Abstract
Dietary metabolomics is a relatively objective approach to identifying new biomarkers of dietary intake and for use alongside traditional methods. However, methods used across dietary feeding studies vary, thus making it challenging to compare results. The objective of this study was to synthesize methodological components of controlled human feeding studies designed to quantify the diet-related metabolome in biospecimens, including plasma, serum, and urine after dietary interventions. Six electronic databases were searched. Included studies were as follows: 1) conducted in healthy adults; 2) intervention studies; 3) feeding studies focusing on dietary patterns; and 4) measured the dietary metabolome. From 12,425 texts, 50 met all inclusion criteria. Interventions were primarily crossover (n = 25) and parallel randomized controlled trials (n = 22), with between 8 and 395 participants. Seventeen different dietary patterns were tested, with the most common being the “High versus Low-Glycemic Index/Load” pattern (n = 11) and “Typical Country Intake” (n = 11); with 32 providing all or the majority (90%) of food, 16 providing some food, and 2 providing no food. Metabolites were identified in urine (n = 31) and plasma/serum (n = 30). Metabolites were quantified using liquid chromatography, mass spectroscopy (n = 31) and used untargeted metabolomics (n = 37). There was extensive variability in the methods used in controlled human feeding studies examining the metabolome, including dietary patterns tested, biospecimen sample collection, and metabolomic analysis techniques. To improve the comparability and reproducibility of controlled human feeding studies examining the metabolome, it is important to provide detailed information about the dietary interventions being tested, including information about included or restricted foods, food groups, and meal plans provided. Strategies to control for individual variability, such as a crossover study design, statistical adjustment methods, dietary-controlled run-in periods, or providing standardized meals or test foods throughout the study should also be considered.
The protocol for this review has been registered at Open Science Framework (https://doi.org/10.17605/OSF.IO/DAHGS).
Keywords: metabolomics, dietary assessment, feeding interventions, dietary metabolome, metabolites, biomarkers, dietary intake
Statement of Significance.
To date, no reviews of the relationship between dietary patterns and the metabolome have specifically focused on methodologies of human feeding studies. The review highlights the need for formal reporting guidelines for diet-related metabolome studies to improve comparability and reproducibility. A checklist of suggested questions for researchers to consider when reporting diet and methodologies used has been provided as a starting point to help guide future research.
Introduction
Dietary metabolomics can be used to quantify the appearance of a range of metabolites in body fluids (blood and urine) after food ingestion and accounts for individual variability in metabolism [1]. The application of dietary metabolomics is promising as a relatively objective approach to identify key biomarkers of dietary intake in the future and for use alongside traditional methods to develop the field of personalized nutrition [2]. To date, there are few validated objective biomarkers of dietary intake. These include 24-h urinary nitrogen as a measure of protein intake; doubly labeled water as a measure of total energy intake; 24-h urinary sodium as a measure of sodium intake; 24-h urinary potassium as measures of dietary potassium intake; and 24-h urinary sucrose and fructose as measures of specific sugars intake [3]. Advances in technology, using metabolomic techniques, such as nuclear magnetic resonance (NMR) and mass spectroscopy methods, have resulted in the detection of a greater number of metabolites in biofluids and are becoming important techniques for the identification of objective measures of dietary intake [4].
Current dietary assessment methods, such as 24-h recalls, food records, and food frequency questionnaires, rely on self-report [3]. These methods provide detailed information on what and when people eat; however, they have inherent misreporting errors, which can be random or systematic and may lead to underestimation of relationships between diet and health [5]. The identification of objective measures of dietary intakes, such as through the dietary metabolome is a promising area with the potential to provide insight into the quantification of dietary intake, individual variation in metabolic response to diet and the role of personalized nutrition in the context of health and disease [2,4,6].
People eat more than single foods or food sources of single nutrients. This has led to interest in dietary patterns and total diet. Dietary patterns aim to capture the complexity of diet by taking into consideration the variety, quality, and frequency of intake [7]. The shift toward dietary patterns as opposed to single foods or nutrients is evident in recent revisions of various national dietary guidelines across the globe [[8], [9], [10]]. Healthier dietary patterns, such as the Dietary Approaches to Stop Hypertension, Mediterranean, and vegetarian-style diets, have been associated with a reduced risk of chronic conditions, such as cardiovascular disease [9,11,12]. The interest in dietary patterns has led researchers to identify biomarkers of dietary patterns to gain insights into diet–disease relationships [13]. Findings from intervention and observational studies have been reviewed [[14], [15], [16]], with findings identifying relationships between plasma, serum and/or urinary metabolites, and dietary patterns, but have not identified any metabolite profiles that distinguish specific dietary patterns [[14], [15], [16]]. Metabolomic profiling, or characterizing the metabolome, involves the detection of small molecules influenced by dietary intake that represent the overall biological system response [17,18]. The most frequently used methods for identifying metabolites are NMR, liquid chromatography–mass spectrometry (LC-MS), or GC-MS [19]. The gold standard approach to metabolite identification is by comparing compounds to a known standard. However, this may not always be feasible, therefore the Metabolomics Standard Initiative has suggested levels of quality metabolite identification [20]. Where metabolites cannot be matched against a known standard, metabolites may be matched using reference databases such as the Human Metabolome Database (HMDB), a freely available electronic database containing >2,230,000 metabolite entries with associated chemical data, clinical data, and molecular biology/biochemistry data [21], METLIN another freely accessible metabolite database with >20,000 metabolite entries [22], and the FooDB (https://foodb.ca/) which is specific to food constituents including those that impact taste, flavor, and color of foods. Metabolites may also be determined by comparison of retention times reported within those in previous publications and databases [23]. However, it is important to consider the matching of retention times in the context of the metabolomic method chosen [24]. For example, LC-MS methods may result in the ability to match compounds with multiple structures. Therefore, to accurately identify the correct metabolite requires expertise in using a process of elimination [24]. Metabolomic profiling can also involve the quantification of various dietary metabolites to inform the degree of representation of such biomarkers in relation to specific dietary traits. Current challenges in measuring and interpreting the dietary metabolome include the lack of standardization in methodologies used between laboratories, limited reproducibility of findings, difficulties with quantification, representation of unknown compounds, and accessibility [25]. These challenges also limit the comparability between studies [25].
Understanding metabolic signatures relative to varying dietary patterns, how they differ between individuals, and how this data can be used to gain greater insight into the relationship between diet and health will inform approaches for personalized nutrition. Differences in research methodologies related to study design, particularly the dietary pattern or feeding intervention component and metabolomic analysis, and therefore study findings have been highlighted as a current challenge in this field [19,25]. Therefore, summarizing research studies is important in terms of developing recommendations regarding study methods going forward. To date, no reviews of the relationship between dietary patterns and the metabolome have specifically focused on the methodologies of human feeding studies. In the context of the current review, controlled feeding studies were those that provided all or partial test foods and/or meals (with or without diet plan materials) to participants for the whole feeding study duration, or studies that were highly prescriptive through provision of strict meal plans/menus and food/nutrient targets. This type of dietary intervention aims to reduce misreporting and facilitates the accuracy of metabolite identification by allowing investigators to exert control over what participants are eating [4]. These studies are the first step toward efforts to identify new biomarkers of dietary intake. Findings from these studies can then be explored in other populations and settings to evaluate translational capacity and standardization of dietary biomarkers [1,4].
Therefore, the aim of the current review was to synthesize the methodological components used in controlled human feeding studies designed to identify the diet-related metabolome in serum, plasma, or urine in response to various dietary patterns. This includes summarizing: 1) the feeding protocol and dietary assessment method(s) used, 2) sample collection and processing, and 3) laboratory methods used for measurement of the dietary metabolome.
Methods
The current scoping review is reported as per PRISMA for Scoping Reviews guidelines [26], including the PRISMA for Scoping Reviews checklist (Supplemental Table 1). The methods of the current review involved identifying the research question, conducting the search, identifying relevant studies for inclusion, mapping the data, collating results, and summarizing findings [27]. The protocol for this review has been registered at Open Science Framework (https://doi.org/10.17605/OSF.IO/DAHGS) and findings are reported as per updated guidelines [28].
Information sources and search strategy
Six key databases were searched for studies up to January 10, 2023 (EDC), including EMBASE, Medline, Cumulative Index to Nursing and Allied Health Literature (CINAHL), PsychINFO, Scopus, and Cochrane, using the following search terms: “Metabolome OR metabolomics OR nutrimetabolomics OR nutrigenomics” AND “Diet OR feeding study OR dietary pattern OR nutrition assessment OR diet∗ assessment”. Limits included full-text publications, humans and the English language. The example search strategy for Medline is provided in Supplemental Table 2.
Study eligibility criteria
A broad research question was set based on criteria specifying the participant, intervention, comparator, outcome, and setting as defined in Table 1. For the current study, feeding studies were defined as either 1) where all foods and/or meals were provided for the entire duration of the intervention or part thereof; 2) where a minimum of at least a full day’s worth of food is provided; 3) where key foods defining a dietary pattern e.g., olive oil, are provided alongside prescription of a dietary pattern; or 4) a highly prescriptive diet with target amounts of certain foods and/or nutrients and meal plans were provided. The following studies were excluded: postprandial interventions, interventions targeting individual foods, interventions assessing the effect of fasting or very low-calorie diets without a weight-stable control, and interventions that did not provide sufficient dietary prescription/instruction.
TABLE 1.
PICOS inclusion and exclusion criteria
| Inclusion | Exclusion | |
|---|---|---|
| Participants |
|
|
| Intervention |
|
|
| Comparator |
|
|
| Outcome |
|
|
| Setting |
|
|
Abbreviations: DASH, Dietary Approaches to Stop Hypertension; PICOS, participant, intervention, comparator, outcome, and setting.
Data selection
All references identified from the search were downloaded into an EndNote X9 (Clarivate Analytics) file and duplicates were removed. These references were then uploaded to Covidence systematic review software (Veritas Health Innovation, www.covidence.org) and any remaining duplicates missed were also removed. All titles and abstracts were screened for inclusion by 2 reviewers (EDC, JJAF, CCC, JS, or RT). If there was a conflict, then a third reviewer (EDC, JJAF, CEC, JS, or RT) made the final decision. Full texts of the studies included at the title and abstract stage were then uploaded to Covidence and screened by at least 2 reviewers (EDC, JJAF, or JS) for inclusion in the review. If a decision could not be made regarding whether a paper should or should not be included, then discussions were first held between the 2 reviewers and if this could still not be resolved, then a third reviewer (CEC) made the final decision.
Data extraction and synthesis
Data extraction of included texts was undertaken by 3 reviewers (EDC, JJAF, and JS). The data extraction template was piloted by 2 reviewers and revised after the extraction of 2 studies. Data were extracted for manuscript details, study population, study design (crossover or parallel), duration, sample size of each group, dietary intervention characteristics and feeding components, dietary assessment methods, and biomarker and metabolite characteristics including metabolome analysis methods, statistical analysis, results, limitations, and author conclusions. Data were synthesized descriptively and reported by utilizing subgroups to describe studies of similar methodological characteristics.
Results
Summary of search
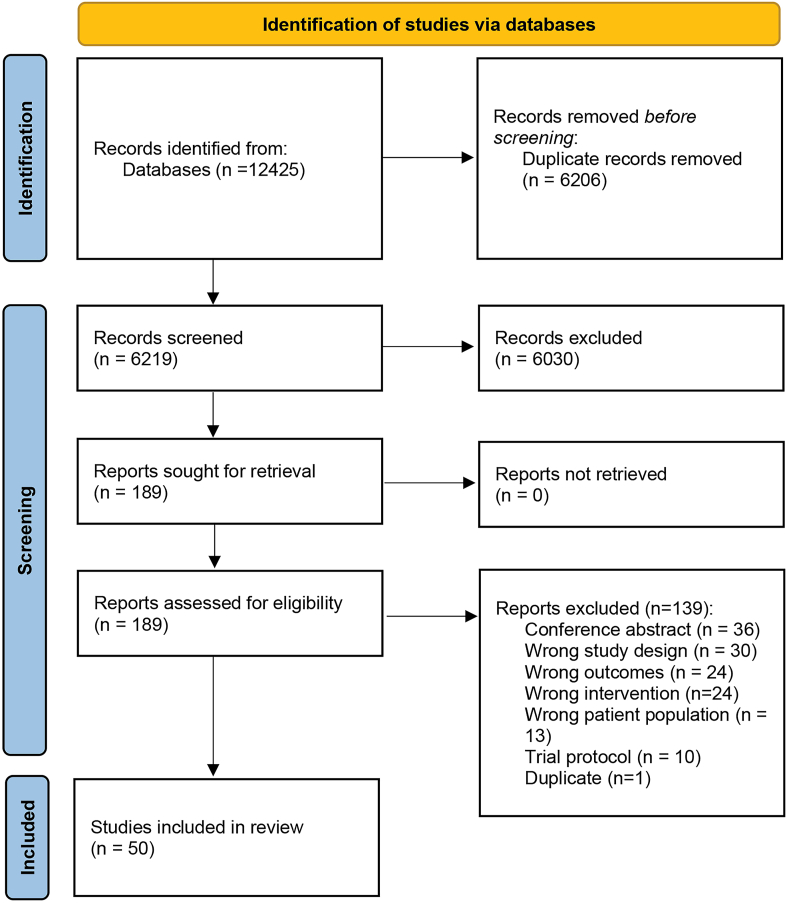
A total of 12,425 texts were identified from the search up until January 2023, of these texts, 50 publications met inclusion criteria and were included in the current review. Figure 1 summarizes the study selection process.
FIGURE 1.
PRISMA flow diagram of included studies.
Characteristics of included studies
The 50 included studies were made up of 26 crossover feeding studies [[29], [30], [31], [32], [33], [34], [35], [36], [37], [38], [39], [40], [41], [42], [43], [44], [45], [46], [47], [48], [49], [50], [51], [52], [53], [54], [55]] (4 without randomization [32,35,52,53]) and 22 randomized controlled trials (RCTs) with parallel groups [[56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73], [74], [75], [76], [77]] and one prepost intervention [78]. The study participants’ age ranged from 18 to 80 y and several studies were conducted on females [32,53,78] or males only [30,45,50,63]. Several studies did not report the number of male and female participants [29,33,37,51,57,59,71,73], of which 3 were unable to report this detail because they were protocol papers [37,57,59]. Studies included between 8 and 395 participants, with a median of 44 participants. Studies were conducted in Europe (n = 34) [30,[32], [33], [34], [35], [36], [37], [38], [39],43,45,46,47,[49], [50], [51],[56], [57], [58], [59], [60], [61],65,67,[69], [70], [71], [72], [73], [74], [75], [76], [77], [78]], the United States (n = 14) [29,31,[40], [41], [42],44,[52], [53], [54], [55],57,62,66,68], Korea (n = 1) [48], and Canada (n = 1) [64]. Study duration ranged from 2 d to 3 y, and 29 studies [29,30,39,40,46,47,[51], [52], [53], [54],[56], [57], [58], [59], [60],[62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73], [74],76] administered a run-in period before the study intervention. Refer to Supplemental Table 3.
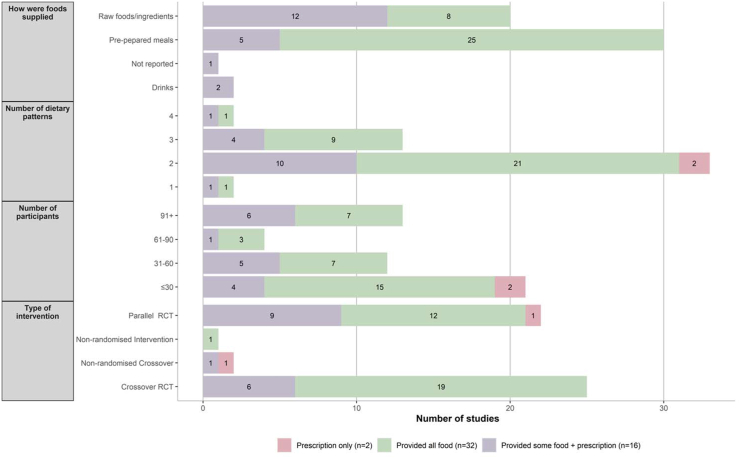
Across the included studies, 17 different dietary patterns were classified. The top 3 dietary patterns investigated were classified as a “High or Low-Glycemic Index or Load Dietary Patterns” (n = 11) [29,31,37,38,43,44,46,55,57,70,76], a “Typical Country Intake Defined Dietary Pattern” (n = 11) [48,51,52,56,60,62,65,66,68,71,72], and a “Macronutrient Defined Dietary Pattern” (n = 10) [29,36,39,40,42,54,69,74,75,77]. Studies tested the impact of between 1 (n = 2) [51,59] and 4 (n = 2) [34,36] dietary patterns, with the majority of interventions comparing 2 dietary patterns (n = 33) [[30], [31], [32], [33],37,38,[41], [42], [43], [44],46,47,49,[52], [53], [54],56,57,60,[62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72],74,[76], [77], [78]] (Figure 2).
FIGURE 2.
Study characteristics according to the amount of food provided. RCT, randomized controlled trial.
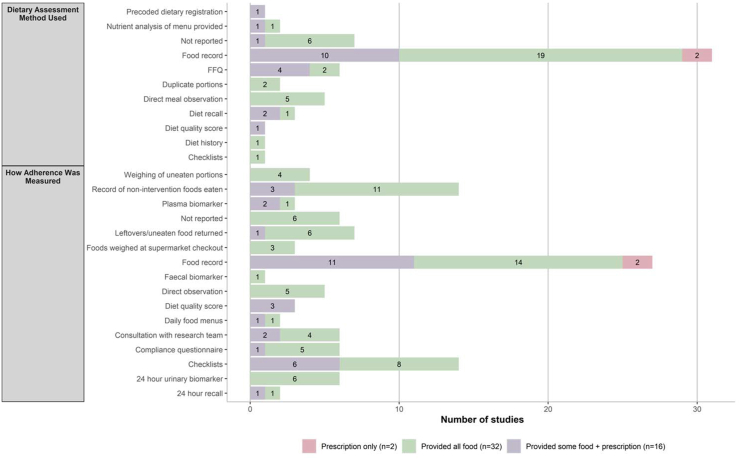
Adherence to dietary interventions was commonly assessed using food records (n = 26) [32,33,35,[37], [38], [39], [40],43,46,52,54,57,[60], [61], [62],[65], [66], [67],[69], [70], [71], [72],74,[76], [77], [78]] (Figure 3). The primary dietary assessment method used was food records (n = 31) [[31], [32], [33],[35], [36], [37], [38], [39], [40], [41],46,52,54,56,57,[60], [61], [62],[64], [65], [66], [67], [68], [69], [70], [71], [72],74,[76], [77], [78]] (Figure 3). Seventeen studies ensured weight maintenance by providing isocaloric diets or adjusting caloric intake periodically during the study in response to weight fluctuation [29,32,37,40,42,44,45,[47], [48], [49], [50],55,57,62,67,68,76].
FIGURE 3.
Dietary assessment methods and adherence monitoring according to the amount of food provided. FFQ, food frequency questionnaire.
Metabolites were identified from urine samples in 31 studies [30,[34], [35], [36], [37],[39], [40], [41], [42], [43],[45], [46], [47], [48],[50], [51], [52], [53],59,60,[62], [63], [64], [65],[67], [68], [69], [70], [71],73,75] and 30 studies quantified metabolites in plasma/serum [29,[31], [32], [33],[37], [38], [39],44,[46], [47], [48], [49],[52], [53], [54], [55], [56], [57], [58], [59], [60], [61],64,66,68,72,74,[76], [77], [78]] (Supplemental Table 3). The majority of studies used a variation of LC-MS (n = 31) [[29], [30], [31], [32], [33],[37], [38], [39],[41], [42], [43], [44],47,49,[51], [52], [53], [54],56,57,[60], [61], [62], [63],[65], [66], [67],71,73,74,76] and used untargeted metabolomics (n = 37) [29,30,[34], [35], [36],38,39,41,43,[45], [46], [47],[49], [50], [51],[53], [54], [55], [56], [57], [58], [59], [60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73],[76], [77], [78]] to identify metabolites in urine and blood biofluids (Supplemental Table 3). Of the studies that used a targeted metabolomics approach (n = 16), [[31], [32], [33],37,40,42,44,48,51,52,58,59,64,67,74,75], less than half reported that they used known standards (n = 7) [31,33,40,42,48,51,67].
Feeding protocol and dietary assessment methods
Of the included studies, 32 provided all or the majority (90%) of food to participants [[29], [30], [31],34,37,[39], [40], [41], [42],[44], [45], [46],48,[50], [51], [52], [53], [54], [55], [56],58,[62], [63], [64], [65], [66],[68], [69], [70], [71], [72],78], 16 provided some but not all food [33,35,36,38,43,47,49,57,[59], [60], [61],67,[73], [74], [75], [76]], and 2 were highly prescriptive but did not provide any food [32,77]. Refer to FIGURE 2, FIGURE 3.
Feeding protocols of studies who provided all food (n = 32)
Studies primarily used a crossover RCT design (n = 19, 54%) [[29], [30], [31],34,37,[39], [40], [41], [42],[44], [45], [46],48,[50], [51], [52], [53], [54], [55]]. The sample size was most frequently between 10 and 20 participants (n = 12 studies) [30,31,34,41,42,45,46,50,51,55,68,78]; however, some studies had larger sample sizes of >100 participants [40,54,56,62,65,66,71,72]. A third (n = 21, 66%) [29,30,39,40,46,[51], [52], [53], [54],56,58,[62], [63], [64], [65], [66],[68], [69], [70], [71], [72]] of studies included a run-in period of ∼2 wk to standardize intake or collect current dietary intake data before their intervention. Total study duration was highly variable and ranged from less than a week [42,63] to 32 wk [29]. However, the duration of feeding periods within interventions was much shorter, with almost a third of studies having a feeding period of 4 wk [29,31,44,48], 3 wk [45], 2 wk [30,39,41,46,50,64,68,69], or ≤1 wk [34,42,[51], [52], [53],63].
A third of the studies that provided all or the majority of food to participants tested only 2 dietary patterns (n = 21) [30,31,37,41,42,44,46,[52], [53], [54],56,[62], [63], [64], [65],[68], [69], [70], [71], [72],78].
Finally, the majority (n = 25, 78%) of studies that supplied food to participants provided it as preprepared meals or snacks [30,31,34,37,[39], [40], [41], [42],[44], [45], [46],48,50,[52], [53], [54], [55],58,62,63,66,[68], [69], [70],78]. Refer to FIGURE 2, FIGURE 3. Food records and recording of nonintervention foods consumed were the most frequently reported methods used to assess adherence to the dietary feeding pattern implemented, and food records were the most frequently used dietary assessment method to assess dietary intake during the intervention.
Feeding protocols of studies that provided some, but not all food (n = 16)
Sixteen studies provided at least some food to participants during the intervention feeding period, mostly these studies were parallel RCTs (n = 9, 56%) [57,[59], [60], [61],67,[73], [74], [75], [76]]. The sample size was highly variable between 10 [36] and 200 [60] participants. Half the studies used a run-in period of between 1 and 4 wk [47,57,59,60,67,73,74,76]. The total intervention period was highly variable from less than a week [35] to 3 y [75], with a mean intervention period of 25 wk.
The majority (n = 11) of studies tested 2 dietary patterns during the intervention [33,38,43,47,49,57,59,60,67,74,76]. Adherence to the intervention and assessment of dietary intakes were frequently assessed using a food record (n = 11, 69%) [33,35,36,38,57,59,60,61,67,74,76]. Three-quarters of studies (n = 12, 75%) provided food as raw foods or ingredients [33,43,49,57,59,60,61,67,[73], [74], [75], [76]].
Feeding protocols of studies that prescribed dietary intake (n = 2)
One crossover RCT [32] and one parallel RCT [77] were highly prescriptive, but did not provide any food. In another study, participants received individualized counseling with a dietitian to implement isocaloric diet plans, which included prescribed targets for macronutrient and fructose intakes, along with lists of allowed and prohibited foods [32]. This study had 12 participants and was conducted for a total of 4 wk, and each intervention arm was 1 wk, with no run-in period. It assessed 2 dietary patterns, specifically low- and high-fructose diets. The other study prescribed meal plans and provided guidance on how to implement the diet [77]. Macronutrient targets were provided to participants to align with either a low-carbohydrate, high-fat diet or a high-carbohydrate, low-fat diet and participants consumed dietary patterns for a total of 8 wk. Adherence and dietary assessment were assessed using food records in both studies [32,77].
Sample collection and processing
Urine (n = 31)
In total, 31 studies collected urine samples [30,[34], [35], [36], [37],[39], [40], [41], [42], [43],[45], [46], [47], [48],[50], [51], [52], [53],59,60,[62], [63], [64], [65],[67], [68], [69], [70], [71],73,75]. These were either 24-h samples (n = 19) [34,[39], [40], [41], [42],45,46,48,50,52,59,60,62,65,[68], [69], [70], [71],73], morning samples (n = 7) [30,[35], [36], [37],47,63,67], spot samples (n = 7) [34,47,[51], [52], [53],63,64], or 4-h samples (n = 1) [43].
Twenty-hour urine samples
Of the 19 studies that collected 24-h urine samples, 1 study reported that participants were fasted for 12 h of the 24-h collection period [34]. How often samples were collected varied between interventions, with samples primarily collected at the end of the intervention period(s) (n = 13) [34,40,41,48,50,52,60,65,68,69,70,71,73] and at baseline (n = 12) [40,46,48,52,59,60,65,[68], [69], [70], [71],73]. Few studies report the materials used to collect urine samples, these included sterile containers (n = 2) [69,70], a 24-h collection container (n = 1) [42], a 2-L container containing 2-g boric acid/L (n = 1) [45], or a plastic container (n = 1) [50]. Collection methods were highly variable with 5 studies reporting samples were stored in a cooler bag or cool conditions during collection [41,65,[69], [70], [71]], 2 measured urinary collection completeness using the recovery rate of para-aminobenzoic acid [69,70], and 1 reported recording total volume and measured pH [41].
Morning urine samples
The majority of morning urine sample collections were reported to be during a fasted period, with fasting times varying from a 10–12-h fast [36,63,67] to an “overnight fast” [35,37]. Samples were commonly collected at the end of the intervention period (n = 7) [30,[35], [36], [37],47,63,67]. No studies reported the materials used to collect samples, but 1 study reported that samples were collected mid-stream [35].
Spot urine samples
Spot samples were mostly collected during a fasted period, with fasting periods again ranging from 10–12-h [34,53,63] to an “overnight fast” [51]. Two studies did not report whether samples were collected during a fasting period or whether the participants were not fasting [47,52]. Samples were commonly collected at the end of the intervention period (n = 5) [47,52,53,63,64], and preintervention (n = 4) [47,53,64]. The materials used to collect were only mentioned in one study; however, details were nonspecific [51]. Alternative methods reported included that spot samples were creatinine corrected [63,64] or that samples were centrifuged before storage [53].
Four-hour urine sample
One study collected a 4-h, nonfasted urine sample [43]. These samples were collected at the start and end of each intervention period. The materials used to collect the samples were not reported.
One study did not report the type of urine sample collected, nor how it was collected, but did report that these samples were collected at baseline, 1 and 3 y [75].
Plasma (n = 25)
Twenty-five studies measured plasma metabolites [29,31,32,[37], [38], [39],44,46,49,[52], [53], [54], [55], [56], [57],[59], [60], [61],64,68,72,74,[76], [77], [78]]. Nineteen of these studies reported that the samples were collected in the morning (n = 19) [31,32,37,38,39,44,49,[53], [54], [55], [56], [57],59,68,72,74,[76], [77], [78]]. Fasting periods were variable between studies, with several studies not reporting this level of detail [29,46,49,61,64].
All studies collected plasma samples at the end of the intervention period(s) [29,31,32,[37], [38], [39],44,46,49,[52], [53], [54], [55], [56], [57],[59], [60], [61],64,68,72,74,[76], [77], [78]]. Several studies did not report what type of blood collection tubes were used to collect plasma samples. Of the few studies that did report the materials used to collect samples, tube types included EDTA tubes (n = 11) [29,32,39,44,49,53,61,64,72,74,76], sodium/lithium heparin (n = 2) [46,57], or a citrate tube (n = 1) [38].
Serum (n = 5)
Five studies analyzed serum metabolites [47,48,58,59,66]. All studies collected samples during a fasted period, of which 4 specified that they collected samples in the morning [47,48,58,59]. All studies collected serum samples at the end of the intervention period(s) and most (n = 3) also reported collecting samples at baseline as well [47,48,58,59]. One study reported EDTA tubes were used to collect whole blood samples but did not specify the materials used to collect serum and the remaining studies did not report the type of blood collection tubes used [47].
Laboratory method for the quantification of dietary metabolome
All samples
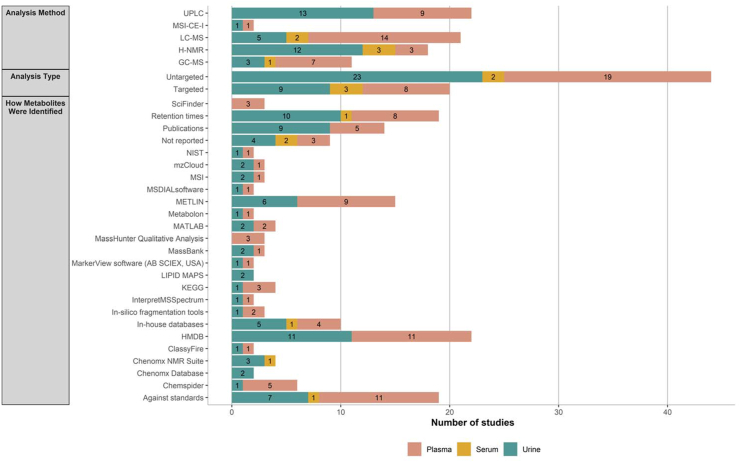
The most used methods to analyze all biofluids were ultrahigh-performance liquid chromatography (n = 18) [30,37,39,[42], [43], [44],51,[53], [54], [55], [56],[60], [61], [62], [63],65,67,71], followed by LC-MS (n = 16) [29,31,32,38,41,44,47,49,52,57,61,66,67,73,74,76], and then NMR (n = 14) [34,35,36,40,[45], [46], [47], [48],50,58,69,70,74,75]. Untargeted metabolomics was frequently used to characterize which metabolites were present in biosamples (n = 37, 73%) [29,30,[34], [35], [36],38,39,41,43,[45], [46], [47],[49], [50], [51],[53], [54], [55], [56], [57],[60], [61], [62], [63], [64], [65], [66], [67], [68], [69], [70], [71], [72], [73],[76], [77], [78]]. Refer to Figure 4.
FIGURE 4.
Databases and analysis used according to biofluid type. ACD, advanced chemistry development; CFM-ID, competitive fragmentation modeling for metabolite identification; FIA–MS, flow injection assay–mass spectroscopy; GC, gas chromatography; HMDB, Human Metabolome Database; H-NMR, proton nuclear magnetic resonance; LC, liquid chromatography; MS, mass spectroscopy; MSI–CE–MS, multi-segment injection–capillary electrophoresis–mass spectrometry; TOCSY, total correlation spectroscopy; UPLC, ultra-high-performance liquid chromatography; KEGG, Kyoto Encyclopedia of Genes and Genomes database; NIST, National Institute of Standards and Technology; MSI, Metabolomics Standard Initiative. ∗In silico fragmentation tools, such as CFM-ID and FingerID.
The most frequently used methods to determine metabolites were The HMDB, with a comparison of retention times against standards and against previous publications (Figure 4).
Urine
The metabolites present in urine were assayed most frequently by either NMR (n = 12) [[34], [35], [36],40,[45], [46], [47], [48],50,69,70,75] or ultrahigh-performance liquid chromatography (n = 13) [30,37,39,42,43,51,53,60,62,63,65,67,71] techniques. Three-quarters (n = 23) of studies analyzing urinary metabolites used an untargeted approach [30,[34], [35], [36],39,41,43,[45], [46], [47],50,51,53,60,[62], [63], [64], [65],[67], [68], [69], [70],73].
Plasma
Plasma metabolites were mostly analyzed using a variation of LC-MS methods (n = 18) [29,31,32,[37], [38], [39],44,49,[52], [53], [54], [55], [56], [57],60,61,74,76] and used an untargeted approach to characterize plasma metabolites (n = 19) [29,38,39,46,49,[53], [54], [55], [56], [57],60,61,64,68,71,72,[76], [77], [78]].
Serum
Serum metabolites were only explored in a handful of studies, these metabolites were identified using NMR (n = 3) [47,48,58], LC-MS (n = 2) [47,66] or GC-MS (n = 1) [66]. Both a targeted (n = 3) [48,58,59] and an untargeted (n = 2) [47,66] approach was used to analyze the serum metabolome.
Discussion
The current scoping review of feeding studies measuring the dietary metabolome included 50 human feeding studies published up until January 2023. These have been primarily parallel or crossover RCTs that examined associations between metabolites measured in common biofluids (i.e., plasma, serum, and/or urine) and dietary intake with all foods provided for intervention periods of ≥2 d, but up until 3 y. A similar number of studies used plasma and urinary samples to measure dietary metabolites. Most studies used untargeted metabolomic laboratory methods to characterize metabolites, an approach that has been used increasingly over the last 5–10 y [79]. Overall, methods used were highly variable between studies, including the type of dietary patterns tested, how biospecimen samples were collected, preanalytic procedures employed, and the analytic techniques used to measure the dietary metabolome. Considering this area of research is progressing relatively fast, the variation in methodologies between studies is not unexpected.
There were few dietary patterns used in the feeding interventions of the studies included in this review. The interest in whole dietary patterns over singular foods and/or nutrients has gained traction in recent years in recognition of the importance of the whole food matrix in terms of nutrient digestion and absorption and hence appearance as metabolites in biological fluids. Similarly, the emergence of newer technologies is able to capture a broad range of metabolites, whether already identified or yet to be, that appear in response to these dietary patterns. In addition, it is logical to choose to explore metabolomic responses in relation to key dietary patterns known to impact health outcomes, such as those identified in the current review. The 2 most evaluated dietary patterns were high-/low-glycemic index (GI) or glycemic load (GL) dietary patterns, and dietary patterns defined by a typical country’s usual intake pattern, typically characterized as an unhealthy dietary pattern compared with a “recommended” diet. A large body of research has demonstrated that low-GI/GL diets have benefits for health, including lowering glycosylated haemoglobin (HbA1c), BMI, plasma lipids and blood pressure [[80], [81], [82]]. Therefore, the identification of metabolites that align with a low-GI/GL dietary pattern is potentially an important area of research with application to use in chronic disease risk assessment and ongoing clinical management. Whereas dietary patterns defined by a country’s typical dietary patterns, usually accord with less healthy dietary intakes, given low rates of alignment with national dietary guideline recommendations [83]. Such dietary patterns are typically energy-dense and nutrient-poor, characterized by high intakes of sodium, added sugars, and saturated fat derived from high intakes of ultraprocessed foods. Identification of metabolites that distinguish unhealthy from healthy dietary patterns is equally important as findings could be used in the future to objectively identify dietary patterns associated with elevated risk of developing specific noncommunicable diseases and hence could potentially be evaluated for use in the context of both prevention and treatment. The emergence of dietary metabolome research may enable future implementation to objectively assess dietary patterns, food groups, or dietary components with greater accuracy, less error, and recall bias [2].
Variations were identified in how similar dietary patterns were defined between different studies, including the ways of reporting the composition of the diets. For example, between studies conducting a Mediterranean-based diet, there were differences in the percentage of energy from macronutrients (e.g., 19% compared with 40% for total fat) or how these patterns were presented (e.g., total gram per macronutrient provided compared with percentage energy for each macronutrient provided) [30,57,58]. In addition, some studies only reported diets in terms of their macronutrient composition, whereas others provided key details on other food, and nutrient targets or provided examples of the meal plans used [30,31,46,75]. It was expected that there would be differences in definitions of the dietary patterns tested, particularly as these may evolve over time or differ between countries. For example, a “typical country” intake was defined by the country conducting the research, such as an “Average Danish” diet compared with a “Typical United States” diet [62,65]. However, to advance this area of research, consistency in the level of detail provided between studies is recommended, sufficient to allow replication, and/or adaptation to regional food supplies if international. Where whole food supply for a particular dietary pattern is provided to participants, it is important to report the types of foods provided, not only macronutrient distributions as this will aid in the interpretation of metabolite findings and help explain differences identified [84,85].
Dietary intake and adherence to feeding interventions were most frequently assessed using food records. This dietary assessment method was consistently used in all feeding protocols. Food records are completed prospectively and therefore can be a useful tool when collecting dietary intake data to identify adherence, particularly when all foods have been provided [5]. Although there is the risk of reactivity bias occurring with food records [3], such that individuals change their intake during the feeding interventions, the provision of food may reduce this and facilitate adherence. Differences in study methods and how foods were provided were apparent between studies that provided different amounts of food or proportions of total daily dietary needs. In studies that provided all foods, the majority used a randomized crossover intervention and provided foods mostly as premade meals and snacks. In contrast, interventions that provided some foods only were mostly parallel RCTs and provided foods as raw ingredients and typically had longer intervention periods. Potential reasons for these differences could be related to minimizing participant burden and preferences, funding limitations and time constraints. When planning future feeding studies, researchers need to consider the strengths and limitations of different study designs and dietary methodologies while balancing this against the feasibility and practicalities of implementing the protocol. However, the use of varying methodologies is necessary to identify a range of valid and reliable dietary metabolites and to advance this area of research, with data ideally contributed to national and international repositories [86].
This review identified that LC-MS is currently the most frequently implemented analytic method used to measure dietary metabolites within feeding studies. Previous studies have reported that the use of MS methods has been more frequent than NMR technologies, with even fewer studies using a combination of MS and NMR technologies [87]. Characteristics inherent to different metabolomic techniques have been reported and potentially influence metabolome detection. For instance, LC-MS and GC-MS are highly sensitive methods that can detect and quantify metabolites present in very small amounts [19,[88], [89], [90]]. LC-MS is best suited for polar and nonpolar metabolites, whereas GC-MS is ideal for volatile and thermally stable metabolites [4,90]. However, these techniques require skilled personnel and complex sample preparation [19,88,89]. On the other hand, NMR spectroscopy is a nondestructive technique that identifies metabolites based on their chemical structure, providing both qualitative and quantitative information, and requires minimal sample preparation [4,19,88,89]. However, NMR is less sensitive than LC-MS and GC-MS and requires larger sample amounts [19,88,89]. Metabolomics approaches can be broadly classified as targeted or untargeted [4]. Targeted metabolomics deals with the identification of selected metabolites via cross-comparison with known standards, whereas untargeted metabolomics mainly focuses on the discovery of novel, yet unknown compounds [4,90]. A study included in this review utilized untargeted metabolomics of urine samples to classify 2 dietary patterns—the New Nordic Diet (NND) and the Average Danish Diet (ADD)—with a low misclassification error rate (19%), demonstrating the effectiveness of untargeted metabolomics as a potential tool to estimate compliance to a specific dietary pattern [65]. Another study employed 1H NMR to analyze the urinary metabolome of 142 centrally obese Danes, revealing unique metabolite markers reflecting changes in protein and carbohydrate metabolism when comparing the same dietary patterns, NND and ADD [71]. For instance, glycine betaine, glucose, trimethylamine N-oxide, and creatinine were increased in the urine of individuals after the NND compared with those after the ADD diet [71]. Although the choice of technique depends on the research question and metabolites being studied, the large diversity of metabolite structures makes it challenging to use only 1 or 2 analytic techniques and the sequential coupling of different techniques has been proven beneficial in improving metabolite identification [79,91]. However, it is acknowledged that this is not always feasible to use multiple techniques. Previous research on metabolites associated with specific foods, food groups, and dietary patterns could be leveraged to aid in planning. A recent critical review by Rafiq et al. [79], for example, evaluated interstudy repeatability by considering the number, study design and consistency of findings and identified 69 metabolites as good candidate biomarkers of food intake.
However, methods describing biosample collection were often very poorly described. For example, less than half of the studies reported the materials used to collect urine [42,45,50,69,70], plasma [29,32,38,39,44,46,49,57,61,64,72,74,76], and/or serum [47] samples. Reporting of plasma samples was more detailed in comparison to urine and serum sample methods. Prior recommendations for the identification of biomarkers of food intake have highlighted the need to consider how sample collection and handling may affect the stability of dietary compounds [91] with preanalytic error having a pronounced effect on sample quality and consequently the blood metabolome identification [92]. For instance, the presence of additives in the sample collection tubes may influence the accuracy of the results. For example, although EDTA and heparin are commonly used, the use of Li+-heparin is not recommended due to the potential for matrix effects caused by increased signals of plastic polymers [92]. Similarly, certain biomarkers of food intake require specific collection conditions for stabilization, such as the use of special tubes for ascorbate and glucose, and urine collection at a pH level of <2 for anthocyanins [91]. Prompt (within 30 min) separation of blood cells from plasma by centrifugation at 4°C is ideal, but storage of whole blood before centrifugation for a maximum of 4 h in an iced water bath is acceptable to minimize the metabolic activity of cells/enzymes and keep metabolite pattern stable [92]. Adjustments to metabolite concentrations may be recommended for spot samples based on creatinine levels [93]. For 24-h urine samples, the inclusion of para-aminobenzoic acid supplementation as a marker for completeness of excretion and kidney function can enhance the precision and reliability of results [94]. In addition, for studies that reported a fasting period for participants, the duration of the fasting period was variable. This could influence potential metabolites identified due to the variable half-lives of different compounds. This is an important detail to consider when trying to understand the degree to which a biomarker reflects intake (exposure), with some metabolites more indicative of immediate rather than habitual or long-term intake [79,91]. Accurate and detailed reporting of methods, including fasting periods and collection materials and procedures used, is important to help improve the standardization, reproducibility, and validity of findings within and between laboratories.
Strengths and limitations
There are several strengths and limitations to the current research review. Strengths include the up-to-date, detailed description and summary of methods used in feeding studies exploring the dietary metabolome in humans. Although previous reviews have focused on the identification of biomarkers [[14], [15], [16]], they have not synthesized methodologies used to characterize them. Summation of the methods used is important to highlight gaps in research and reporting, to inform future clinical methodology design in the area of feeding studies and the dietary metabolome, and to improve research and reproducibility of future research studies. An additional strength is the objectivity of metabolomic data in this area of nutrition research, which currently relies heavily on self-reported methods to quantify dietary exposures and to evaluate diet–disease relationships. Advances in this area are important to enhance understanding of what people eat, individual variability in terms of food metabolism, variability in response to nutrition interventions, and how this impacts health outcomes. The limitations identified are that the majority of included studies are secondary analyses of stored samples. For example, 1 study used samples that were >20 y old [62]. This could impact the identification of metabolites, as there is uncertainty regarding how metabolites decompose with time during lengthy storage periods [91]. Approximately half the studies in the current review did not provide detailed methods on how biosamples were collected, which again may impact how stable metabolites are during storage and limits understanding of optimal sample collection and handling procedures in future studies.
Recommendations for future studies
Future studies should consider providing detailed descriptions of methodologies, including information on sample collection materials and procedures, as variations may influence the identification of metabolites. Considering this is still an emerging field of research with varying study designs, dietary interventions and methodological approaches will inform the progression of this field of research. Future studies should consider the strengths and limitations of the different methodological approaches, including study design, types of dietary interventions, method of administrating dietary interventions, and analytic techniques when designing future interventions and tailor the approach to the specific study objectives. Collaborations and data sharing are important to allow for greater insights into the identification of the dietary metabolome; this will help advancement in this field [86]. Working to identify and validate candidate markers of dietary components as well as whole dietary patterns is necessary and the use of standardized methods, such as the criteria outlined by Dragsted et al. [91], is recommended. Future research should explore associations between metabolites and categories of specific foods, food groups, or dietary patterns as outlined in a prior critical review [79] to further establish potential candidate biomarkers.
The current review calls for formal reporting guidelines, similar to STROBE-nut, are necessary for controlled human feeding studies examining the metabolome as various methodologies and analytic approaches are currently being used. To facilitate manuscript preparation, peer review, and comparative analysis of results across different studies, it is recommended that a formal consensus is reached for reporting guidelines. This will guide clinical protocol design and create a standard for the minimum detail required in reporting methods. Until such a consensus is reached, a list of suggested questions to consider when reporting diet and methodologies used in controlled human feeding studies examining the metabolome to help guide future research (Supplemental Table 4) has been compiled. Two areas requiring emphasis for improvement in the reporting of methods for controlled human feeding studies examining the metabolome are summarized below:
-
1.
To improve comparability and reproducibility, studies should describe the dietary interventions being tested in conjunction with a detailed description of the specific foods and food groups included or restricted as part of the specific dietary pattern, and this should also include the meal plans, including details of specific food/beverage portion sizes and frequency of inclusion/consumption.
-
2.
When investigating the effects of diet in human metabolomic studies, strategies to control for interindividual variability are essential. One such strategy is to use crossover study designs, which can help to reduce interindividual variability as participants serve as their own controls, accounting for confounders such as age, sex, genetics, microbiome, and lifestyle variables. However, logistic and resource constraints may limit this strategy in studies of longer duration. In such cases, researchers can use statistical methods to adjust for individual differences and measure metabolomic data at multiple time points throughout the study. To mitigate interindividual variability and reduce the confounding effects of habitual dietary intake, a controlled diet run-in period can be employed in the study design. This involves providing participants with standardized meals for 1–3 d at baseline or during washout periods, using precise meal plans, portion sizes, or a set of nutrient compositions to establish a common baseline. A similar approach could be used in parallel study designs, where standardized test meals or foods are provided at multiple timepoints across the study. In addition to reducing interindividual variability, providing standardized meals has the added advantage of addressing recall or self-report bias when reporting dietary intake because the diet has been supplied.
Conclusion
Understanding the dietary metabolome is important for providing a greater understanding of diet–disease relationships, as well as the progression of the personalized nutrition field. The current review identified that feeding studies using urine and/or plasma biofluids for the identification of metabolites are occurring and that this is evolving. It was identified that the methods used to date were highly variable, including in the dietary patterns explored, sample collection and analysis techniques. This highlights the need for national and international discussions to identify research priorities to progress this area of research more efficiently.
Acknowledgments
We would like to thank research librarian Nicole Brown for support with the search strategy and Dr Rachael Taylor for assistance with the title and abstract screening.
Author contributions
The authors’ responsibilities were as follows – EDC, JJAF, CEC: designed and conducted the research, wrote the manuscript, and had primary responsibility for final content; JS: contributed to data analysis; EDC, JJAF, CEC, JS: contributed to data collection and reviewed and edited the manuscript; and all authors: read and approved the final manuscript.
Conflict of interest
JJAF also works part-time for Sanitarium the Health Food Company; this company had no input into the current study. All other authors declare no conflicts of interest.
Funding
CEC is supported by a National Health and Medical Research Council (NHMRC) Investigator Grant.
Data availability
The data used in this review are available from the corresponding author upon reasonable request.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.advnut.2023.08.010.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Guertin K.A., Moore S.C., Sampson J.N., Huang W.Y., Xiao Q., Stolzenberg-Solomon R.Z., et al. Metabolomics in nutritional epidemiology: identifying metabolites associated with diet and quantifying their potential to uncover diet-disease relations in populations. Am. J. Clin. Nutr. 2014;100(1):208–217. doi: 10.3945/ajcn.113.078758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O'Gorman A., Brennan L. The role of metabolomics in determination of new dietary biomarkers. Proc. Nutr. Soc. 2017;76(3):295–302. doi: 10.1017/S0029665116002974. [DOI] [PubMed] [Google Scholar]
- 3.Kirkpatrick S.I., Baranowski T., Subar A.F., Tooze J.A., Frongillo E.A. Best practices for conducting and interpreting studies to validate self-report dietary assessment methods. J. Acad. Nutr. Diet. 2019;119(11):1801–1816. doi: 10.1016/j.jand.2019.06.010. [DOI] [PubMed] [Google Scholar]
- 4.Guasch-Ferré M., Bhupathiraju S.N., Hu F.B. Use of metabolomics in improving assessment of dietary intake. Clin. Chem. 2018;64(1):82–98. doi: 10.1373/clinchem.2017.272344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Subar A.F., Freedman L.S., Tooze J.A., Kirkpatrick S.I., Boushey C., Neuhouser M.L., et al. Addressing current criticism regarding the value of self-report dietary data. J. Nutr. 2015;145(12):2639–2645. doi: 10.3945/jn.115.219634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brennan L., Hu F.B. Metabolomics-based dietary biomarkers in nutritional epidemiology-current status and future opportunities. Mol. Nutr. Food Res. 2019;63(1) doi: 10.1002/mnfr.201701064. [DOI] [PubMed] [Google Scholar]
- 7.Hu F.B. Dietary pattern analysis: a new direction in nutritional epidemiology. Curr. Opin. Lipidol. 2002;13(1):3–9. doi: 10.1097/00041433-200202000-00002. [DOI] [PubMed] [Google Scholar]
- 8.Food and Agriculture Organization of the United Nations . 2022. Food-based Dietary Guidelines — Denmark.https://www.fao.org/nutrition/education/food-dietary-guidelines/regions/countries/denmark/en/ [Internet] [updated 2020; cited 03/09/2023]. Available from: [Google Scholar]
- 9.U.S. Department of Agriculture and U.S. Department of Health and Human Services . 2020. Dietary Guidelines for Americans, 2020–2025.DietaryGuidelines.gov [Internet] updated December 2020; cited 03/09/2023]. Available from: [Google Scholar]
- 10.Government of Canda . 2022. Canada’s Dietary Guidelines.https://food-guide.canada.ca/en/guidelines/section-1-foundation-healthy-eating/ [Internet] [updated June 10, 2022]. Available from: [Google Scholar]
- 11.Sanches Machado d'Almeida K., Ronchi Spillere S., Zuchinali P., Corrêa Souza G. Mediterranean diet and other dietary patterns in primary prevention of heart failure and changes in cardiac function markers: a systematic review. Nutrients. 2018;10(1):58. doi: 10.3390/nu10010058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim H., Caulfield L.E., Garcia-Larsen V., Steffen L.M., Coresh J., Rebholz C.M. Plant-based diets are associated with a lower risk of incident cardiovascular disease, cardiovascular disease mortality, and all-cause mortality in a general population of middle-aged adults. J. Am. Heart Assoc. 2019;8(16) doi: 10.1161/JAHA.119.012865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schulz C.A., Oluwagbemigun K., Nöthlings U. Advances in dietary pattern analysis in nutritional epidemiology. Eur. J. Nutr. 2021;60(8):4115–4130. doi: 10.1007/s00394-021-02545-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Clarke E.D., Rollo M.E., Pezdirc K., Collins C.E., Haslam R.L. Urinary biomarkers of dietary intake: a review. Nutr. Rev. 2020;78(5):364–381. doi: 10.1093/nutrit/nuz048. [DOI] [PubMed] [Google Scholar]
- 15.Liang S., Nasir R.F., Bell-Anderson K.S., Toniutti C.A., O’Leary F.M., Skilton M.R. Biomarkers of dietary patterns: a systematic review of randomized controlled trials. Nutr. Rev. 2022;80(8):1856–1895. doi: 10.1093/nutrit/nuac009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andraos S., Beck K.L., Jones M.B., Han T.-L., Conlon C.A., de Seymour J.V. Characterizing patterns of dietary exposure using metabolomic profiles of human biospecimens: a systematic review. Nutr. Rev. 2022;80(4):699–708. doi: 10.1093/nutrit/nuab103. [DOI] [PubMed] [Google Scholar]
- 17.Tzoulaki I., Ebbels T.M., Valdes A., Elliott P., Ioannidis J.P. Design and analysis of metabolomics studies in epidemiologic research: a primer on -omic technologies. Am. J. Epidemiol. 2014;180(2):129–139. doi: 10.1093/aje/kwu143. [DOI] [PubMed] [Google Scholar]
- 18.Jones D.P., Park Y., Ziegler T.R. Nutritional metabolomics: progress in addressing complexity in diet and health. Annu. Rev. Nutr. 2012;32:183–202. doi: 10.1146/annurev-nutr-072610-145159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang X.W., Li Q.H., Xu Z.D., Dou J.J. Mass spectrometry-based metabolomics in health and medical science: a systematic review. RSC Adv. 2020;10(6):3092–3104. doi: 10.1039/c9ra08985c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sumner L.W., Amberg A., Barrett D., Beale M.H., Beger R., Daykin C.A., et al. Proposed minimum reporting standards for chemical analysis Chemical Analysis Working Group (CAWG) Metabolomics Standards Initiative (MSI) Metabolomics. 2007;3(3):211–221. doi: 10.1007/s11306-007-0082-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wishart D.S., Tzur D., Knox C., Eisner R., Guo A.C., Young N., et al. HMDB: the Human Metabolome Database. Nucleic Acids Res. 2007;35(Database issue):D521–D526. doi: 10.1093/nar/gkl923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guijas C., Montenegro-Burke J.R., Domingo-Almenara X., Palermo A., Warth B., Hermann G., et al. METLIN: a technology platform for identifying knowns and unknowns. Anal. Chem. 2018;90(5):3156–3164. doi: 10.1021/acs.analchem.7b04424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Meshref S., Li Y., Feng Y.L. Prediction of liquid chromatographic retention time using quantitative structure-retention relationships to assist non-targeted identification of unknown metabolites of phthalates in human urine with high-resolution mass spectrometry. J. Chromatogr. A. 2020;1634:461691. doi: 10.1016/j.chroma.2020.461691. [DOI] [PubMed] [Google Scholar]
- 24.Stanstrup J., Neumann S., Vrhovšek U. PredRet: prediction of retention time by direct mapping between multiple chromatographic systems. Anal. Chem. 2015;87(18):9421–9428. doi: 10.1021/acs.analchem.5b02287. [DOI] [PubMed] [Google Scholar]
- 25.Ahmed S., de la Parra J., Elouafi I., German B., Jarvis A., Lal V., et al. Foodomics: a data-driven approach to revolutionize nutrition and sustainable diets. Front. Nutr. 2022;9:874312. doi: 10.3389/fnut.2022.874312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tricco A.C., Lillie E., Zarin W., O'Brien K.K., Colquhoun H., Levac D., et al. PRISMA Extension for Scoping Reviews (PRISMA-ScR): checklist and explanation. Ann. Intern. Med. 2018;169(7):467–473. doi: 10.7326/M18-0850. [DOI] [PubMed] [Google Scholar]
- 27.Arksey H., O'Malley L. Scoping studies: towards a methodological framework. Int. J. Soc. Res. Methodol. 2005;8(1):19–32. [Google Scholar]
- 28.Levac D., Colquhoun H., O'Brien K.K. Scoping studies: advancing the methodology. Implement. Sci. 2010;5:69. doi: 10.1186/1748-5908-5-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.T. Esko, J.N. Hirschhorn, H.A. Feldman, Y.-H.H. Hsu, A.A. Deik, C.B. Clish, et al., Metabolomic profiles as reliable biomarkers of dietary composition, Am. J. Clin. Nutr. 105 (3) (2107) 547–554. [DOI] [PMC free article] [PubMed]
- 30.Barber C., Mego M., Bendezu R.A., Masihy M., Guarner F., Azpiroz F., et al. Differential effects of western and Mediterranean-type diets on gut microbiota: a metagenomics and metabolomics approach. Nutrients. 2021;13(8):2638. doi: 10.3390/nu13082638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barton S., Navarro S.L., Buas M.F., Schwarz Y., Gu H., Djukovic D., et al. Targeted plasma metabolome response to variations in dietary glycemic load in a randomized, controlled, crossover feeding trial in healthy adults. Food Funct. 2015;6(9):2949–2956. doi: 10.1039/c5fo00287g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez-Granda A., Damms-Machado A., Basrai M., Bischoff S.C. Changes in plasma acylcarnitine and lysophosphatidylcholine levels following a high-fructose diet: a targeted metabolomics study in healthy women. Nutrients. 2018;10(9):1254. doi: 10.3390/nu10091254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galié S., García-Gavilán J., Papandreou C., Camacho-Barcía L., Arcelin P., Palau-Galindo A., et al. Effects of Mediterranean diet on plasma metabolites and their relationship with insulin resistance and gut microbiota composition in a crossover randomized clinical trial. Clin. Nutr. 2021;40(6):3798–3806. doi: 10.1016/j.clnu.2021.04.028. [DOI] [PubMed] [Google Scholar]
- 34.Garcia-Perez I., Posma J.M., Gibson R., Chambers E.S., Hansen T.H., Vestergaard H., et al. Objective assessment of dietary patterns by use of metabolic phenotyping: a randomised, controlled, crossover trial. Lancet Diabetes Endocrinol. 2017;5(3):184–195. doi: 10.1016/S2213-8587(16)30419-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walsh M.C., Brennan L., Pujos-Guillot E., Sébédio J., Scalbert A., Fagan A., et al. Influence of acute phytochemical intake on human urinary metabolomic profiles. Am. J. Clin. Nutr. 2007;86(6):1687–1693. doi: 10.1093/ajcn/86.5.1687. [DOI] [PubMed] [Google Scholar]
- 36.González-Guardia L., Yubero-Serrano E.M., Delgado-Lista J., Perez-Martinez P., Garcia-Rios A., Marin C., et al. Effects of the Mediterranean diet supplemented with coenzyme Q10 on metabolomic profiles in elderly men and women. J. Gerontol. A Biol. Sci. Med. Sci. 2015;70(1):78–84. doi: 10.1093/gerona/glu098. [DOI] [PubMed] [Google Scholar]
- 37.Guglielmetti S., Bernardi S., Del Bo C., Cherubini A., Porrini M., Gargari G., et al. Effect of a polyphenol-rich dietary pattern on intestinal permeability and gut and blood microbiomics in older subjects: study protocol of the MaPLE randomised controlled trial. BMC Geriatr. 2020;20(1):77. doi: 10.1186/s12877-020-1472-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johansson-Persson A., Barri T., Ulmius M., Önning G., Dragsted L.O. LC-QTOF/MS metabolomic profiles in human plasma after a 5-week high dietary fiber intake. Anal. Bioanal. Chem. 2013;405(14):4799–4809. doi: 10.1007/s00216-013-6874-5. [DOI] [PubMed] [Google Scholar]
- 39.Khodorova N.V., Rietman A., Rutledge D.N., Schwarz J., Piedcoq J., Pilard S., et al. Urinary medium-chained acyl-carnitines sign high caloric intake whereas short-chained acyl-carnitines sign high -protein diet within a high-fat, hypercaloric diet in a randomized crossover design dietary trial. Nutrients. 2021;13(4):1191. doi: 10.3390/nu13041191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Loo R.L., Zou X., Appel L.J., Nicholson J.K., Holmes E. Characterization of metabolic responses to healthy diets and association with blood pressure: application to the Optimal Macronutrient Intake Trial for Heart Health (OmniHeart), a randomized controlled study. Am. J. Clin. Nutr. 2018;107(3):323–334. doi: 10.1093/ajcn/nqx072. [DOI] [PubMed] [Google Scholar]
- 41.May D.H., Navarro S.L., Ruczinski I., Jason H., Ogata Y., Schwarz Y., et al. Metabolomic profiling of urine: response to a randomised, controlled feeding study of select fruits and vegetables, and application to an observational study. Br. J. Nutr. 2013;110(10):1760–1770. doi: 10.1017/S000711451300127X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McNairn M., Brito A., Dillard K., Heath H., Pantaleon M., Fanter R., et al. Postprandial dried blood spot-based nutritional metabolomic analysis discriminates a high-fat, high-protein meat-based diet from a high carbohydrate vegan diet: a randomized controlled crossover trial. J. Acad. Nutr. Diet. 2021;121(5):931–941.e2. doi: 10.1016/j.jand.2020.10.024. [DOI] [PubMed] [Google Scholar]
- 43.Roager H.M., Vogt J.K., Kristensen M., Hansen L.B.S., Ibrugger S., Maerkedahl R.B., et al. Whole grain-rich diet reduces body weight and systemic low-grade inflammation without inducing major changes of the gut microbiome: a randomised cross-over trial. Gut. 2019;68:83–93. doi: 10.1136/gutjnl-2017-314786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Navarro S.L., Tarkhan A., Shojaie A., Randolph T.W., Gu H., Djukovic D., et al. Plasma metabolomics profiles suggest beneficial effects of a low-glycemic load dietary pattern on inflammation and energy metabolism. Am. J. Clin. Nutr. 2019;110(4):984–992. doi: 10.1093/ajcn/nqz169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stella C., Beckwith-Hall B., Cloarec O., Holmes E., Lindon J.C., Powell J., et al. Susceptibility of human metabolic phenotypes to dietary modulation. J. Proteome Res. 2006;5(10):2780–2788. doi: 10.1021/pr060265y. [DOI] [PubMed] [Google Scholar]
- 46.Ross A.B., Pere-Trepat E., Montoliu I., Martin F.P.J., Collino S., Moco S., et al. A whole-grain-rich diet reduces urinary excretion of markers of protein catabolism and gut microbiota metabolism in healthy men after one week. J. Nutr. 2013;143(6):766–773. doi: 10.3945/jn.112.172197. [DOI] [PubMed] [Google Scholar]
- 47.Schmedes M., Aadland E.K., Sundekilde U.K., Jacques H., Lavigne C., Graff I.E., et al. Lean-seafood intake decreases urinary markers of mitochondrial lipid and energy metabolism in healthy subjects: metabolomics results from a randomized crossover intervention study. Mol. Nutr. Food. Res. 2016;60(7):1661–1672. doi: 10.1002/mnfr.201500785. [DOI] [PubMed] [Google Scholar]
- 48.Shin J.H., Jung S., Kim S.A., Kang M.S., Kim M.S., Joung H., et al. Differential effects of typical Korean versus American-style diets on gut microbial composition and metabolic profile in healthy overweight Koreans: a randomized crossover trial. Nutrients. 2019;11(10):2450. doi: 10.3390/nu11102450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tovar J., de Mello V.D., Nilsson A., Johansson M., Paananen J., Lehtonen M., et al. Reduction in cardiometabolic risk factors by a multifunctional diet is mediated via several branches of metabolism as evidenced by nontargeted metabolite profiling approach. Mol. Nutr. Food Res. 2017;61(2) doi: 10.1002/mnfr.201600552. [DOI] [PubMed] [Google Scholar]
- 50.Zheng H., Yde C.C., Clausen M.R., Kristensen M., Lorenzen J., Astrup A., et al. Metabolomics investigation to shed light on cheese as a possible piece in the French paradox puzzle. J. Agric. Food Chem. 2015;63(10):2830–2839. doi: 10.1021/jf505878a. [DOI] [PubMed] [Google Scholar]
- 51.Lloyd A.J., Willis N.D., Wilson T., Zubair H., Chambers E., Garcia-Perez I., et al. Addressing the pitfalls when designing intervention studies to discover and validate biomarkers of habitual dietary intake. Metabolomics. 2019;15(5):72. doi: 10.1007/s11306-019-1532-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Guthrie L., Spencer S.P., Perelman D., Van Treuren W., Han S., Yu F.B., et al. Impact of a 7-day homogeneous diet on interpersonal variation in human gut microbiomes and metabolomes. Cell Host Microbe. 2022;30(6):863–874.e4. doi: 10.1016/j.chom.2022.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li J., George Markowitz R.H., Brooks A.W., Mallott E.K., Leigh B.A., Olszewski T., et al. Individuality and ethnicity eclipse a short-term dietary intervention in shaping microbiomes and viromes. PLOS Biol. 2022;20(8) doi: 10.1371/journal.pbio.3001758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim H., Lichtenstein A.H., White K., Wong K.E., Miller E.R., Coresh J., et al. Plasma metabolites associated with a protein-rich dietary pattern: results from the OmniHeart trial. Mol. Nutr. Food Res. 2022;66(6) doi: 10.1002/mnfr.202100890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Huang N.K., Matthan N.R., Matuszek G., Lichtenstein A.H. Plasma metabolite response to simple, refined and unrefined carbohydrate-enriched diets in older adults-randomized controlled crossover trial. Metabolites. 2022;12(6):547. doi: 10.3390/metabo12060547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Acar E., Gürdeniz G., Khakimov B., Savorani F., Korndal S.K., Larsen T.M., et al. Biomarkers of individual foods, and separation of diets using untargeted LC–MS-based plasma metabolomics in a randomized controlled trial. Mol. Nutr. Food Res. 2019;63(1) doi: 10.1002/mnfr.201800215. [DOI] [PubMed] [Google Scholar]
- 57.Bergia R.E., Biskup I., Giacco R., Costabile G., Gray S., Wright A., et al. The MEDGICarb-Study: design of a multi-center randomized controlled trial to determine the differential health-promoting effects of low- and high-glycemic index Mediterranean-style eating patterns. Contemp. Clin. Trials Commun. 2020;19:100640. doi: 10.1016/j.conctc.2020.100640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Burton-Pimentel K.J., Pimentel G., Hughes M., Michielsen C.C., Fatima A., Vionnet N., et al. Discriminating dietary responses by combining transcriptomics and metabolomics data in nutrition intervention studies. Mol. Nutr. Food Res. 2021;65(4) doi: 10.1002/mnfr.202000647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dawczynski C., Cullen P.M., Schlattmann P., Lorkowski S. A study protocol of a randomized trial evaluating the effect of using defined menu plans within an intensive personal nutritional counseling program on cardiovascular risk factors: the MoKaRi (modulation of cardiovascular risk factors) trial. Contemp. Clin. Trials Commun. 2021;22:100761. doi: 10.1016/j.conctc.2021.100761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gürdeniz G., Uusitupa M., Hermansen K., Savolainen M.J., Schwab U., Kolehmainen M., et al. Analysis of the SYSDIET Healthy Nordic Diet randomized trial based on metabolic profiling reveal beneficial effects on glucose metabolism and blood lipids. Clin. Nutr. 2022;41(2):441–451. doi: 10.1016/j.clnu.2021.12.031. [DOI] [PubMed] [Google Scholar]
- 61.Hanhineva K., Lankinen M.A., Padret A., Schwab U., Kolehmainen M., Paananen J., et al. Nontargeted metabolite profiling discriminates diet-specific biomarkers for consumption of whole grains, fatty fish, and bilberries in a randomized controlled trial. J. Nutr. 2015;145(1):7–17. doi: 10.3945/jn.114.196840. [DOI] [PubMed] [Google Scholar]
- 62.Kim H., Lichtenstein A.H., Wong K.E., Appel L.J., Coresh J., Rebholz C.M. Urine metabolites associated with the Dietary Approaches to Stop Hypertension (DASH) diet: results from the DASH-Sodium trial. Mol. Nutr. Food Res. 2021;65(3) doi: 10.1002/mnfr.202000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu X., Yin P., Shao Y., Wang Z., Wang B., Lehmann R., et al. Which is the urine sample material of choice for metabolomics-driven biomarker studies? Anal. Chim. Acta. 2020;1105:120–127. doi: 10.1016/j.aca.2020.01.028. [DOI] [PubMed] [Google Scholar]
- 64.Wellington N., Shanmuganathan M., de Souza R.J., Zulyniak M.A., Azab S., Bloomfield J., et al. Metabolic trajectories following contrasting prudent and western diets from food provisions: identifying robust biomarkers of short-term changes in habitual diet. Nutrients. 2019;11(10):2407. doi: 10.3390/nu11102407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Andersen M.-B.S., Rinnan A., Manach C., Poulsen S.K., Pujos-Guillot E., Larsen T.M., et al. Untargeted metabolomics as a screening tool for estimating compliance to a dietary pattern. J. Proteome Res. 2014;13(3):1405–1418. doi: 10.1021/pr400964s. [DOI] [PubMed] [Google Scholar]
- 66.Rebholz C.M., Lichtenstein A.H., Zheng Z., Appe L.J., Coresh J. Serum untargeted metabolomic profile of the Dietary Approaches to Stop Hypertension (DASH) dietary pattern. Am. J. Clin. Nutr. 2018;108(2):243–255. doi: 10.1093/ajcn/nqy099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Meslier V., Laiola M., Roager H.M., De Filippis F., Roume H., Quinquis B., et al. Mediterranean diet intervention in overweight and obese subjects lowers plasma cholesterol and causes changes in the gut microbiome and metabolome independently of energy intake. Gut. 2020;69(7):1258–1268. doi: 10.1136/gutjnl-2019-320438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pourafshar S., Nicchitta M., Tyson C.C., Svetkey L.P., Corcoran D., Bain J., et al. Urine and plasma metabolome of healthy adults consuming the Dietary Approaches to Stop Hypertension diet: a pilot study. Circulation. 2020;141(Suppl 1) doi: 10.1161/circ.141.suppl_1.MP43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rasmussen L.G., Winning H., Savorani F., Toft H., Larsen T.M., Dragsted L.O., et al. Assessment of the effect of high or low protein diet on the human urine metabolome as measured by NMR. Nutrients. 2012;4(2):112–131. doi: 10.3390/nu4020112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rasmussen L.G., Winning H., Savorani F., Ritz C., Engelsen S.B., Astrup A., et al. Assessment of dietary exposure related to dietary GI and fibre intake in a nutritional metabolomic study of human urine. Genes Nutr. 2012;7(2):281–293. doi: 10.1007/s12263-011-0250-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Trimigno A., Khakimov B., Savorani F., Poulsen S.K., Astrup A., Dragsted L.O., et al. Human urine 1H NMR metabolomics reveals alterations of the protein and carbohydrate metabolism when comparing habitual Average Danish diet vs. healthy New Nordic diet. Nutrition. 2020;79–80:110867. doi: 10.1016/j.nut.2020.110867. [DOI] [PubMed] [Google Scholar]
- 72.Khakimov B., Poulsen S.K., Savorani F., Acar E., Gürdeniz G., Larsen T.M., et al. New Nordic diet versus average Danish diet: a randomized controlled trial revealed healthy long-term effects of the new Nordic diet by GC-MS blood plasma metabolomics. J. Proteome Res. 2016;15(6):1939–1954. doi: 10.1021/acs.jproteome.6b00109. [DOI] [PubMed] [Google Scholar]
- 73.Ulaszewska M.M., Trost K., Stanstrup J., Tuohy K.M., Franceschi P., Chong M.F.F., et al. Urinary metabolomic profiling to identify biomarkers of a flavonoid-rich and flavonoid-poor fruits and vegetables diet in adults: the FLAVURS trial. Metabolomics. 2016;12(2):1–22. [Google Scholar]
- 74.Ulven S.M., Christensen J.J., Nygård O., Svardal A., Leder L., Ottestad I., et al. Using metabolic profiling and gene expression analyses to explore molecular effects of replacing saturated fat with polyunsaturated fat—a randomized controlled dietary intervention study. Am. J. Clin. Nutr. 2019;109(5):1239–1250. doi: 10.1093/ajcn/nqy356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Vazquez-Fresno R., Llorach R., Urpi-Sarda M., Lupianez-Barbero A., Lupianez-Barbero A., Estruch R., Corella D., et al. Metabolomic pattern analysis after Mediterranean diet intervention in a nondiabetic population: a 1- and 3-year follow-up in the PREDIMED study. J. Proteome Res. 2015;14(1):531–540. doi: 10.1021/pr5007894. [DOI] [PubMed] [Google Scholar]
- 76.Vitale M., Hanhineva K., Koistinen V., Auriola S., Paananen J., Costabile G., et al. Putative metabolites involved in the beneficial effects of wholegrain cereal: nontargeted metabolite profiling approach. Nutr. Metab. Cardiovasc. Dis. 2021;31(4):1156–1165. doi: 10.1016/j.numecd.2020.12.022. [DOI] [PubMed] [Google Scholar]
- 77.McCullough D., Harrison T., Boddy L.M., Enright K.J., Amirabdollahian F., Schmidt M.A., et al. The effect of dietary carbohydrate and fat manipulation on the metabolome and markers of glucose and insulin metabolism: a randomised parallel trial. Nutrients. 2022;14(18):3691. doi: 10.3390/nu14183691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Chorell E., Ryberg M., Larsson C., Sandberg S., Mellberg C., Lindahl B., et al. Plasma metabolomic response to postmenopausal weight loss induced by different diets. Metabolomics. 2016;12(5):85. [Google Scholar]
- 79.Rafiq T., Azab S.M., Teo K.K., Thabane L., Anand S.S., Morrison K.M., et al. Nutritional metabolomics and the classification of dietary biomarker candidates: a critical review. Adv. Nutr. 2021;12(6):2333–2357. doi: 10.1093/advances/nmab054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Evans C.E., Greenwood D.C., Threapleton D.E., Gale C.P., Cleghorn C.L., Burley V.J. Glycemic index, glycemic load, and blood pressure: a systematic review and meta-analysis of randomized controlled trials. Am. J. Clin. Nutr. 2017;105(5):1176–1190. doi: 10.3945/ajcn.116.143685. [DOI] [PubMed] [Google Scholar]
- 81.Chiavaroli L., Lee D., Ahmed A., Cheung A., Khan T.A., Blanco S., et al. Effect of low glycaemic index or load dietary patterns on glycaemic control and cardiometabolic risk factors in diabetes: systematic review and meta-analysis of randomised controlled trials. BMJ. 2021;374:n1651. doi: 10.1136/bmj.n1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chiavaroli L., Kendall C.W.C., Braunstein C.R., Blanco Mejia S., Leiter L.A., Jenkins D.J.A., et al. Effect of pasta in the context of low-glycaemic index dietary patterns on body weight and markers of adiposity: a systematic review and meta-analysis of randomised controlled trials in adults. BMJ (Open) 2018;8(3) doi: 10.1136/bmjopen-2017-019438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Leme A.C.B., Hou S., Fisberg R.M., Fisberg M., Haines J. Adherence to food-based dietary guidelines: a systemic review of high-income and low- and middle-income countries. Nutrients. 2021;13(3):1038. doi: 10.3390/nu13031038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Alemán-Jiménez C., Domínguez-Perles R., Medina S., Prgomet I., López-González I., Simonelli-Muñoz A., et al. Pharmacokinetics and bioavailability of hydroxytyrosol are dependent on the food matrix in humans. Eur. J. Nutr. 2021;60(2):905–915. doi: 10.1007/s00394-020-02295-0. [DOI] [PubMed] [Google Scholar]
- 85.Di Lorenzo C., Colombo F., Biella S., Stockley C., Restani P. Polyphenols and human health: the role of bioavailability. Nutrients. 2021;13(1):273. doi: 10.3390/nu13010273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Scalbert A., Brennan L., Manach C., Andres-Lacueva C., Dragsted L.O., Draper J., et al. The food metabolome: a window over dietary exposure. Am. J. Clin. Nutr. 2014;99(6):1286–1308. doi: 10.3945/ajcn.113.076133. [DOI] [PubMed] [Google Scholar]
- 87.Bhinderwala F., Wase N., DiRusso C., Powers R. Combining mass spectrometry and NMR improves metabolite detection and annotation. J. Proteome Res. 2018;17(11):4017–4022. doi: 10.1021/acs.jproteome.8b00567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Schrimpe-Rutledge A.C., Codreanu S.G., Sherrod S.D., McLean J.A. Untargeted metabolomics strategies-challenges and emerging directions. J. Am. Soc. Mass Spectrom. 2016;27(12):1897–1905. doi: 10.1007/s13361-016-1469-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Wishart D.S. Metabolomics: applications to food science and nutrition research. Trends Food Sci. Technol. 2008;19(9):482–493. [Google Scholar]
- 90.Naureen Z., Cristoni S., Donato K., Medori M.C., Samaja M., Herbst K.L., et al. Metabolomics application for the design of an optimal diet. J. Prev. Med. Hyg. 2022;63(2):E142–E149. doi: 10.15167/2421-4248/jpmh2022.63.2S3.2755. Suppl 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Dragsted L.O., Gao Q., Scalbert A., Vergères G., Kolehmainen M., Manach C., et al. Validation of biomarkers of food intake-critical assessment of candidate biomarkers. Genes Nutr. 2018;13:14. doi: 10.1186/s12263-018-0603-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Yin P., Lehmann R., Xu G. Effects of pre-analytical processes on blood samples used in metabolomics studies. Anal. Bioanal. Chem. 2015;407(17):4879–4892. doi: 10.1007/s00216-015-8565-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Heavner D.L., Morgan W.T., Sears S.B., Richardson J.D., Byrd G.D., Ogden M.W. Effect of creatinine and specific gravity normalization techniques on xenobiotic biomarkers in smokers’ spot and 24-h urines. J. Pharm. Biomed. Anal. 2006;40(4):928–942. doi: 10.1016/j.jpba.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 94.John K.A., Cogswell M.E., Campbell N.R., Nowson C.A., Legetic B., Hennis A.J., et al. Accuracy and usefulness of select methods for assessing complete collection of 24-Hour hour urine: a systematic review. J. Clin. Hypertens. (Greenwich). 2016;18(5):456–467. doi: 10.1111/jch.12763. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used in this review are available from the corresponding author upon reasonable request.