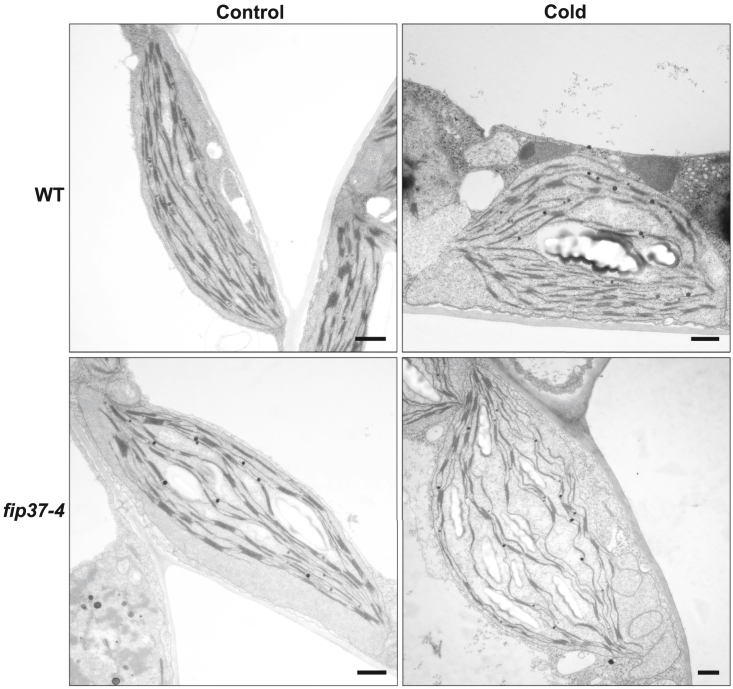
CurT1 complexes are highly enriched in grana stacks and are required for the curvature, and thus the architecture, of the thylakoid membrane (Armbruster et al., 2013). Because levels of the CurT1 protein appeared to be severely reduced in cold-acclimated fip37-4 mutants compared with the WT, we attempted to examine the ultrastructure of the chloroplast. For this, chloroplasts from 14-day-old seedlings were analyzed by transmission electron microscopy before and after 4 days of cold treatment. Under standard conditions, chloroplasts from both WT and fip37-4 had intact plastids and parallel-running thylakoid membranes with well-defined grana stacks, indicating that reduced levels of FIP37 at 22°C had no obvious effect on chloroplast ultrastructure (Figure 6). However, after cold treatment, thylakoids in fip37-4 were organized in a wavy manner, and grana stacks consisted of fewer membrane layers, similar to those of curt1 mutants, whereas the WT maintained a similar ultrastructure compared with standard conditions (Figure 6). Reduced levels of CurT1 supported the ultrastructural changes and strongly indicated that FIP37, in addition to its association with photosynthesis and cold acclimation, may potentially be involved in a broader range of chloroplast functions.
Figure 6.
Chloroplast ultrastructure of WT and fip37-4 grown under standard and cold conditions.
Representative transmission electron micrographs of WT and fip37-4 chloroplasts. Plants were cultivated on Murashige and Skoog medium for 10 days under standard conditions and then placed in the cold for 4 days. Scale bar: 1 μm.
FIP37 deficiency induces ROS formation in the cold
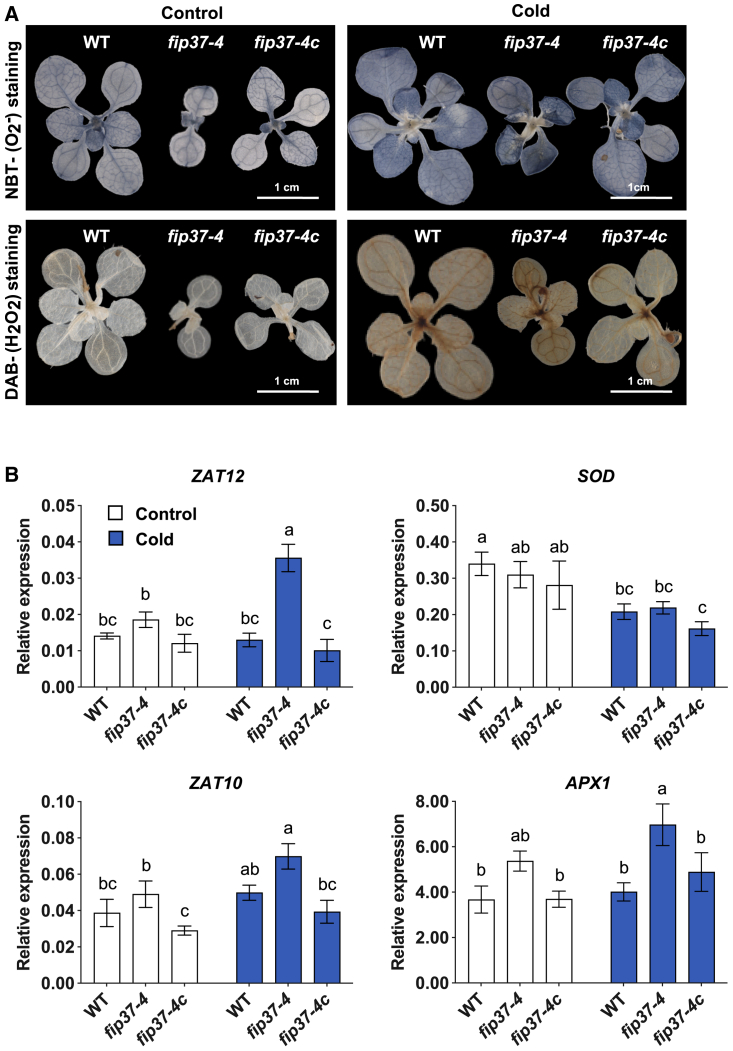
Chloroplasts can produce reactive oxygen species (ROS) under hostile environmental conditions via both photosystems (Li and Kim, 2021), and we therefore examined ROS formation after cold treatment (Figure 7A). Nitro blue tetrazolium (NBT) and 3,3′-diaminobenzidine (DAB) stains, which detect O2− and H2O2, respectively, revealed no differences among the WT, fip37-4, and fip37-4c under standard conditions at 22°C. However, transfer to cold resulted in more intense staining with both dyes in the fip37-4 mutant compared with the WT and fip37-4c, indicating an increased oxidative burst (Figure 7A).
Figure 7.
Cold treatment leads to increased accumulation of ROS in fip37-4.
(A) NBT and DAB staining of plants before or after 4 days of cold treatment. NBT staining is indicative of superoxide (O2−) production and DAB staining of H2O2 production. Scale bar: 1 cm.
(B) Expression of ROS-related genes (ZAT12, ZAT10, SOD1, and APX1) analyzed by qRT–PCR. Bars denote the means of 3 independent biological replicates with the corresponding standard deviations. Different letters within individual titles represent significant differences between tested temperatures (22°C and 4°C) and genotypes (WT, fip37-4, and fip37-4c) according to two-way ANOVA with post hoc Tukey’s HSD (p ≤ 0.05).
Because of the oxidative stress to which the mutants were exposed in the cold, expression of ROS-related genes was monitored by qRT–PCR. In the cold, expression of the ROS-responsive transcription factors ZAT12 and ZAT10 was significantly increased in fip37-4 (Figure 7B). The transcript level of superoxide dismutase showed an increase in all tested lines but no marked differences between WT and fip37-4 in the cold. Expression of ascorbate peroxidase1 was upregulated under both standard and cold conditions in the mutant (Figure 7). Overall, these results suggest that ROS formation in plants with lower FIP37 levels increased under cold conditions, although the ability to respond to oxidative stress with the induction of cold-responsive genes was not impaired.
Anthocyanin production is absent in cold-treated fip37-4
Anthocyanins are secondary metabolites that accumulate in response to pathogen attack and cold, as well as UV and strong light, suggesting a possible role in plant tolerance to various environmental conditions (Winkel-Shirley, 2002; Xu and Rothstein, 2018). However, the molecular mechanism by which anthocyanins protect photosynthetic leaves against external stressors is still enigmatic (Araguirang and Richter 2022). The purple leaf phenotype observed in the WT and fip37-4c was based on anthocyanin accumulation, which was clearly absent in fip37-4 in the cold (Supplemental Figure 5A). The anthocyanin content was calculated to be reduced to less than 20% in fip37-4 compared with the WT and fip37-4c under cold conditions (Supplemental Figure 5B). Increased ROS production is thought to suppress flavonoid biosynthesis, and synthesis of sucrose and triose phosphate is known to be a prerequisite for anthocyanin production mediated by transcription factors (Richter et al., 2020). Reduced photosynthetic efficiency and increased ROS production in fip37-4 could therefore be responsible for the lack of anthocyanins in the cold.
The fip37-4 mutant is responsive to cold
To investigate whether a lack of induction of cold-responsive genes could explain the fip37-4 phenotype, we further tested the expression of known genes associated with cold shock response and cold acclimation. C-repeat-binding factors play important roles in plant acclimation to cold (Liu et al., 2019). These factors bind to the C-repeat responsive element motif found in promoters of cold-responsive (COR) genes such as COR15A and RD29A. Cold-triggered induction of C-repeat-binding factor 2, COR15A, and RD29A expression in fip37-4 was comparable to that in the WT, albeit somewhat delayed (Supplemental Figure 6). Thus, fip37-4 did not lose its ability to induce the expression of genes important for cold acclimation, suggesting that reduced m6A methylation has little or no effect on the cold response of the studied genes.
In conclusion, our results suggest that downregulation of FIP37 has no particular effect on photosynthesis under standard conditions but is crucial for efficient photosynthesis and other chloroplast functions related to plant growth, viability, and fitness during cold acclimation.
Discussion
The identification of molecular components that mediate plant acclimation to adverse environmental conditions is an important and timely task in plant biology and breeding programs. Regardless of their nature, effects of perturbations on plants always take place on multiple levels, such as growth, development, and photosynthetic functions (Li et al., 2022). This implies that acclimation factors that allow plants to better cope with stress must also act at multiple levels. It is therefore tempting to speculate that methylation of different mRNAs at defined positions makes a significant contribution to such multifactorial responses during acclimation processes (Manavski et al., 2021a). Indeed, RNA methylation introduced by m6A writer proteins is emerging as an essential posttranscriptional mechanism of gene regulation in plant responses to (a)biotic stresses and developmental stages. The potential role of writers, erasers, and readers in plant stress response has been addressed previously (Liu et al., 2020; Lu et al., 2020: Sun et al., 2020; Manavski et al., 2021a; Shao et al., 2021; Qin et al., 2022; Zhang et al., 2022). The results presented here demonstrate that expression of all 42 chloroplast-associated epitranscriptomic players studied was responsive to cold, heat, and high light acclimation. Reversibility of the expression changes upon deacclimation strongly suggests that these factors have important modulating roles under these particular conditions (Figure 1A). The expression signature of most relevant down- and upregulated epitranscriptomic factors after NF treatment could not be properly modified by gun mutants, indicating that GUN factors rarely interfere with retrograde communication regarding RNA modifications (Figure 1B). Overall, these data provide evidence for the hierarchy of regulation of posttranscriptional RNA modification, in which the chloroplast plays a central role as an environmental sensor and trigger for mainly gun-independent retrograde signaling.
Consistent with upregulated gene expression of writer complex components in the cold, m6A marks of cellular mRNAs also increased, indicating a reshaping of the cellular m6A methylome during cold acclimation (Figures 1C–1E). The ability of the chloroplast itself to act on the cellular m6A transcriptome through retrograde signals suggests an important link between the perception of external stimuli and the cellular m6A methylome (Figure 1B). Here, we showed that FIP37, as a central subunit of the main nuclear m6A writer complex, plays an important role in regulation of multifaceted and dynamic processes related to photosynthesis, chloroplast development, and chloroplast structure during cold treatment. Moreover, we observed that the cold sensitivity of fip37-4, in terms of Fv/Fm, was independent of age, size, and developmental stage.
Interestingly, the writer mutant vir-1 has a clear and sudden effect on PSII accumulation after high light treatment (Zhang et al., 2022), whereas cold affects PSI in fip37-4. This reflects a rapid response to the generally faster occurrence of sudden high light compared with slower temperature decreases in nature and again suggests defined strategies of the cellular m6A RNA methylome for coping with different stressors. The qualitatively and temporally distinct effects of cold and high light treatment on m6A writer mutants also suggest that the activity and/or targets of the m6A writer complex change according to environmental cues. Thus, the cellular m6A methylome provides not only a regulatory platform but also an opportunity to interact with the multifaceted effects of cold and other stressors on plant processes, often primarily related to photosynthesis and chloroplast organization.
The fact that m6A writer mutants showed essentially no marked effects on photosynthesis under standard conditions provides evidence for an important and distinct role of the cellular m6A epitranscriptome in the cold and high light. Clearly, writer-induced m6A marks and their effects on the fate of mRNAs that preferentially encode chloroplast proteins positively influence photosynthesis on multiple levels under unfavorable conditions.
Evidence for a beneficial impact of m6A RNA methylation on plant performance under drought stress has only recently been described. When the methyltransferase MTB of watermelon was overexpressed in tobacco, transgenic plants showed increased drought tolerance and decreased photoinhibition, most likely due to enhanced scavenging of ROS species under drought (He et al., 2021). Furthermore, when apple plants were exposed to drought, expression of the methyltransferase MdMTA and m6A levels were both induced to promote stability and translation efficiency of transcripts involved in lignin deposition and ROS scavenging (Hou et al., 2022).
The influence of methylation on photosynthesis is also supported by the fact that mRNAs associated with chloroplast functions are frequently methylated (Shen et al., 2016; Manavski et al., 2021a; Qin et al., 2022). Moreover, photosynthetic leaves have the highest extent of m6A methylation among different organs (Wan et al., 2015). In addition, m6A demethylases, such as ALKBH9B (AT2G17970) and ALKBH10B (AT4G02940), were downregulated in the cold (Figure 1A). Depending on the stressor, epitranscriptomic players can be either up- or downregulated. For instance, expression of the RNA demethylase ALKBH6 increased under salt stress (NaCl) but decreased under cold (Huong et al., 2020). Interestingly, alkbh6 mutants grew more slowly and had a lower survival rate at high temperature compared with the WT (Huong et al., 2020). These results are consistent with our expression analysis of m6A erasers and confirm the downregulation of other Arabidopsis m6A demethylases at low temperatures (Figure 1A). Together, these results strongly suggest that m6A players may have specific roles under distinct environmental conditions.
S-adenosylmethionine synthetase (SAMS) is an enzyme that uses ATP and methionine to catalyze the biosynthesis of S-adenosylmethionine, the substrate for methylation of numerous secondary metabolites and macromolecules as well as DNA and RNA writer enzymes. Interestingly, SAMS is known to confer resistance to drought, cold, and oxidative stress in overexpression lines (He et al., 2019; Choi et al., 2022). Both the enzymatic activity and gene expression of SAMS increased in the cold compared with control conditions, showing a correlation with expression of the m6A writer components (Figures 1B and 1C). Under drought, oxidative stress, and low temperatures, SAMS-overexpressing Nicotiana benthamiana lines displayed lower ion leakage than the WT, suggesting that SAMS proteins are important for stress resistance (Choi et al., 2022). However, no correlations with methylation of RNAs or other molecules or photosynthesis were evaluated, and this would be an interesting task for future research.
Photosynthetic parameters were consistent with downregulation of proteins of PSI, the Cytb6f complex, and the CET in fip37-4 in the cold (Figures 2, 3, 4, and 5 and Supplemental Figure 4). However, slight changes in the levels of cytoplasmic and plastid transcripts of PSI, PSII, and Cytb6f in the mutant did not correlate with protein amounts (Figures 4 and 5). This implies that m6A-mediated posttranscriptional processes, such as export of RNA from the nucleus, translation, and/or protein stability, account for the cold sensitivity of the mutant. Protein deficiencies in cold-treated fip37-4 mutants compared with the WT were consistent with accumulation of cytosolic monosomes (Figure 5B). Indeed, in vivo labeling experiments clearly showed a general decrease in translation efficiency in the cytosol of cold-treated fip37-4 (Figure 5B). This demonstrates the mechanism by which FIP37 acts on cytoplasmic RNAs that mainly encode chloroplast proteins, including rbcS, and thus explains the reduced levels of chloroplast proteins. Polysome-seq analysis recently confirmed that the translation efficiency of approximately one-third of the genes in the Arabidopsis genome was dysregulated in response to cold in hypomorphic mutants of the writer complex (Wang et al., 2023). Given that cytoplasmic transcripts encoding chloroplast proteins were highly enriched in m6A marks compared with transcripts with other functions, translational defects in these transcripts in m6A writer mutants primarily affected chloroplasts in the cold (Figure 5).
Reduced biomass production, retarded growth of primary roots, and accumulation of ROS during cold treatment were recently described in the Arabidopsis mta mutant (Govindan et al., 2022). Root length was not significantly altered when fip37-4 was grown at 22°C for 3 weeks (Supplemental Figures 7A–7C). However, when plants were cultured at 4°C for a longer period, the root length of the mutant was, on average, about 4 times shorter than that of the WT, and growth was retarded in the mutant (Supplemental Figures 7A–7C). This hypersensitivity to cold could well be explained by the deleterious effect of the mutations on photosynthesis and other chloroplast functions described here. The unaltered ability of the mutant to induce expression of genes required for cold response and ROS scavenging shows that FIP37 is not required for this induction at the level of RNA accumulation.
CURT1 protein levels were also downregulated, and thylakoid curvature was altered accordingly in fip37-4 in the cold. The nuclear-encoded CURT1 proteins are chloroplast localized (Armbruster et al., 2013), and the number of m6A sites in RNAs of CurT1A–CurtT1D vary between the WT and fip37-4 (Qin et al., 2022). Therefore, m6A marks also seem to have important functions in the organization of the thylakoid membrane (Figure 6).
Regardless of the genetic compartment, RNA methylation can strongly influence chloroplast functions and stabilize photosynthesis under unfavorable environmental conditions. All these findings characterized cellular m6A-methylation marks as a central, dynamic, and complex platform that governs gene expression in all plant genetic compartments. They also established a link between m6A RNA methylation and chloroplast functions that enables plants to respond positively to cold and other challenging conditions at multiple levels.
Methods
Plant materials and growth conditions
We used the Arabidopsis thaliana accession Col-0 as the WT control and the previously described HAKAI knockout and knockdown lines for FIP37 (fip37-4), MTA (mta), VIRILIZER (vir-1), and the corresponding complemented lines vir-1c and fip37-4c (Shen et al., 2016; Růžička et al., 2017; Parker et al., 2020). Plants were cultivated on half-strength Murashige and Skoog medium supplemented with 1.5% sucrose or on soil under long-day standard conditions (16 h at 22°C and 80 μmol photons m−2 s−1/8 h at 18°C in the dark) using light-emitting diode cabinets (LED-41 HIL2, Percival Scientific, Perry, IA, USA). Surface-sterilized seeds were stratified at 4°C for 2 days and then cultivated for 10–21 days under standard conditions. For high light acclimation, the light intensity was increased to 450 μmol photons m−2 s−1. For cold and heat treatments, plants were grown at constant temperatures of 4°C and 32°C, respectively. Acclimation treatments were applied after standard growth conditions for up to 4 days. To test deacclimation, plants were transferred to standard conditions for 4 additional days after heat, cold, and high light treatments.
Transcript analysis
Transcriptome data were retrieved from Garcia-Molina et al. (2020) and Koussevitzky et al. (2007) (GEO: GSE125950 and GSE12887, respectively). Heatmaps were elaborated with the package pheatmap integrated in RStudio (https://www.bioconductor.org). Gene expression analyses using 10-day-old seedlings, total RNA isolation, electrophoresis, gel blotting, and hybridization with radioactive [32P]-dCTP-labeled probes were performed as described previously (Manavski et al., 2015). Primers for PCR probes and oligonucleotides are listed in Supplemental Table 2.
Protein analysis
Immunoblot analyses were performed with total proteins isolated from WT and fip37-4 seedlings in extraction buffer (100 mM NaCl, 50 mM Tris–HCl [pH 7.5], 0.5% [v/v] Triton X-100, 1 mM DTT) as described previously (Schmid et al., 2019). Equal amounts of total protein (10 μg) were loaded onto 10% polyacrylamide (w/v) Mini-PROTEAN SDS-PAGE gels (Bio-Rad); fractionated by electrophoresis and blotted onto PVDF membranes; blocked with 5% (w/v) milk powder prepared in TBS-T (50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 0.1% [v/v] Tween 20); and incubated overnight with primary antibodies specific for PsaA, PsaD, PsaO, D1, D2, PsbO, LHCB6, Cytb6, Cytf, AtpB, NdhB (Agrisera, https://www.agrisera.com), PGR5 (Munekage et al., 2002), and CurT1 (Armbruster et al., 2013). After subsequent washing steps, membranes were incubated with the appropriate peroxidase-conjugated secondary antibody. Immunoblots were developed with the SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Scientific) using the Fusion FX7 GelDoc (PeqLab).
In vivo labeling was essentially performed as described previously (Manavski et al., 2021b), except that instead of leaf discs, 4-day-old cold-treated seedlings were used. Prior to incubation with [35S] methionine at a final concentration of 50 mCi (specific activity >1000 Ci/mmol), seedlings were incubated for 30 min in the dark in labeling buffer supplemented with either 250 μg/ml chloramphenicol or 50 μg/ml cycloheximide. Labeled proteins were blotted onto a PVDF membrane and visualized by phosphorimaging (Amersham, Typhoon laser scanner).
Blue native PAGE
Isolation and solubilization of thylakoids were performed as described previously (Meurer et al., 2017). Proteins normalized to chlorophyll content were loaded onto a NativePAGE 3%–12%, Bis-Tris, 1 mm, Mini Protein Gel (Thermo Scientific) and separated at 200 V in the cold. Proteins were then either transferred onto PVDF membranes or stained with ROTIBlue quick (Roth).
Preparation of nucleosides for m6A quantification using LC–MS
For LC–MS, total RNA was extracted from 10-day-old WT seedlings before and after 4 days of cold, heat, or high light treatment. DNase treatment was performed using 10 μg total RNA (Zymo, RNA isolation kit). For poly(A)-enriched mRNAs, the NEBNext Poly(A) mRNA Magnetic Isolation kit (New England Biolabs) was used, and RNA integrity was confirmed with a Bioanalyzer (Agilent RNA 6000 Pico). RNAs were then concentrated in a speed vacuum centrifuge (Eppendorf Concentrator 5301) and precipitated overnight using 5 μl 5 M ammonium acetate and 125 μl cold 100% ethanol. Prior to LC–MS, samples were centrifuged for 1 h at 4°C, and the pellets were resuspended in RNase-free water. RNA was digested to single nucleosides using 2 U alkaline phosphatase, 0.2 U phosphodiesterase I (VWR, Radnor, PA, USA), 2 U benzonase in 5 mM Tris buffer (pH 8), and 1 mM MgCl2. Furthermore, 0.5 μg tetrahydrouridine (Merck, Darmstadt, Germany), 1 μM butylated hydroxytoluene, and 0.1 μg pentostatin were added in order to avoid deamination and oxidation of the nucleosides. After incubation for 2 h at 37°C, 10 μl LC–MS buffer (5 mM NH4OAc [pH 5.3]) was added, and LC–MS was performed.
LC–MS of hydrolyzed nucleosides
LC–MS experiments were performed on an Agilent 1290 Infinity II equipped with a Phenomenex Synergi 2.5 μm Fusion-RP 100 Å column (100 × 2 mm) coupled to an Agilent 6470 Triple Quad equipped with an ESI ion source. Ten microliters of each sample were injected without prior filtering. Chromatographic separation was carried out at 35°C with a flow rate of 0.35 ml/min using a linear gradient of two solvents: 5 mM ammonium acetate (pH 5.3) as solvent A and acetonitrile as solvent B (gradient: 0–1 min kept at 0% B, 1–5 min increase to 10% B, 5–7 min increase to 40% B, 7–8 min kept at 40% B, 8–8.5 min decrease to 0% B, 8.5–11 min kept at 0% B). Quantification was performed using calibration curves of synthetic standards and stable isotope-labeled internal standards (20 ng in 1 μl was automatically added by the instrument per sample) for each nucleoside (Heiss et al., 2021) using Agilent MassHunter software (v.9.0.647.0). To obtain the calibration curves, a solution containing synthetic standards of all nucleosides was serially diluted by a factor of 1:2 (12 calibration levels). The highest injected amounts were 100 (C, U, G, A) or 5 pmol (m6A).
Photosynthetic measurements
Photosynthetic measurements were performed on seedlings using an IMAGING or DUAL PAM instrument (Heinz Walz), essentially as previously described (Lezhneva and Meurer 2004). Dark-adapted leaves were used to determine the maximum quantum yield of PSII (Fv/Fm). Φ(II), Φ(NPQ), and Φ(NO) were calculated at steady state with leaves adapted to actinic light intensities of 80 μmol photons m−2 s−1. Saturating light pulses with a duration of 800 ms and a light intensity of 6000 μmol photons m−2 s−1 were used. The quantum yield of PSI Φ(I), as well as acceptor Φ(NA) and donor side Φ(ND) limitations, were measured as described previously (Plöchinger et al., 2016).
Transmission electron microscopy
For transmission electron microscopy, 10-day-old seedlings of WT and fip37-4 cultivated on Murashige and Skoog medium supplemented with sucrose were analyzed before and after 4 days of cold treatment. Plants were stored in the dark for 16 h before fixation to avoid starch accumulation. Primary leaves were cut into 1-mm2 pieces and fixed for 1–3 days at 4°C in cacodylate buffer (75 mM sodium cacodylate, 2 mM MgCl2 [pH 7], supplemented with 2.5% glutaraldehyde) as described previously (Garcia-Molina et al., 2020). After postfixation with 1% (w/v) OsO4 for 1 h, en bloc staining with 1% (w/v) uranyl acetate in 20% (v/v) acetone and dehydration in a graded acetone series were performed, and the plant material was embedded in Spurr’s resin. Ultrathin sections were contrasted with lead citrate and examined with a Zeiss EM 912 transmission electron microscope (Zeiss, Oberkochen, Germany) operating at 80 kV in the zero-loss mode. Images were acquired with a Tröndle 2k × 2k slow-scan CCD camera (TRS, Moorenweis, Germany). Images were processed with FIJI v.2.1 (https://fiji.sc).
ROS measurements
In situ detection of superoxide and H2O2 was performed with NBT and DAB, respectively, as described previously (Fryer et al., 2002; Wohlgemuth et al., 2002). In brief, Arabidopsis plants were vacuum infiltrated with 0.1% NBT, 50 mM potassium phosphate buffer (pH 7.8), and 10 mM sodium azide for 20 min, then incubated for 1 h at room temperature. Plants were then boiled in 95% ethanol for 15 min to remove chlorophyll. For H2O2 detection, plants were vacuum infiltrated with 5 mM DAB–HCl (pH 3) for 20 min and incubated in the same solution for at least 8 h overnight. Plants were then boiled in ethanol:acetic acid:glycerol (3:1:1) solution under the hood until they turned transparent and were later photographed.
qRT–PCR
For qRT–PCR, rosettes of at least five plants grown in vitro were harvested and homogenized in liquid nitrogen. RNA extraction was performed with the NucleoSpin RNA Plant Kit (Macherey-Nagel, Düren, Germany) according to the manufacturer’s protocol. RNA purity and concentration were determined using a NanoDrop spectrophotometer. Total RNA was transcribed into cDNA using the qScript cDNA Synthesis Kit (Quantabio, Beverly, MA, USA). The qPCR was performed in 10 μl using the Quantabio SYBR Green quantification kit on the PFX96 system (BioRad, Hercules, CA, USA) with the specific primers listed in Supplemental Table 2. After 3 min of denaturation at 95°C, 39 cycles were performed (95°C 10 s, 60°C 40 s). Actin (At2g37620) was used as a reference for transcript normalization. The expression of each gene was calculated in relation to the reference. The relative transcript levels of the different genes were evaluated by scoring the runs according to the following formula: copy number = E(reference)CT/E(GOI)CT (E, primer efficiency factor; CT, number of cycles; GOI, gene of interest) essentially as described previously (Vandesompele et al., 2002). Mean values and standard deviations were calculated from at least three biological replicates in technical duplicates.
RNA gel blot and polysome loading analysis
RNA gel blots were prepared as described previously (Stoppel et al., 2011). Nylon membranes were hybridized overnight with either radioactively labeled PCR products or 80-mer probes at 65°C or 55°C, respectively, as described previously (Manavski et al., 2015). Sequences of the primers and 80-mers are listed in Supplemental Table 2. Polysome loading analysis was performed essentially as described previously (Meurer et al., 2002).
Chlorophyll and anthocyanin measurements
Chlorophyll measurements were performed as described previously (Porra and Scheer, 2019). Anthocyanin content was calculated by measuring the absorption of the aqueous phase at 530 and 657 nm (Neff and Chory, 1998).
Funding
This research was supported by the Deutsche Akademischer Austauschdienst (91692277 to A.M.V.) and the Deutsche Forschungsgemeinschaft (TRR 175 projects B07 to D.L., B08 to T.M., and A03 to J.M.).
Author contributions
A.M.V., N.M., L.-M.S., P.T.R., C.S., L.B., G.A., A.G.-M., and J.M. performed the research. A.M.V., N.M., T.M., D.L., S.K., and J.M. analyzed the data. A.M.V. and J.M. designed the work and wrote the article with the contribution of all coauthors.
Acknowledgments
We thank Gordon G. Simpson (University of Dundee) for kindly providing fip37-4, vir-1, and vir-1c lines, Hao Yu (University of Singapore) for fip37-4c lines, and Rupert G. Fray (University of Nottingham) for hakai and mta seeds. We also thank Andreas Klingl for providing the electron microscopy facility. No conflict of interest is declared.
Published: June 7, 2023
Footnotes
Published by the Plant Communications Shanghai Editorial Office in association with Cell Press, an imprint of Elsevier Inc., on behalf of CSPB and CEMPS, CAS.
Supplemental information is available at Plant Communications Online.
Accession numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries under locus numbers AT3G54170 (FIP37), AT5G01160 (HAKAI), AT3G05680 (VIR), AT4G10760 (MTA), and AT4G09980 (MTB).
Supplemental information
References
- Anderson S., Kramer M., Gosai S., Yu X., Vandivier L., Nelson A., Anderson Z., Beilstein M., Fray R., Lyons E., et al. N6-Methyladenosine inhibits local ribonucleolytic cleavage to stabilize mRNAs in arabidopsis. Cell Rep. 2018;25:1146–1157. doi: 10.1016/j.celrep.2018.10.020. [DOI] [PubMed] [Google Scholar]
- Armbruster U., Labs M., Pribil M., Viola S., Xu W., Scharfenberg M., Hertle A., Rojahn U., Jensen P., Rappaport F., et al. Arabidopsis CURVATURE THYLAKOID1 proteins modify thylakoid architecture by inducing membrane curvature. Plant Cell. 2013;25:2661–2678. doi: 10.1105/tpc.113.113118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P., Machnicka M.A., Purta E., Piątkowski P., Bagiński B., Wirecki T.K., de Crécy-Lagard V., Ross R., Limbach P.A., Kotter A., et al. Modomics: a database of RNA modification pathways. Nucleic Acids Research. 2021;46:303–307. doi: 10.1093/nar/gkab1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinnusamy V., Zhu J., Zhu J.-K. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007;12:444–451. doi: 10.1016/j.tplants.2007.07.002. [DOI] [PubMed] [Google Scholar]
- Choi H.L., Seo J.W., Hwang M.H., Yu C.Y., Seong E.S. A cold stress-responsive gene, provides resistance to environmental stress in T2-generation transgenic plants. Transgenic Res. 2022;31:381–389. doi: 10.1007/s11248-022-00307-9. [DOI] [PubMed] [Google Scholar]
- Crawford T., Lehotai N., Strand Å. The role of retrograde signals during plant stress responses. J. Exp. Bot. 2018;69:2783–2795. doi: 10.1093/jxb/erx481. [DOI] [PubMed] [Google Scholar]
- Crosatti C., Rizza F., Badeck F.W., Mazzucotelli E., Cattivelli L. Harden the chloroplast to protect the plant. Physiol. Plantarum. 2013;147:55–63. doi: 10.1111/j.1399-3054.2012.01689.x. [DOI] [PubMed] [Google Scholar]
- Fryer M.J., Oxborough K., Mullineaux P.M., Baker N.R. Imaging of photo-oxidative stress responses in leaves. J. Exp. Bot. 2002;53:1249–1254. doi: 10.1093/jexbot/53.372.1249. [DOI] [PubMed] [Google Scholar]
- Garcia-Molina A., Kleine T., Schneider K., Mühlhaus T., Lehmann M., Leister D. Translational components contribute to acclimation responses to high light, heat, and cold in arabidopsis. iScience. 2020;23:101331. doi: 10.1016/j.isci.2020.101331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govindan G., Sharma B., Li Y.F., Armstrong C.D., Merum P., Rohila J.S., Gregory B.D., Sunkar R. mRNA N6-methyladenosine is critical for cold tolerance in Arabidopsis. Plant J. 2022;111:1052–1068. doi: 10.1111/tpj.15872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He M.-W., Wang Y., Wu J.-Q., Shu S., Sun J., Guo S.-R. Isolation and characterization of S-Adenosylmethionine synthase gene from cucumber and responsive to abiotic stress. Plant Physiol. Biochem. 2019;141:431–445. doi: 10.1016/j.plaphy.2019.06.006. [DOI] [PubMed] [Google Scholar]
- He Y., Li Y., Yao Y., Zhang H., Wang Y., Gao J., Fan M. Overexpression of watermelon m6A methyltransferase ClMTB enhances drought tolerance in tobacco by mitigating oxidative stress and photosynthesis inhibition and modulating stress-responsive gene expression. Plant Physiol. Biochem. 2021;168:340–352. doi: 10.1016/j.plaphy.2021.10.007. [DOI] [PubMed] [Google Scholar]
- Heiss M., Borland K., Yoluç Y., Kellner S. Quantification of modified nucleosides in the context of NAIL-MS. Methods Mol. Biol. 2021;2298:279–306. doi: 10.1007/978-1-0716-1374-0_18. [DOI] [PubMed] [Google Scholar]
- Hou N., Li C., He J., Liu Y., Yu S., Malnoy M., Mobeen Tahir M., Xu L., Ma F., Guan Q. MdMTA-mediated m6A modification enhances drought tolerance by promoting mRNA stability and translation efficiency of genes involved in lignin deposition and oxidative stress. New Phytol. 2022;234:1294–1314. doi: 10.1111/nph.18069. [DOI] [PubMed] [Google Scholar]
- Hu J., Cai J., Park S.J., Lee K., Li Y., Chen Y., Yun J.-Y., Xu T., Kang H. N6-Methyladenosine mRNA methylation is important for salt stress tolerance in Arabidopsis. Plant J. 2021;106:1759–1775. doi: 10.1111/tpj.15270. [DOI] [PubMed] [Google Scholar]
- Huong T.T., Ngoc L.N.T., Kang H. Functional characterization of a putative RNA demethylase ALKBH6 in arabidopsis growth and abiotic stress responses. Int. J. Mol. Sci. 2020;21:6707. doi: 10.3390/ijms21186707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleine T., Nägele T., Neuhaus H.E., Schmitz-Linneweber C., Fernie A.R., Geigenberger P., Grimm B., Kaufmann K., Klipp E., Meurer J., et al. Acclimation in plants - the green hub consortium. Plant J. 2021;106:23–40. doi: 10.1111/tpj.15144. [DOI] [PubMed] [Google Scholar]
- Koussevitzky S., Nott A., Mockler T.C., Hong F., Sachetto-Martins G., Surpin M., Lim J., Mittler R., Chory J. Signals from chloroplasts converge to regulate nuclear gene expression. Science. 2007;316:715–719. doi: 10.1126/science.1114655. [DOI] [PubMed] [Google Scholar]
- Lezhneva L., Meurer J. The nuclear factor HCF145 affects chloroplast psaA-psaB-rps14 transcript abundance in Arabidopsis thaliana. Plant J. 2004;38:740–753. doi: 10.1111/j.1365-313X.2004.02081.x. [DOI] [PubMed] [Google Scholar]
- Li M., Kim C. Chloroplast ROS and stress signaling. Plant Commun. 2022;3:100264. doi: 10.1016/j.xplc.2021.100264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J.-Y., Yang C., Tian Y.-Y., Liu J.-X. Regulation of chloroplast development and function at adverse temperatures in plants. Plant Cell Physiol. 2022;63:580–591. doi: 10.1093/pcp/pcac022. [DOI] [PubMed] [Google Scholar]
- Liang Z., Riaz A., Chachar S., Ding Y., Du H., Gu X. Epigenetic modifications of mRNA and DNA in plants. Mol. Plant. 2020;13:14–30. doi: 10.1016/j.molp.2019.12.007. [DOI] [PubMed] [Google Scholar]
- Liu Y., Dang P., Liu L., He C. Cold acclimation by the CBF-COR pathway in a changing climate: lessons from. Plant Cell Rep. 2019;38:511–519. doi: 10.1007/s00299-019-02376-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G., Wang J., Hou X. Transcriptome-wide N6-methyladenosine (m6A) methylome profiling of heat stress in pak-choi (Brassica rapa ssp. chinensis) Plants. 2020;9:1080. doi: 10.3390/plants9091080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu L., Zhang Y., He Q., Qi Z., Zhang G., Xu W., Yi T., Wu G., Li R. MTA, an RNA m6A methyltransferase, enhances drought tolerance by regulating the development of trichomes and roots in poplar. Int. J. Mol. Sci. 2020;21:2462. doi: 10.3390/ijms21072462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo G.Z., MacQueen A., Zheng G., Duan H., Dore L.C., Lu Z., Liu J., Chen K., Jia G., Bergelson J., et al. Unique features of the m6A methylome in Arabidopsis thaliana. Nat. Commun. 2014;5:5630–5638. doi: 10.1038/ncomms6630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavski N., Torabi S., Lezhneva L., Arif M.A., Frank W., Meurer J. HIGH CHLOROPHYLL FLUORESCENCE145 binds to and stabilizes the psaA 5`UTR via a newly defined repeat motif in embryophyta. Plant Cell. 2015;27:2600–2615. doi: 10.1105/tpc.15.00234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavski N., Schmid L.M., Meurer J. RNA-stabilization factors in chloroplasts of vascular plants. Essays Biochem. 2018;62:51–64. doi: 10.1042/EBC20170061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavski N., Vicente A., Chi W., Meurer J. The chloroplast epitranscriptome: factors, sites, regulation, and detection methods. Genes. 2021;12:1121–1122. doi: 10.1042/EBC20170061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manavski N., Mathieu S., Rojas M., Méteignier L.V., Brachmann A., Barkan A., Hammani K. In vivo stabilization of endogenous chloroplast RNAs by customized artificial pentatricopeptide repeat proteins. Nucleic Acids Res. 2021;49:5985–5997. doi: 10.1093/nar/gkab390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J., Meierhoff K., Westhoff P. Isolation of high-chlorophyll-fluorescence mutants of Arabidopsis thaliana and their characterization by spectroscopy, immunoblotting and northern hybridisation. Planta. 1996;198:385–396. doi: 10.1007/BF00620055. [DOI] [PubMed] [Google Scholar]
- Meurer J., Lezhneva L., Amann K., Gödel M., Bezhani S., Sherameti I., Oelmüller R. A peptide chain release factor 2 affects the stability of UGA-containing transcripts in Arabidopsis chloroplasts. Plant Cell. 2002;14:3255–3269. doi: 10.1105/tpc.006809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meurer J., Schmid L.M., Stoppel R., Leister D., Brachmann A., Manavski N. PALE CRESS binds to plastid RNAs and facilitates the biogenesis of the 50S ribosomal subunit. Plant J. 2017;92:400–413. doi: 10.1111/tpj.13662. [DOI] [PubMed] [Google Scholar]
- Munekage Y., Hojo M., Meurer J., Endo T., Tasaka M., Shikanai T. PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell. 2002;110:361–371. doi: 10.1016/s0092-8674(02)00867-x. [DOI] [PubMed] [Google Scholar]
- Nakaminami K., Seki M. RNA regulation in plant cold stress response. Adv. Exp. Med. Biol. 2018;1081:23–44. doi: 10.1007/978-981-13-1244-1_2. [DOI] [PubMed] [Google Scholar]
- Neff M.M., Chory J. Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol. 1998;118:27–35. doi: 10.1104/pp.118.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker M.T., Knop K., Sherwood A.V., Schurch N.J., Mackinnon K., Gould P.D., Hall A.J., Barton G.J., Simpson G.G. Nanopore direct RNA sequencing maps the complexity of Arabidopsis mRNA processing and m6A modification. Elife. 2020;9:1496588-35. doi: 10.7554/eLife.49658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plöchinger M., Torabi S., Rantala M., Tikkanen M., Suorsa M., Jensen P.-E., Aro E.M., Meurer J. The low molecular weight protein PsaI stabilizes the light-harvesting complex II docking site of photosystem I. Plant Physiol. 2016;172:450–463. doi: 10.1104/pp.16.00647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porra R.J., Scheer H. Towards a more accurate future for chlorophyll a and b determinations: the inaccuracies of Daniel Arnon's assay. Photosynth. Res. 2019;140:215–219. doi: 10.1007/s11120-018-0579-8. [DOI] [PubMed] [Google Scholar]
- Qin H., Ou L., Gao J., Chen L., Wang J.W., Hao P., Li X. DENA: training an authentic neural network model using Nanopore sequencing data of Arabidopsis transcripts for detection and quantification of N6-methyladenosine on RNA. Genome Biol. 2022;23 doi: 10.1186/s13059-021-02598-3. 25-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter A.S., Tohge T., Fernie A.R., Grimm B. The genomes uncoupled-dependent signalling pathway coordinates plastid biogenesis with the synthesis of anthocyanins. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2020;375:20190403. doi: 10.1098/rstb.2019.0403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Araguirang G.E., Richter A.S. Activation of anthocyanin biosynthesis in high light - what is the initial signal? New Phytol. 2022;236:2037–2043. doi: 10.1111/nph.18488. [DOI] [PubMed] [Google Scholar]
- Růžička K., Zhang M., Campilho A., Bodi Z., Kashif M., Saleh M., Eeckhout D., El-Showk S., Li H., Zhong S., et al. Identification of factors required for m6A mRNA methylation in Arabidopsis reveals a role for the conserved E3 ubiquitin ligase HAKAI. New Phytol. 2017;215:157–172. doi: 10.1111/nph.14586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmid L.M., Ohler L., Möhlmann T., Brachmann A., Muiño J.M., Leister D., Meurer J., Manavski N. PUMPKIN, the sole plastid UMP kinase, associates with group II introns and alters their metabolism. Plant Physiol. 2019;179:248–264. doi: 10.1104/pp.18.00687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenkert S., Fernie A.R., Geigenberger P., Leister D., Möhlmann T., Naranjo B., Neuhaus H.E. Chloroplasts are key players to cope with light and temperature stress. Trends Plant Sci. 2022;27:577–587. doi: 10.1016/j.tplants.2021.12.004. [DOI] [PubMed] [Google Scholar]
- Scutenaire J., Deragon J.M., Jean V., Benhamed M., Raynaud C., Favory J.J., Merret R., Bousquet-Antonelli C. The YTH domain protein ECT2 is an m6A reader required for normal trichome branching in Arabidopsis. Plant Cell. 2018;30:986–1005. doi: 10.1105/tpc.17.00854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y., Wong C.E., Shen L., Yu H. N6-methyladenosine modification underlies messenger RNA metabolism and plant development. Curr. Opin. Plant Biol. 2021;63:102047. doi: 10.1016/j.pbi.2021.102047. [DOI] [PubMed] [Google Scholar]
- Shen L., Liang Z., Gu X., Chen Y., Teo Z.W.N., Hou X., Cai W.M., Dedon P.C., Liu L., Yu H. N6-Methyladenosine RNA modification regulates shoot stem cell fate in arabidopsis. Dev. Cell. 2016;38:186–200. doi: 10.1016/j.devcel.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen L. Functional interdependence of N6-methyladenosine methyltransferase complex subunits in Arabidopsis. Plant Cell. 2023;35:1901–1916. doi: 10.1093/plcell/koad070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi H., Chai P., Jia R., Fan X. Novel insight into the regulatory roles of diverse RNA modifications: Re-defining the bridge between transcription and translation. Mol. Cancer. 2020;19:78. doi: 10.1186/s12943-020-01194-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppel R., Lezhneva L., Schwenkert S., Torabi S., Felder S., Meierhoff K., Westhoff P., Meurer J. Recruitment of a ribosomal release factor for light- and stress-dependent regulation of petB transcript stability in Arabidopsis chloroplasts. Plant Cell. 2011;23:2680–2695. doi: 10.1105/tpc.111.085324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Bie X.M., Wang N., Zhang X.S., Gao X.-Q. Genome-wide identification and expression analysis of YTH domain-containing RNA-binding protein family in common wheat. BMC Plant Biol. 2020;20:351. doi: 10.1186/s12870-020-02505-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theocharis A., Clément C., Barka E.A. Physiological and molecular changes in plants grown at low temperatures. Planta. 2012;235:1091–1105. doi: 10.1007/s00425-012-1641-y. [DOI] [PubMed] [Google Scholar]
- Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3 doi: 10.1186/gb-2002-3-7-research0034. RESEARCH0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y., Tang K., Zhang D., Xie S., Zhu X., Wang Z., Lang Z. Transcriptome-wide high-throughput deep m6A-seq reveals unique differential m6A methylation patterns between three organs in Arabidopsis thaliana. Genome Biol. 2015;16:272. doi: 10.1186/s13059-015-0839-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Tang K., Zhang D., Wan Y., Wen Y., Lu Q., Wang L. High-throughput m6A-seq reveals RNA m6A methylation patterns in the chloroplast and mitochondria transcriptomes of Arabidopsis thaliana. PLoS One. 2017;12:e0185612. doi: 10.1371/journal.pone.0185612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zhuang H., Fan W., Zhang X., Dong H., Yang H., Cho J. m6A RNA methylation impairs gene expression variability and reproductive thermotolerance in Arabidopsis. Genome Biol. 2022;23:244. doi: 10.1186/s13059-022-02814-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Wang H., Xu Z., Jiang S., Shi Y., Xie H., Wang S., Hua J., Wu Y. m6A mRNA modification promotes chilling tolerance and modulates gene translation efficiency in Arabidopsis. Plant Physiol. 2023;192:1466–1482. doi: 10.1093/plphys/kiad112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. Biosynthesis of flavonoids and effects of stress. Curr. Opin. Plant Biol. 2002;5:218–223. doi: 10.1016/s1369-5266(02)00256-x. [DOI] [PubMed] [Google Scholar]
- Wohlgemuth H., Mittelstrass K., Kschieschan S., Bender J., Weigel H.-J., Overmyer K., Kangasjärvi J., Sandermann H., Langebartels C. Activation of an oxidative burst is a general feature of sensitive plants exposed to the air pollutant ozone. Plant Cell Environ. 2002;25:717–726. doi: 10.1046/j.1365-3040.2002.00859.x. [DOI] [Google Scholar]
- Wong C.E., Zhang S., Xu T., Zhang Y., Teo Z.W.N., Yan A., Shen L., Yu H. Shaping the landscape of N6-methyladenosine RNA methylation in Arabidopsis. Plant Physiol. 2023;191:2045–2063. doi: 10.1093/plphys/kiad010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Rothstein S.J. ROS-Induced anthocyanin production provides feedback protection by scavenging ROS and maintaining photosynthetic capacity in Arabidopsis. Plant Signal. Behav. 2018;13:e1451708. doi: 10.1080/15592324.2018.1451708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Sharma B., Gregory B.D. The impact of epitranscriptomic marks on posttranscriptional regulation in plants. Brief. Funct. Genomics. 2021;20:113–124. doi: 10.1093/bfgp/elaa021. [DOI] [PubMed] [Google Scholar]
- Zhang M., Zeng Y., Peng R., Dong J., Lan Y., Duan S., Chang Z., Ren J., Luo G., Liu B., et al. N6-methyladenosine RNA modification regulates photosynthesis during photodamage in plants. Nat. Commun. 2022;13:7441. doi: 10.1038/s41467-022-35146-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.