Abstract
Many gram-negative bacteria synthesize N-acyl homoserine lactone autoinducer molecules as quorum-sensing signals which act as cell density-dependent regulators of gene expression. We have investigated the in vivo source of the acyl chain and homoserine lactone components of the autoinducer synthesized by the LuxI homolog, TraI. In Escherichia coli, synthesis of N-(3-oxooctanoyl)homoserine lactone by TraI was unaffected in a fadD mutant blocked in β-oxidative fatty acid degradation. Also, conditions known to induce the fad regulon did not increase autoinducer synthesis. In contrast, cerulenin and diazoborine, specific inhibitors of fatty acid synthesis, both blocked autoinducer synthesis even in a strain dependent on β-oxidative fatty acid degradation for growth. These data provide the first in vivo evidence that the acyl chains in autoinducers synthesized by LuxI-family synthases are derived from acyl-acyl carrier protein substrates rather than acyl coenzyme A substrates. Also, we show that decreased levels of intracellular S-adenosylmethionine caused by expression of bacteriophage T3 S-adenosylmethionine hydrolase result in a marked reduction in autoinducer synthesis, thus providing direct in vivo evidence that the homoserine lactone ring of LuxI-family autoinducers is derived from S-adenosylmethionine.
In recent years, it has become evident that a large number of gram-negative bacteria regulate various physiological processes in a cell density-dependent manner by synthesizing N-acyl homoserine lactones (N-acyl HSLs), referred to as autoinducers (AIs). These quorum-sensing molecules are thought to freely diffuse across the cell membrane accumulating in the growth medium and in the cell (at an equivalent concentration), as the cell density increases (31). When the intracellular AI concentration becomes high enough, the AI acts as a gene regulator by complexing with its cognate transcriptional regulator protein (reviewed in references 20 and 43). The list of processes regulated in this way includes luminescence in Vibrio fischeri (15) and Vibrio harveyi (9), Ti plasmid conjugal transfer in Agrobacterium tumefaciens (63), swarming in Serratia liquefaciens (17), synthesis of poly-3-hydroxybutyrate in V. harveyi (53), and the expression of virulence determinants in a variety of plant and animal pathogens (reviewed in references 20 and 43). The model system for understanding N-acyl HSL-regulated processes is the induction of luminescence in V. fischeri. In this system, expression of the lux operon is induced in response to the transcriptional activator LuxR binding to its cognate Lux box DNA sequence when complexed with an N-acyl HSL AI [N-(3-oxohexanoyl)HSL] synthesized by the enzyme LuxI (20).
With the exception of the AinS enzyme of V. fischeri (22) and the LuxLM system of V. harveyi (4), all identified N-acyl HSL AI synthases are LuxI homologs. Expression of luxI and homologous genes from a variety of species has been shown to be necessary and sufficient to direct the synthesis of the N-acyl HSL AI(s) in the parent organisms or in Escherichia coli (LuxI [18], LasI [41], TraI [30], EsaI [6], YenI [56], etc.). This implies that the substrates for N-acyl HSL synthesis by the LuxI family of enzymes are intermediates of metabolic pathways common to (at least) gram-negative bacteria.
To date, while in vitro studies have suggested that the substrates for AI synthesis are S-adenosylmethionine (AdoMet) and acyl-acyl carrier proteins (acyl-ACPs), these results have yet to be fully confirmed in vivo. Eberhard et al. (16) found that crude extracts of V. fischeri provided with AdoMet and N-(3-oxohexanoyl) coenzyme A (CoA) were able to generate N-(3-oxohexanoyl)HSL, the primary AI made by this species (15). No evidence of AI synthesis was found when N-(3-oxohexanoyl)-CoA was replaced by malonyl-CoA, acetyl-CoA, citrate, and NADPH. These findings suggested that the acyl chains of N-acyl HSLs are obtained as the CoA intermediates of the fatty acid β-oxidation pathway.
Subsequently, using purified recombinant LuxI and TraI (homolog from A. tumefaciens) we (46) and others (38), respectively, have shown that N-acyl HSL AIs are synthesized in vitro from AdoMet and their corresponding acyl-ACPs, rather than from CoAs. This implies that in vivo the acyl chains of N-acyl HSLs synthesized by LuxI-type synthases are derived from intermediates in the fatty acid biosynthesis pathway, rather than from β-oxidation. Consistent with this possibility, Moré et al. (38) found that a dialyzed E. coli extract supplied with His-tagged TraI regained the ability to synthesize the Agrobacterium AI (AAI) lost as a result of dialysis when the extract was supplied with malonyl-CoA and NADPH or NADH and had reduced AI synthesis activity (50%) when treated with the fatty acid synthesis inhibitor cerulenin. The findings of Eberhard et al. (16) would then be assumed to be due to an enzymatic activity in the V. fischeri extract which converted the added acyl-CoA into an acyl-ACP. This type of acyl transfer reaction is known to occur in E. coli and is catalyzed by the enzyme 3-ketoacyl-ACP synthase I (1). In fact, these workers offered this as a possible explanation of their results. However, it is unclear why the V. fischeri extracts (16), unlike the E. coli extracts (38), were unable to produce detectable AI when provided with AdoMet, malonyl-CoA, and NADPH.
While it could be argued that the above-mentioned in vitro studies provide sufficient evidence that the acyl chains in AIs are derived from acyl-ACPs, it should be noted that the Vmax values of the purified LuxI and TraI enzymes used in these studies were very low (approximately 1 mol of AI synthesized/mol of enzyme/min). This may be due to either the inherently low activity of these enzymes (38), a reduction in activity resulting from the presence of the N-terminal purification tags (46), or inherent instability, resulting in a large proportion of the affinity-purified enzymes being inactive (46). Alternatively, the low Vmax raises the possibility that other reactions may occur in vivo.
Regarding the source of the HSL ring in AIs, in vitro studies with crude extracts of V. fischeri (16) or purified LuxI (46) or TraI (38) have all found that AI synthesis is dependent on the presence of AdoMet, rather than any of the intermediates of the threonine-methionine biosynthesis pathway that are structurally related to HSL. Furthermore, using amino acid-starved E. coli mutants blocked at various steps in the threonine-methionine biosynthesis pathway and carrying the luxI gene on a multicopy plasmid, Hanzelka and Greenberg (26) found that methionine or AdoMet is required for AI synthesis by LuxI. They also found that cycloleucine, a compound known to inhibit the methionine adenosyltransferase reaction (12, 34), produced a significant reduction in AI synthesis, leading to the conclusion that the HSL ring of AI was probably derived from AdoMet. However, there appear to be other effects of cycloleucine on E. coli metabolism, since the branched-chain amino acids valine and isoleucine and also leucine have been found to overcome growth inhibition of E. coli by cycloleucine (3).
In this study, we investigated the role of the fatty acid synthesis and degradation pathways in supplying acyl chains for AI synthesis in vivo. We used specific inhibitors of fatty acid biosynthesis and E. coli mutants blocked in the β-oxidation pathway to demonstrate that, in vivo, the acyl chain of N-(3-oxooctanoyl)HSL synthesized by TraI is supplied entirely by the fatty acid biosynthesis pathway. In addition, we provide further in vivo evidence that the HSL ring is derived from AdoMet by showing that decreases in the AdoMet pool caused by expression of the bacteriophage T3 AdoMet hydrolase gene (which specifically degrades AdoMet [23]) resulted in a significant reduction in the synthesis of N-(3-oxooctanoyl)HSL by TraI.
MATERIALS AND METHODS
Materials and general methods.
Common antibiotics, cerulenin, nalidixic acid, S-adenosylhomocysteine, AdoMet, and isopropyl-β-d-thiogalactopyranoside (IPTG) were obtained from Sigma. Ammonium hydroxide, formic acid, ethyl ether, and various other miscellaneous chemicals were obtained from Fisher. Bacto Tryptone, yeast extract, and agar were obtained from Difco. Ethyl acetate was obtained from Mallinckrodt Chemical, trichloroacetic acid was obtained from Aldrich, S-[methyl-3H]adenosyl-l-methionine was obtained from DuPont, and diazoborine was a kind gift from Friederike Turnowsky, University of Graz, Graz, Austria, while N-[3-oxooctanoyl]HSL was a kind gift of S. K. Farrand, Department of Microbiology, University of Illinois. Oligonucleotides used were T3SAM5P, 5′ AACATGAGGCATATGCTCATGATTTTCACTAAAG 3′, and T3SAM3P, 5′ CATCGTGCGGATCCTTGAGTTTAAC 3′.
Plasmid DNA was isolated, manipulated, and transformed into E. coli strains by standard procedures (44).
Bacterial strains, plasmids, media, and culture conditions.
The main bacterial strains and plasmids used in this study are listed in Table 1. Strain constructions with P1vir transductional crosses were carried out by conventional methods (14). The E. coli strains were grown at the specified temperature in either rich broth (RB; 1% Bacto Tryptone, 0.1% yeast extract, 0.5% NaCl), supplemented with 0.1% oleate where required, or minimal salts medium E (14) supplemented for the various auxotrophies as specified in the work of Davis et al. (14). The A. tumefaciens bioassay reporter strain NT1(pDC141E33) was grown in AB minimal mannitol liquid medium (5) supplemented with kanamycin (100 μg/ml) and carbenicillin (100 μg/ml) at 30°C. For maintenance of plasmids, the media for E. coli strains were supplemented with ampicillin (100 μg/ml), chloramphenicol (20 μg/ml), and spectinomycin (100 μg/ml).
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristic(s)a | Source or reference |
|---|---|---|
| E. coli strains | ||
| JM83 | ara Δ(lac-proAB) rpsL φ80lacZΔM15 | 62 |
| JM83-TI | JM83(pSEP1, pMS421) | This study |
| JMFR-TI | JM83; fadR::Tn10(pSEP1, pMS421) | This study |
| JMD6-TI | JM83; fadD88 zea::Tn10(pSEP1, pMS421) | This study |
| JMAC-TI | JM83; Δ(aroP-aceEF) zac::Tn10(pSEP1, pMS421) | This study |
| C41(DE3) | Mutant of BL21(DE3) with reduced T7 RNA polymerase activity | 36 |
| C41T3-TI | C41(DE3) (pT3SE8, pSEP1, pMS421) | This study |
| C41PC-TI | C41(DE3) (pPCBLE1, pSEP1, pMS421) | This study |
| LEPRT7 | T7 gene 1 under control of pR and cIts857 inserted in the λatt site of LE392 | 19 |
| LEPT3-TI | LEPRT7 (pT3SE8, pSEP1, pMS421) | This study |
| LEPPC-TI | LEPRT7 (pPCBLE1, pSEP1, pMS421) | This study |
| A. tumefaciens NTI(pDCI41E33) | Indicator strain for detecting AAI | S. K. Farrand |
| Plasmids | ||
| pET16b | His-tagged T7 expression vector | Novagen |
| pSEP1 | traI gene downstream of the lac promoter of pUC19; Apr | S. K. Farrand |
| pMS421 | lacIq plasmid; Spr | 24 |
| pETT3S | T3 AdoMet hydrolase gene in the NcoI-BamHI sites of pET16b; Apr | This study |
| pT3SLE8 | 1.1-kb BspHI fragment of pETT3S in the TcrBspHI site of pLysE; Cmr | This study |
| pc104 | C-terminal 312 bp of YPC1 downstream of the T7 promoter of pET16b; Apr | 58 |
| pPCBLE1 | 1.6-kb BspHI fragment of pc104 in the TcrBspHI site of pLysE; Cmr | This study |
Apr, ampicillin resistant; Cmr, chloramphenicol resistant; Spr, spectinomycin resistant; Tcr, tetracycline resistance gene.
Plasmid constructions.
A dilute stock of bacteriophage T3 (gift of Ian J. Molineux, University of Texas, Austin) was used as a template for PCR amplification of the T3 AdoMet hydrolase gene with the 5′ T3SAM5P and 3′ T3SAM3P primers. The T3SAM5P primer introduced a 5′ BspHI site overlapping the initiating ATG, and T3SAM3P introduced a BamHI site downstream of the TAA termination codon. The 503-bp BspHI/BamHI-digested PCR fragment containing the entire coding sequence was cloned into the NcoI and BamHI sites of pET16b (Novagen) behind the T7 promoter (pETT3S). pT3SLE8 was obtained by cloning the entire T7 promoter-terminator–T3 AdoMet hydrolase region of pETT3S as a 1.1-kb BspHI fragment into the BspHI site within the tetracycline resistance gene of pLysE (51). The control plasmid pPCBLE1 was similarly obtained from pLysE by using the corresponding 1.6-kb BspHI fragment of a pET16b derivative plasmid (pc104), containing a 980-bp insert encoding the C-terminal 104-amino-acid biotin domain of the yeast PYC1 gene (58) in the NcoI and BamHI sites in place of the T3 AdoMet hydrolase gene.
AI extraction from growth media.
Samples used to determine the level of AI present in the growth medium were prepared by extraction of cell-free medium with an equal volume of ethyl acetate. The ethyl acetate was then evaporated under nitrogen, and the residue was dissolved in 0.1 volume of Milli-Q water and stored at −20 or −80°C.
Quantitation of the AI concentration in the growth medium.
Duplicate or triplicate 1-μl aliquots of the samples were spotted at sites spaced across the entire area of a 20-by-20-cm silica gel reversed-phase thin-layer chromatography (TLC) plate (UNIPLATE; Analtech) such that the TLC plate was used as a sample adsorption surface matrix rather than as a chromatography plate. A bioassay was then performed by overlaying the TLC plate with molten agar containing 5-bromo-4-chloro-3-indolyl-β-d-galactoside and the A. tumefaciens indicator strain NT1(pDC141E33) as previously described (47). A dilution series of five or six AAI standards from 20 to 0.078 pg were also spotted onto each plate. These standards were used to construct a standard curve by plotting the measured diameter of the blue spots against the log10 of the AAI concentration, and the AAI concentrations in the samples were calculated from the average diameters of the sample spots.
Preparation of cell extracts for intracellular AdoMet determinations.
Cell extracts were prepared essentially as described by Satishchandran et al. (45). Cells were harvested by centrifugation from 5 to 15 ml of the appropriate culture of known optical density, immediately resuspended in 200 μl, lysed by the addition of trichloroacetic acid to 10%, and stored at −80°C. Immediately prior to high-performance liquid chromatography analysis, the samples were thawed, 20 μl of [3H-methyl]-AdoMet (approximately 1,600 cpm) was added as an internal standard, and the trichloroacetic acid was removed by three extractions with an equal volume of water-saturated ether.
Determination of the intracellular AdoMet concentration.
Intracellular AdoMet levels were determined by a modification of the method of Satishchandran et al. (45). Isocratic ion-exchange high-performance liquid chromatography was performed with UV detection at 254 nm with a Waters analytical 4.6-by-250-mm Sherisorb 5S SCX column attached to a Beckman System Gold 125 solvent system, a 168 UV detector, and a 171 radioisotope detector. The elution buffer was prepared as a fourfold-concentrated solution containing 33.5 ml of concentrated NH4OH per liter, adjusted to pH 4.0 with formic acid. With a flow rate of 1.5 ml/min, AdoMet eluted as a well-resolved peak at 9.1 min, easily distinguished from S-adenosylhomocysteine and 5′-methylthioadenosine, which eluted at 3.5 and 5.3 min, respectively. AdoMet concentrations were calculated from the integrated peak areas of duplicate samples, by using a standard curve constructed with duplicate standards of S-adenosylhomocysteine at seven different concentrations such that the standards contained amounts ranging from 0.3125 to 20 nmol.
RESULTS
Mutants defective in fatty acid degradation are not blocked in AAI synthesis.
The fatty acid degradation pathway in E. coli is dependent upon the conversion of the fatty acids into acyl-CoA thioesters by the action of the fadD gene product. Consequently, fadD mutants are blocked in fatty acid degradation (32, 40). Therefore, if the acyl chains of N-acyl HSL AI molecules were derived in vivo from acyl-CoAs, an E. coli fadD strain containing the IPTG-inducible traI gene on pSEP1 should be unable to synthesize AIs.
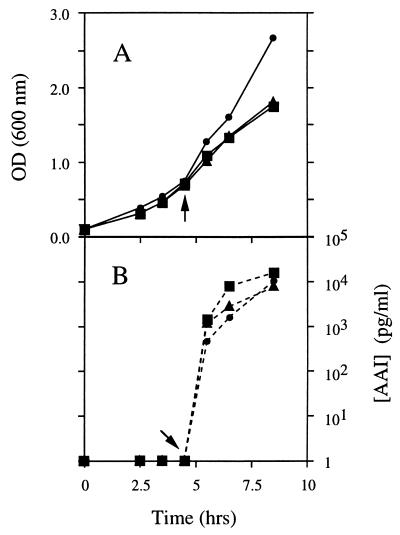
To investigate this prediction, a fadD null mutation (fadD88 zea::Tn10) was transduced into an otherwise wild-type strain (JM83) carrying a plasmid with the traI gene under lac control (pSEP1) and a lacIq plasmid (pMS421) to ensure repression of the lac promoter in the absence of IPTG. The amount of AAI present in the medium after various periods of induction with IPTG was determined for this strain (JMD6-TI) and compared with that for the parental strain carrying the same plasmids (JM83-TI) (Fig. 1). There was no significant difference in the amounts of AAI present in the medium from the fadD mutant strain and in that from the wild-type parental strain, indicating that AI synthesis does not require β-oxidation. We have also found that addition of oleic acid (Fig. 1) or disruption of the fadR gene (data not shown), both of which result in induction of the β-oxidation pathway (32, 48, 49, 60), failed to increase AI synthesis.
FIG. 1.
Effect of fatty acids and a fadD disruption on AAI synthesis. Shake-flask RB cultures of the parental strain JM83-TI (▪) and a fadD derivative of this strain (JMD6-TI [▴]) were grown at 37°C from a starting OD600 of 0.1. The optical density was monitored at regular intervals; after 1 h, the strain JM83-TI culture was divided into two flasks, and oleate was added to one of the flasks (•) to a final concentration of 0.1%. The expression of the TraI enzyme in each of the cultures was induced at mid-log phase by the addition of IPTG to 1 mM (indicated by the arrows), and samples for AAI assays were removed after 0, 1, 2, and 4 h of induction. (A) Optical density. (B) Concentration of AAI detected in medium from each of the cultures.
AI synthesis is blocked by specific inhibitors of fatty acid synthesis.
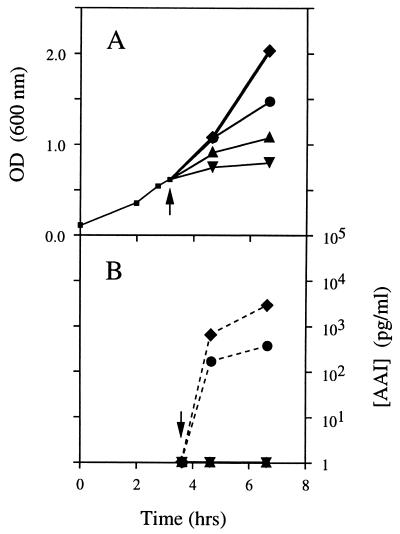
We have tested two specific inhibitors of fatty acid synthesis, diazoborine and cerulenin, for their effects on AAI synthesis (Fig. 2). In the presence of either diazoborine or cerulenin, there was no detectable AAI present in the cultures after either 1 or 3 h of TraI expression. Diazoborine inhibits fatty acid synthesis (25) by blocking the active site of the FabI enzyme (the NADH-dependent enoyl-ACP reductase) (7, 57), whereas cerulenin is a specific and irreversible inhibitor of the β-ketoacyl-ACP synthases I and II (fabB and fabF gene products, respectively) (13, 59). In contrast, when strain JM83-TI was treated with an inhibitor of metabolic processes other than fatty acid synthesis (nalidixic acid, a DNA gyrase inhibitor [21, 52]) as a control, after 3 h of induction with IPTG high concentrations of AAI (>500 pg/ml) were detected in the growth medium. However, this AAI concentration was less than that detected in the untreated culture. The cause of this lower AAI synthesis is not known, but one possibility is that it may be an indirect consequence of a reduced ATP pool, resulting from the constant active efflux of nalidixic acid by the AcrAB efflux pump (39).
FIG. 2.
Effect of diazoborine or cerulenin on AAI synthesis. A shake-flask RB culture of strain JM83-TI was grown at 30°C while the optical density was monitored at regular intervals. At mid-log phase, the culture was divided into five separate flasks. Three of the flasks contained an inhibitor: diazoborine (100 μg/ml [▴]), cerulenin (100 μg/ml [▾]), or nalidixic acid (100 μg/ml [•]). After these cultures had been exposed to the inhibitors for 30 min, IPTG was added (0.5 mM; indicated by the arrows) to each of these flasks and to one of the flasks lacking an inhibitor (⧫). The final flask (▪) was used as the minus-IPTG control. Duplicate samples were taken for AAI bioassays after 0, 1, and 4 h of IPTG induction. (A) Optical density. (B) Concentration of AAI detected in medium from each of the cultures.
The growth curves showed that cerulenin and diazoborine were both potent inhibitors of growth, producing a more rapid cessation of growth than that caused by nalidixic acid. To ensure that the observed effect on AAI synthesis was actually due to the inhibition of fatty acid synthesis per se rather than being a side product of blocking growth, the effects of diazoborine and nalidixic acid on AAI production by another strain, JMAC-TI, were also investigated.
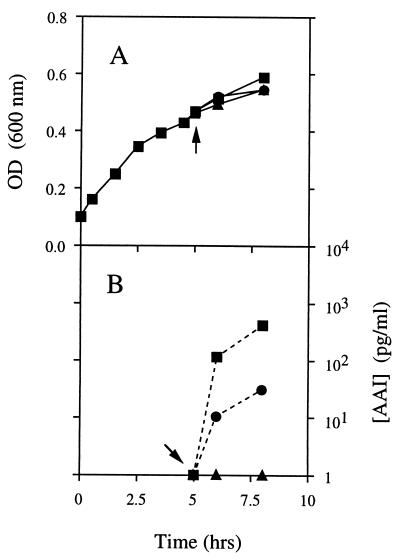
Strain JMAC-TI was constructed by transducing an aceEF mutation [(aroP-aceEF)zac::Tn10 (11)] into strain JM83-TI. As a result, JMAC-TI lacked pyruvate dehydrogenase activity and was therefore dependent on acetate or fatty acids to generate acetyl-CoA for growth. When supplemented with fatty acids in rich medium, the cells grew sufficiently slowly that there was little difference in growth rate upon addition of diazoborine or nalidixic acid (Fig. 3). However, there was once again no detectable AAI in the medium of diazoborine-treated cells, while AAI was detected in the media from both untreated cultures and cultures treated with nalidixic acid. Since this strain was dependent on a functional β-oxidation pathway for growth to produce acetyl-CoA from oleic acid, the cells must have contained acyl-CoAs. Hence, this result shows that, even when acyl-CoAs were present in vivo, inhibition of fatty acid synthesis rendered the cells unable to synthesize N-acyl HSLs.
FIG. 3.
Effect of diazoborine on AAI synthesis in strain JMAC-TI. An RB-oleate (0.1%) culture of JMAC-TI was grown at 37°C from a starting OD600 of 0.1, and the OD600 was monitored until mid-log phase. The culture was then divided into three flasks, containing either no inhibitor (▪), diazoborine (100 μg/ml [▴]), or nalidixic acid (100 μg/ml [•]). After 15 min of incubation with the inhibitors, IPTG was added (indicated by the arrows) to a final concentration of 0.5 mM, and the concentrations of AAI in the media were determined after 0, 1, and 3 h of IPTG induction. (A) Optical density. (B) Concentration of AAI detected in medium from each of the cultures.
We used specific inhibitors rather than mutants of the fatty acid synthetic pathway in this study for the following reasons. Firstly, null mutants in the early steps of the fatty acid synthetic pathway are lethal to E. coli (35). Secondly, although we found that strains containing temperature-sensitive mutations in accB, accD, and fabI all exhibited an inability to synthesize AIs by either LuxI or TraI at the nonpermissive temperature (data not shown), interpretation of these data was complicated by the fact that wild-type strains having a fully functional fatty acid synthesis pathway similarly exhibit much-reduced AI production by TraI and LuxI at 42°C, presumably due to the reduced activity of the AI synthases at this temperature.
Reduction in the intracellular AdoMet pool size by T3 AdoMet hydrolase expression results in decreased AI synthesis.
The approach we used to investigate the effect of the AdoMet pool size on AI synthesis was to assay AAI synthesis in strains expressing TraI from the pSEP1 plasmid in the presence of an enzyme that specifically degrades AdoMet (T3 AdoMet hydrolase [EC 3.3.1.2] [23]). We tested two different AdoMet expression constructs in the course of this work.
The T7 expression plasmid pETT3S was found to be toxic to strain BL21(DE3) even in the presence of a LacI-overexpressing plasmid (pMS421 [24]), presumably due to the background expression being sufficient to severely reduce intracellular AdoMet pools. The toxicity was not relieved by the introduction of the pLysS plasmid (which reduces T7 polymerase activity by expressing T7 lyzozyme [51]) but was significantly reduced by the presence of the pLysE plasmid (which provides a higher level of T7 lyzozyme expression [51]). To enable the T7 promoter-T3 AdoMet hydrolase and T7 lysozyme (pLysE) functions to be stably maintained in the presence of pSEP1 and pMS421 (to shut down the expression of TraI from the lac promoter of pSEP1), we constructed a composite plasmid (pT3SLE8) combining both functions on the pLysE plasmid by cloning the entire T7 promoter-T3 AdoMet hydrolase region of pETT3S as a 1.1-kb BspHI fragment into the BspHI site in the tetracycline resistance gene of pLysE. As a negative control for the effects resulting from expression of the T3 AdoMet hydrolase gene from the T7 promoter, a plasmid analogous to pT3SLE8 (pPCBLE1) was constructed to express a protein of similar size unrelated to AdoMet metabolism (the yeast PYC1 C-terminal 104-amino-acid biotin domain peptide [58]).
When pT3SLE8 was transformed into strain C41(DE3) [a mutant of BL21(DE3) selected for its ability to maintain and express toxic proteins (36)], the resultant strain (C41T3-TI) grew a little slower than the parental strain but did not exhibit the plasmid instability of pETT3S in BL21(DE3). The control plasmid pPCBLE1 did not noticeably slow the growth of strain C41. To determine the effect of these plasmids on AAI production, pSEP1, pMS421, and pT3SLE8 (strain C41T3-TI) or pPCBLE1 (strain C41PC-TI) were cotransformed into C41(DE3). IPTG was added to induce the expression of TraI from the lac promoter of pSEP1 and T7 polymerase expression and, thus, either T3 AdoMet hydrolase expression (strain C41T3-TI) or YPC1 biotin domain expression (strain C41PC-TI). The intracellular AdoMet and extracellular AAI concentrations were then determined at various time points before and after IPTG addition so as to determine the effect of changes in the AdoMet pools on AAI synthesis (Fig. 4). As strain C41PC-TI has a shorter doubling time than C41T3-TI, this strain was induced at a lower optical density than C41T3-TI cultures a and b so that any increased accumulation of AAI observed in this culture relative to that in the C41T3-TI cultures could not be attributed to its higher optical density.
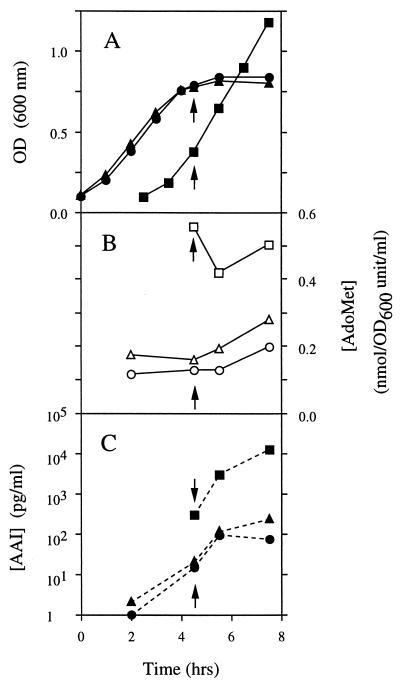
FIG. 4.
Effect of AdoMet hydrolase expression in strain C41 on AAI synthesis. Shake-flask RB cultures of a positive control strain (C41PC-TI [▪ and □]) and two isolates of C41 containing the T3 AdoMet hydrolase expression plasmid pACETT3 (C41T3-TIa [▴ and ▾] and C41T3-TIb [• and ○]) were grown at 37°C from a starting OD600 of 0.1. The optical density was monitored at regular intervals (A), and the cells were induced with 0.5 mM IPTG (indicated by the arrows) after being washed twice with minimal E medium to remove any AAI resulting from background TraI expression. Duplicate 15-ml culture samples were used to assay the intracellular AdoMet levels (B) and the AAI concentrations in the media (C) after 0, 1, and 3 h of IPTG induction.
In the two cultures of the AdoMet hydrolase-expressing strain C41T3-TI (a and b), the intracellular AdoMet concentrations at the time IPTG was added (indicated by the arrows) were 3.5- and 4.3-fold lower, respectively, than that of the control strain C41PC-TI (Fig. 4), presumably due to a basal expression of the AdoMet hydrolase gene. At the same time, the amount of AAI detected in the growth media of these strains was 14- to 20-fold lower, respectively, than that present in the control strain. Similarly, after 1 h of IPTG induction of both TraI and T7 RNA polymerase, the intracellular AdoMet concentrations of C41T3-TI isolates a and b were 2.2- and 3.3-fold lower, respectively, than that of the control strain C41PC-TI, whereas the concentrations of AAI detected in the media were 5.75- to 6.3-fold lower.
A similar experiment was performed with strain LEPRT7 (19) in which T7 RNA polymerase is induced by shift of the culture from 30 to 37°C. Strain LEPRT7 carrying either the AdoMet hydrolase expression plasmid, pT3SLE8 (LEPT3-TI), or the control expression plasmid, pPCBLE1 (LEPPC-TI), was grown to mid-log phase at 30°C in the presence of IPTG to induce the expression of TraI, washed twice to remove AAI accumulated in the medium, and then transferred to fresh medium containing 0.5 mM IPTG at 37°C (IPTG was added to failitate continued TraI expression). The intracellular AdoMet and extracellular AAI concentrations were then determined after 0, 0.5, 2, and 4 h of growth at 37°C (Fig. 5).
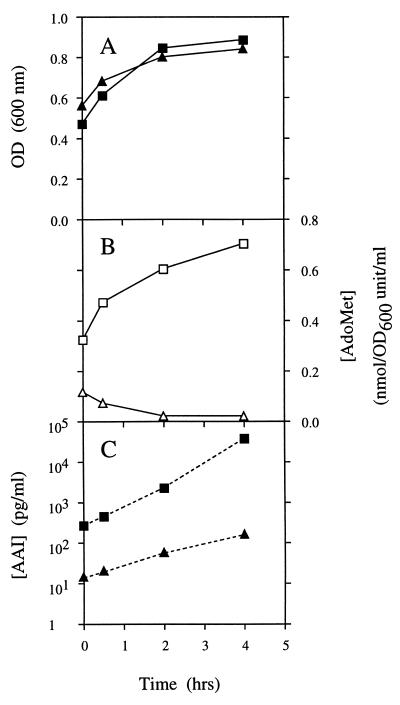
FIG. 5.
Effect of AdoMet hydrolase expression in strain LEPRT7 on AAI synthesis. Shake-flask RB cultures of a positive control strain (LEPPC-TI [▪ and □]) and an isolate of LEPRT7 containing the T3 AdoMet hydrolase expression plasmid pACETT3 (LEPT3-TI [▴ and ▾]) were grown at 30°C from a starting OD600 of 0.1. The cultures were grown to mid-log phase in the presence of 0.5 mM IPTG. The cells were washed twice with minimal E salts medium, resuspended in fresh RB medium containing 0.5 mM IPTG, and incubated at 37°C, and the optical density was monitored at regular intervals (A). Duplicate 15-ml samples were used to assay the intracellular AdoMet levels (B) and the AAI concentrations in the medium (C) after 0, 0.5, 2, and 4 h of heat induction.
At the start of the 37°C incubation period, the intracellular AdoMet concentration in the LEPT3-TI cells was almost threefold lower than that in the control LEPPC-TI cells, presumably due to a low level of T7 polymerase expression that occurred during growth at 30°C. The AAI concentration in the medium of the T3 AdoMet hydrolase strain was also lower, by more than 10-fold. During the course of the 4 h of growth at 37°C, the AdoMet concentration in the control increased from 0.33 to 0.7 nmol per unit of optical density at 600 nm (OD600) per ml while the OD600 of the culture increased from 0.47 to 0.88. The AAI concentration in the medium also increased, by over 2 orders of magnitude. In contrast, 4 h of heat induction of the AdoMet hydrolase strain resulted in a five- to sixfold decrease in intracellular AdoMet levels such that the AdoMet concentration was 33-fold less than that in the control strain. Similarly, while there was some accumulation of AAI in the medium over the 4-h period, this accumulation was at least 10-fold lower than that in the control strain. As a result, the final AAI concentration in the AdoMet hydrolase strain was more than 100-fold lower than that in the control strain, despite the fact that the optical densities of the two strains were very similar.
The experiments using T3 AdoMet hydrolase expression to decrease the AdoMet pool size in strains C41 and LEPRT7 suggest that the intracellular AdoMet concentration is limiting for AAI production. From the number of CFU per OD600 unit (approximately 1.2 × 109) determined for the parental uninduced strains and the assumption of an average cytoplasmic volume of 2.1 × 10−15 liters (61), after 1 h of T3 AdoMet hydrolase expression the intracellular AdoMet concentrations in strain C41T3-TI a and b were calculated to be 76 and 50 μM, respectively. Similarly, in strain LEPT3-TI after 2 h of T3 AdoMet hydrolase expression the intracellular AdoMet concentration was calculated to be 9.1 μM. These concentrations are consistent with AdoMet being limiting for AAI synthesis if we assume that the Km of native TraI for AdoMet in vivo is approximately 48 μM, the figure determined for His-tagged TraI in vitro (38).
Mutations in the threonine-methionine pathway that block homoserine synthesis do not decrease AI synthesis.
As homoserine is present in the cell as an intermediate of the threonine-methionine biosynthesis pathway, we investigated the possibility that either homoserine, homoserine phosphate, or O-succinyl homoserine from this pathway is the source of the HSL ring for N-acyl HSL synthesis in vivo. We assayed the AAI production in rich medium for mutant E. coli strains lacking activity in either aspartate kinase (strain GT164 [55]), aspartate semialdehyde dehydrogenase (strain CGSC 5081 [27] and strain G6MD3), homoserine dehydrogenase (strain CGSC 5075 [54]), homoserine kinase (strain CGSC 5076 [54]), or homoserine succinyltransferase (strains AB1932 and CGSC 6481). The bioassays indicated that there were no significant differences between the amounts of AAI produced by these strains upon IPTG induction of TraI expression and those produced by strains not blocked in this pathway (data not shown).
DISCUSSION
All the known AIs produced by gram-negative bacteria as density-dependent gene regulators can be classified structurally as N-acyl HSLs. As the name suggests, these molecules have two structural components, an HSL ring and an acyl group. In this study, we have investigated the source of these two components in vivo.
The most likely metabolic pathways which could supply the various acyl groups found in N-acyl HSL AIs are the fatty acid synthesis pathway and/or the β-oxidative fatty acid degradation pathway. In E. coli, fatty acid synthesis can be considered to be essentially a housekeeping function, although the fabA gene and thus synthesis of unsaturated fatty acids are subject to repression in the presence of long-chain fatty acids (28). In contrast, the β-oxidative fatty acid degradation (fad) pathway is expressed only in the presence of long-chain (>C12) fatty acids in the medium (8, 32, 40).
To determine whether the β-oxidation pathway contributes to N-acyl HSL synthesis, we investigated the effect of a fadD null mutation on AAI synthesis by the A. tumefaciens TraI enzyme in E. coli. Long-chain fatty acids are transported into E. coli by the combined actions of the fadL and fadD gene products. FadD catalyzes the conversion of the incoming fatty acids to their CoA thioesters, thus activating the acids for the other enzymes of the β-oxidation pathway (32). Furthermore, both in vitro and in vivo evidence have shown that FadD is the only E. coli acyl-CoA synthase (8, 32, 33, 40). Consequently, fadD mutants are unable to generate the necessary CoA thioesters required for β-oxidation and are thus totally blocked in fatty acid degradation. We found that the amount of AAI synthesized by E. coli was not affected by the fadD mutation (Fig. 1). From these data, we conclude that the acyl chains of AAI are not supplied by the β-oxidative fatty acid degradation pathway in vivo. In contrast, inhibition of fatty acid synthesis by either diazoborine or cerulenin (7, 25, 57) even in a strain dependent on β-oxidation for growth blocked synthesis of AAI. This agrees with the partial inhibition of AAI synthesis by cerulenin observed in vitro by Moré et al. (38). Furthermore, this cessation of AAI synthesis that we observed was due to the inhibition of fatty acid synthesis per se rather than to some other process affected by the inhibition of cell growth, as evident from the AAI synthesis observed during growth inhibition by nalidixic acid.
These data provide the first in vivo evidence indicating that the acyl group in N-acyl HSL AIs produced by the LuxI-type family of enzymes is derived from acyl-ACP thioesters generated in the course of fatty acid synthesis rather than from CoA thioesters generated by β-oxidation. This conclusion agrees with our previous in vitro findings with a purified maltose binding protein-LuxI fusion protein (46) and with those of Moré et al. (38) with His-tagged TraI. Both of these enzymes were able to synthesize their N-acyl HSL products in the presence of the appropriate acyl-ACP, but not if the acyl-ACP was replaced by the corresponding acyl-CoA.
Cao and Meighen had previously shown that cerulenin caused an inhibition of AI [N-(β-hydroxybutanoyl)HSL] synthesis in V. harveyi (10). They also found that the β-hydroxy group of this AI was in the D (now called R) conformation and that stimulation of AI synthesis occurred mainly with the addition of the 3R-isomer of butyric acid to the medium, consistent with the acyl group coming from fatty acid synthesis. However, these conclusions may not be relevant to the mechanism of AI synthesis by the LuxI family of enzymes for the following reasons. First, although there are two AI synthesis-detection systems in V. harveyi, neither synthase shows any discernible sequence homology to LuxI, and V. fischeri luxI and luxR hybridization probes failed to detect any luxI or luxR homologs in this species (4, 37). Also, transposon mutagenesis suggested that two genes (luxL and luxM) are required for the synthesis of N-(3-hydroxybutanyl)HSL in V. harveyi, although polar effects of luxL on luxM were not ruled out (4). Finally, the (3R)-hydroxybutanoyl acyl group of the V. harveyi AI is the monomer unit of poly-[(3R)-hydroxybutyrate] (PHB), and this species of bacterium has recently been found to carry out AI-dependent PHB synthesis (53). Hence, this acyl group may well be supplied by the pool of (3R)-hydroxylbutanoyl–CoA, the precursor for PHB synthesis in prokaryotes, obtained from the condensation of two molecules of acetyl-CoA followed by stereospecific reduction of the keto group (50), rather than by fatty acid synthesis per se.
Our finding that E. coli strains with mutations in the threonine-methionine biosynthesis pathway blocked in the synthesis of either homoserine, homoserine phosphate, or O-succinyl homoserine remain able to synthesize AAI (data not shown) agrees with the finding of Hanzelka and Greenberg (26) that only mutants blocked in the synthesis of methionine, rather than of homoserine or HSL, are unable to synthesize AI when expressing LuxI. These results also agree with our previous in vitro findings with a purified maltose binding protein-LuxI fusion protein (46) and with those of Moré et al. (38) with His-tagged TraI. Both of these enzymes synthesized the N-acyl HSL product in the presence of AdoMet, but not if AdoMet was replaced by homoserine or HSL.
If AdoMet is indeed the substrate that supplies the HSL ring for N-acyl HSL synthesis in in vivo alterations in the cells, AdoMet levels should affect the synthesis of N-acyl HSL AIs. We investigated this prediction by assaying AAI synthesis in the presence of an enzyme that specifically degrades AdoMet, thus lowering the intracellular AdoMet pool size. This approach has been used successfully before to study AdoMet-requiring processes in E. coli (12, 29) and is more specific than the use of methionine analogs (cycloleucine and ethionine) to inhibit AdoMet synthesis, as these analogs are incorporated in proteins in the place of methionine and/or provoke general amino acid starvation (2, 42). Furthermore, the fact that branched-chain amino acids protect against cycloleucine-induced growth inhibition indicates that cycloleucine has other effects on E. coli metabolism (3).
Using the T3 AdoMet hydrolase expression approach, we found that E. coli cells with a reduced AdoMet pool size have a greatly reduced capacity to synthesize AAI (Fig. 5), confirming the in vitro studies. The finding that small changes in intracellular AdoMet levels produced proportionally larger changes in AAI synthesis by TraI is consistent with our measurements of the intracellular AdoMet concentrations and the Km of TraI for this substrate, which is reported to be 48 μM (38). Therefore, it seems likely that AI synthesis is limited by the intracellular AdoMet concentration in E. coli. It would be of interest to test if AdoMet limits AI synthesis in the native organisms.
ACKNOWLEDGMENTS
We are grateful to S. K. Farrand for supplying us with N-[3-oxooctanoyl]HSL to use as a standard, for the plasmid pSEP1, and the A. tumefaciens indicator strain NT1(pDC141E33) and to Friederike Turnowsky, University of Graz, Graz, Austria, for kindly supplying us with diazoborine.
This work was supported by National Institutes of Health grant AI15650.
REFERENCES
- 1.Alberts A W, Bell R M, Vagelos P R. Acyl carrier protein. XV. Studies of β-ketoacyl-acyl carrier protein synthetase. J Biol Chem. 1972;247:3190–3198. [PubMed] [Google Scholar]
- 2.Alix J-H. Molecular aspects of the in vivo and in vitro effects of ethionine, an analog of methionine. Microbiol Rev. 1982;46:281–295. doi: 10.1128/mr.46.3.281-295.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baker H, Frank O, DeAngelis B, Baker E. Branched-chain amino acids overcome cycloleucine growth inhibition in B12 and non-B12-requiring microorganisms. Int J Vitam Nutr Res. 1991;61:319–324. [PubMed] [Google Scholar]
- 4.Bassler B L, Wright M, Showalter R E, Silverman M E. Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol. 1993;9:773–786. doi: 10.1111/j.1365-2958.1993.tb01737.x. [DOI] [PubMed] [Google Scholar]
- 5.Beck von Bodman S, Hayman G T, Farrand S K. Opine catabolism and conjugal transfer of the nopaline Ti plasmid pTiC58 are coordinately regulated by a single repressor. Proc Natl Acad Sci USA. 1992;89:643–647. doi: 10.1073/pnas.89.2.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beck von Bodman S B, Farrand S K. Capsular polysaccharide biosynthesis and pathogenicity in Erwinia stewartii require induction by an N-acylhomoserine lactone autoinducer. J Bacteriol. 1995;177:5000–5008. doi: 10.1128/jb.177.17.5000-5008.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bergler H, Wallner P, Ebeling A, Leitinger B, Fuchsbichler S, Aschauer H, Kollenz G, Hogenauer G, Turnowsky F. Protein EnvM is the NADH-dependent enoyl-ACP reductase (FabI) of Escherichia coli. J Biol Chem. 1994;269:5493–5496. [PubMed] [Google Scholar]
- 8.Black P N, DiRusso C C. Molecular and biochemical analysis of fatty acid transport, metabolism, and gene regulation in Escherichia coli. Biochim Biophys Acta. 1994;1210:123–145. doi: 10.1016/0005-2760(94)90113-9. [DOI] [PubMed] [Google Scholar]
- 9.Cao J, Meighen E A. Purification and structural identification of an autoinducer for the luminescence system of Vibrio harveyi. J Biol Chem. 1989;264:21670–21676. [PubMed] [Google Scholar]
- 10.Cao J G, Meighen E A. Biosynthesis and stereochemistry of the autoinducer controlling luminescence in Vibrio harveyi. J Bacteriol. 1993;175:3856–3862. doi: 10.1128/jb.175.12.3856-3862.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang Y-Y, Cronan J E J. Mapping nonselectable genes of Escherichia coli by using transposon Tn10: location of a gene affecting pyruvate oxidase. J Bacteriol. 1982;151:1279–1289. doi: 10.1128/jb.151.3.1279-1289.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cronan J E, Jr, Reed R, Taylor F R, Jackson M B. Properties and biosynthesis of cyclopropane fatty acids in Escherichia coli. J Bacteriol. 1979;138:118–121. doi: 10.1128/jb.138.1.118-121.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.D’Agnolo G, Rosenfeld I S, Awaya J, Omura S, Vagelos P R. Inhibition of fatty acid synthesis by the antibiotic cerulenin. Specific inactivation of beta-ketoacyl-acyl carrier protein synthetase. Biochim Biophys Acta. 1973;326:155–166. doi: 10.1016/0005-2760(73)90241-5. [DOI] [PubMed] [Google Scholar]
- 14.Davis R W, Botstein D, Roth J R. Advanced bacterial genetics: a manual for genetic engineering. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1980. [Google Scholar]
- 15.Eberhard A, Burlingame A L, Eberhard C, Kenyon G L, Nealson K H, Oppenheimer N J. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry. 1981;20:2444–2449. doi: 10.1021/bi00512a013. [DOI] [PubMed] [Google Scholar]
- 16.Eberhard A, Longin T, Widrig C A, Stranick S J. Synthesis of the lux gene autoinducer in Vibrio fischeri is positively autoregulated. Arch Microbiol. 1991;155:294–297. [Google Scholar]
- 17.Eberl L, Winson M K, Sternberg C, Stewart G S A B, Christiansen G, Chhabra S R, Bycroft B W, Williams P, Molin S, Givskov M. Involvement of N-acyl-l-homoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol Microbiol. 1996;20:127–136. doi: 10.1111/j.1365-2958.1996.tb02495.x. [DOI] [PubMed] [Google Scholar]
- 18.Engebrecht J, Silverman M. Identification of genes and gene products necessary for bacterial bioluminescence. Proc Natl Acad Sci USA. 1984;81:4154–4158. doi: 10.1073/pnas.81.13.4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fedorova N D, Peredelchuk M Y, Kirpichnikov M P, Bennett G N. Escherichia coli strain for thermoinducible T7 RNA polymerase-driven expression. Gene. 1996;177:267–268. doi: 10.1016/0378-1119(96)00294-6. [DOI] [PubMed] [Google Scholar]
- 20.Fuqua C, Winans S C, Greenberg E P. Census and consensus in bacterial ecosystems: the LuxR-LuxI family of quorum-sensing transcriptional regulators. Annu Rev Microbiol. 1996;50:727–751. doi: 10.1146/annurev.micro.50.1.727. [DOI] [PubMed] [Google Scholar]
- 21.Gellert M, Mizuuchi K, O’Dea M H, Itoh T, Tomizawa J-I. Nalidixic acid resistance: a second genetic character involved in DNA gyrase activity. Proc Natl Acad Sci USA. 1977;74:4772–4776. doi: 10.1073/pnas.74.11.4772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gilson L, Kuo A, Dunlap P V. AinS and a new family of autoinducer synthesis proteins. J Bacteriol. 1995;177:6946–6951. doi: 10.1128/jb.177.23.6946-6951.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gold M, Hausmann R, Maitra U, Hurwitz J. The enzymatic methylation of RNA and DNA. VIII. Effects of bacteriophage infection on the activity of the methylating enzymes. Proc Natl Acad Sci USA. 1964;52:292–297. doi: 10.1073/pnas.52.2.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grana D, Gardella T, Susskind M M. The effects of mutations in the ant promoter of phage P22 depend on context. Genetics. 1988;120:319–327. doi: 10.1093/genetics/120.2.319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grassberger M A, Turnowsky F, Hildebrandt J. Preparation and antibacterial activities of new 1,2,3-diazaborine derivatives and analogues. J Med Chem. 1984;27:947–953. doi: 10.1021/jm00374a003. [DOI] [PubMed] [Google Scholar]
- 26.Hanzelka B L, Greenberg E P. Quorum sensing in Vibrio fischeri: evidence that S-adenosylmethionine is the amino acid substrate for autoinducer synthesis. J Bacteriol. 1996;178:5291–5294. doi: 10.1128/jb.178.17.5291-5294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hatfield D, Hofnung M, Schwartz M. Genetic analysis of the maltose A region in Escherichia coli. J Bacteriol. 1969;98:559–567. doi: 10.1128/jb.98.2.559-567.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Henry M F, Cronan J E., Jr Escherichia coli transcription factor that both activates fatty acid synthesis and represses fatty acid degradation. J Mol Biol. 1991;222:843–849. doi: 10.1016/0022-2836(91)90574-p. [DOI] [PubMed] [Google Scholar]
- 29.Hughes J A, Brown L R, Ferro A J. Expression of the cloned coliphage T3 S-adenosylmethionine hydrolase gene inhibits DNA methylation and polyamine biosynthesis in Escherichia coli. J Bacteriol. 1987;169:3625–3632. doi: 10.1128/jb.169.8.3625-3632.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hwang I, Li P L, Zhang L, Piper K R, Cook D M, Tate M E, Farrand S K. TraI, a LuxI homologue, is responsible for production of conjugation factor, the Ti plasmid N-acylhomoserine lactone autoinducer. Proc Natl Acad Sci USA. 1994;91:4639–4643. doi: 10.1073/pnas.91.11.4639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kaplan H B, Greenberg E P. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. J Bacteriol. 1985;163:210–214. doi: 10.1128/jb.163.3.1210-1214.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein K, Steinberg R, Fiethen B, Overath P. Fatty acid degradation in Escherichia coli. An inducible system for the uptake of fatty acids and further characterization of old mutants. Eur J Biochem. 1971;19:442–450. doi: 10.1111/j.1432-1033.1971.tb01334.x. [DOI] [PubMed] [Google Scholar]
- 33.Knoll L J, Gordon J I. Use of strains containing fad mutations plus a triple plasmid system to study import of myristate, its activation by Saccharomyces cerevisiae acyl-CoA synthetase and its utilization by S. cerevisiae myristoyl-CoA-protein N-myristoyltransferase. J Biol Chem. 1993;268:4281–4290. [PubMed] [Google Scholar]
- 34.Lombardini J, Coulter A W, Talalay P. Analogues of methionine as substrates and inhibitors of the methionine adenosyltransferase reaction. Mol Pharmacol. 1970;6:481–499. [PubMed] [Google Scholar]
- 35.Magnuson K, Jackowski S, Rock C O, Cronan J E., Jr Regulation of fatty acid biosynthesis in Escherichia coli. Microbiol Rev. 1993;57:522–542. doi: 10.1128/mr.57.3.522-542.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miroux B, Walker J E. Over-production of proteins in Escherichia coli: mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 37.Miyamoto C M, Graham A F, Meighen E A. Nucleotide sequence of the luxC gene and the upstream DNA from the bioluminescent system of Vibrio harveyi. Nucleic Acids Res. 1988;16:1551–1562. doi: 10.1093/nar/16.4.1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Moré M I, Finger L D, Stryker J L, Fuqua C, Eberhard A, Winans S C. Enzymatic synthesis of a quorum-sensing autoinducer through use of defined substrates. Science. 1996;272:1655–1658. doi: 10.1126/science.272.5268.1655. [DOI] [PubMed] [Google Scholar]
- 39.Okusu H, Ma D, Nikaido H. AcrAB efflux pump plays a major role in antibiotic resistance phenotype of Escherichia coli multiple-antibiotic-resistance (Mar) mutants. J Bacteriol. 1996;178:306–308. doi: 10.1128/jb.178.1.306-308.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Overath P, Pauli G, Schairer H U. Fatty acid degradation in Escherichia coli. An inducible acyl-CoA synthetase, the mapping of old mutations, and the isolation of regulatory mutants. Eur J Biochem. 1969;7:559–574. [PubMed] [Google Scholar]
- 41.Pearson J P, Gray K M, Passador L, Tucker K D, Eberhard A, Iglewski B A, Greenberg E P. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc Natl Acad Sci USA. 1994;91:197–201. doi: 10.1073/pnas.91.1.197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pine M J. Comparative physiological effects of incorporated amino acid analogs in Escherichia coli. Antimicrob Agents Chemother. 1978;13:676–685. doi: 10.1128/aac.13.4.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salmond G P, Bycroft B W, Stewart G S, Williams P. The bacterial ’enigma’: cracking the code of cell-cell communication. Mol Microbiol. 1995;16:615–624. doi: 10.1111/j.1365-2958.1995.tb02424.x. [DOI] [PubMed] [Google Scholar]
- 44.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 45.Satishchandran C, Taylor J C, Markham G D. Novel Escherichia coli K-12 mutants impaired in S-adenosylmethionine synthesis. J Bacteriol. 1990;172:4489–4496. doi: 10.1128/jb.172.8.4489-4496.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schaefer A L, Val D L, Hanzelka B L, Cronan J E, Jr, Greenberg E P. Generation of cell-to-cell signals in quorum sensing: acyl homoserine lactone synthase activity of a purified Vibrio fischeri LuxI protein. Proc Natl Acad Sci USA. 1996;93:9505–9509. doi: 10.1073/pnas.93.18.9505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shaw P D, Ping G, Daly S L, Cha C, Cronan J E, Jr, Rinehart K L, Farrand S K. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. Proc Natl Acad Sci USA. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Simons R W, Egan P A, Chute H T, Nunn W D. Regulation of fatty acid degradation in Escherichia coli: isolation and characterization of strains bearing insertion and temperature-sensitive mutations in gene fadR. J Bacteriol. 1980;142:621–632. doi: 10.1128/jb.142.2.621-632.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Simons R W, Hughes K T, Nunn W D. Regulation of fatty acid degradation in Escherichia coli: dominance studies with strains merodiploid in gene fadR. J Bacteriol. 1980;143:726–730. doi: 10.1128/jb.143.2.726-730.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steinbuchel A, Schlegel H G. Physiology and molecular genetics of poly(β-hydroxyalkanoic acid) synthesis in Alcaligenes eutrophus. Mol Microbiol. 1991;5:535–542. doi: 10.1111/j.1365-2958.1991.tb00725.x. [DOI] [PubMed] [Google Scholar]
- 51.Studier F W. Use of bacteriophage T7 lysozyme to improve an inducible T7 expression system. J Mol Biol. 1991;219:37–44. doi: 10.1016/0022-2836(91)90855-z. [DOI] [PubMed] [Google Scholar]
- 52.Sugino A, Peebles C L, Kreuzer K N, Cozzarelli N R. Mechanism of action of nalidixic acid: purification of Escherichia coli nalA gene product and its relationship to DNA gyrase and a novel nicking-closing enzyme. Proc Natl Acad Sci USA. 1977;74:4767–4771. doi: 10.1073/pnas.74.11.4767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun W, Cao J-G, Teng K, Meighen E A. Biosynthesis of poly-3-hydroxybutyrate in the luminescent bacterium, Vibrio harveyi, and regulation by the lux autoinducer N-(3-hydroxybutanoyl)homoserine lactone. J Biol Chem. 1994;269:20785–20790. [PubMed] [Google Scholar]
- 54.Theze J, Margarita D, Cohen G N, Borne F, Patte J C. Mapping of the structural genes of the three aspartokinases and of the two homoserine dehydrogenases of Escherichia coli K-12. J Bacteriol. 1974;117:133–143. doi: 10.1128/jb.117.1.133-143.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Theze J, Saint-Girons I. Threonine locus of Escherichia coli K-12: genetic structure and evidence for an operon. J Bacteriol. 1974;118:990–998. doi: 10.1128/jb.118.3.990-998.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Throup J P, Camara M, Briggs G S, Winson M K, Chhabra S R, Bycroft B W, Williams P, Stewart G S A B. Characterisation of the yenI/yenR locus from Yersinia enterocolitica mediating the synthesis of two N-acylhomoserine lactone signal molecules. Mol Microbiol. 1995;17:345–356. doi: 10.1111/j.1365-2958.1995.mmi_17020345.x. [DOI] [PubMed] [Google Scholar]
- 57.Turnowsky F, Fuchs K, Jeschek C, Hogenauer G. envM genes of Salmonella typhimurium and Escherichia coli. J Bacteriol. 1989;171:6555–6565. doi: 10.1128/jb.171.12.6555-6565.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Val D L, Chapman-Smith A, Walker M E, Cronan J E, Wallace J C. Polymorphism of the yeast pyruvate carboxylase 2 gene and protein: effects on protein biotinylation. Biochem J. 1995;312:817–825. doi: 10.1042/bj3120817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vance D, Goldberg I, Mitsuhashi O, Bloch K, Omura S, Nomura S. Inhibition of fatty acid synthetases by the antibiotic cerulenin. Biochem Biophys Res Commun. 1972;48:649–656. doi: 10.1016/0006-291x(72)90397-x. [DOI] [PubMed] [Google Scholar]
- 60.Weeks G, Shapiro M, Burns R O, Wakil S J. Control of fatty acid metabolism. I. Induction of the enzymes of fatty acid oxidation in Escherichia coli. J Bacteriol. 1969;97:827–837. doi: 10.1128/jb.97.2.827-836.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Woldringh C L, Nanninga N. Structure of the nucleoid and cytoplasm in the intact cell. In: Naninga N, editor. Molecular cytology of Escherichia coli. Orlando, Fla: Academic Press; 1985. pp. 161–198. [Google Scholar]
- 62.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 63.Zhang L, Murphy P J, Kerr A, Tate M E. Agrobacterium conjugation and gene regulation by N-acyl-l-homoserine lactones. Nature. 1993;362:446–448. doi: 10.1038/362446a0. [DOI] [PubMed] [Google Scholar]