Abstract
An ammonium transporter of Azospirillum brasilense was characterized. In contrast to most previously reported putative prokaryotic NH4+ transporter genes, A. brasilense amtB is not part of an operon with glnB or glnZ which, in A. brasilense, encode nitrogen regulatory proteins PII and PZ, respectively. Sequence analysis predicts the presence of 12 transmembrane domains in the deduced AmtB protein and classifies AmtB as an integral membrane protein. Nitrogen regulates the transcription of the amtB gene in A. brasilense by the Ntr system. amtB is the first gene identified in A. brasilense whose expression is regulated by NtrC. The observation that ammonium uptake is still possible in mutants lacking the AmtB protein suggests the presence of a second NH4+ transport mechanism. Growth of amtB mutants at low ammonium concentrations is reduced compared to that of the wild type. This suggests that AmtB has a role in scavenging ammonium at low concentrations.
Azospirillum species are nitrogen-fixing organisms (diazotrophs), capable of forming an associative relationship with the roots of several economically important cereals (68). Many studies have indicated that Azospirillum promotes plant growth, but the exact mechanism of growth promotion has not been fully characterized. Like most organisms, Azospirillum uses ammonium salts as a preferred nitrogen source (53). In the absence of combined nitrogen and under microaerobiosis conditions, the nitrogenase enzyme complex is synthesized and converts atmospheric N2 to NH4+.
Unprotonated NH3 is predicted to diffuse out of bacterial cells due to a concentration gradient across the plasma membrane (36). The pH gradient (generally slightly more alkaline inside the bacteria) enhances this process. Therefore, an active ammonium uptake system is required to retain the intracellular fixed nitrogen, acquired at high energy cost by the nitrogenase. Hartmann and Kleiner (24) have shown that ammonium uptake in Azospirillum spp. is energy dependent, follows the Michaelis-Menten kinetics, and is repressed by ammonium. No functional characterization of genetic components of this system has yet been reported.
Recently, genes encoding ammonium transporter proteins and putative ammonium transporter proteins have been reported for Saccharomyces cerevisiae (43), Arabidopsis thaliana (52), and Lycopersicon esculentum (tomato) (39). In Bacillus subtilis the nrgA gene, whose corresponding amino acid sequence is homologous to those of NH4+ transporter proteins, is part of the dicistronic nrgAB operon (76). nrgB possibly encodes a nitrogen regulatory protein homologous to PII proteins, but the biochemical functions of the nrgA and nrgB gene products in B. subtilis have not been reported. The nrgAB operon is highly expressed during nitrogen-limited growth.
More recently, Siewe et al. (61) characterized the first reported prokaryotic NH4+ transporter gene (amt) in Corynebacterium glutamicum. However, in 1986, Jayakumar et al. (28) had already reported on an amtA gene in Escherichia coli which complemented a mutant with less than 10% of the parental CH3NH3+ uptake activity. The complete amtA sequence was published by Fabiny et al. (19). An analysis of the deduced amino acid sequence of the product of amtA, AmtA, predicted that the protein was a cytoplasmic component of an ammonium transport system. In 1992, however, Neuwald et al. (51) reported that the amtA gene corresponded to cysQ, a gene needed for cysteine synthesis in E. coli. Later on, Van Heeswijk et al. (70) isolated an amtB gene in E. coli K-12. The amtB gene product is homologous to transmembrane NH4+ transporters, but its functional characterization has not yet been reported. The E. coli amtB gene is cotranscribed with glnK, located upstream of amtB. glnK encodes a second PII-like protein (6, 70). The glnK gene product and the glnB gene product (PII) are known to play a role in the reversible adenylylation of glutamine synthetase (GS) in response to the nitrogen status of the cells. In addition, PII stimulates the kinase-phosphatase enzyme, NtrB, to dephosphorylate the phosphorylated transcriptional activator NtrC. Phosphorylated NtrC is necessary to activate transcription from several RpoN-dependent promoters (reviewed in reference 65; 41, 46).
Two PII homologs have been identified in Azospirillum brasilense (14, 15). glnB is part of the nitrogen-regulated, but NtrC-independent, glnBA operon, and its product is required for nitrogen fixation. In contrast to what is found for other species, PII (glnB gene product) is not involved in the ammonium control of GS activity by adenylylation. The level of glnA expression is, however, lower in glnB mutant strains than in the wild-type strain (14, 15). The second PII-like protein of A. brasilense, PZ, is not functionally equivalent to PII. glnB-null mutants exhibit a Nif− phenotype that is not complemented by structural gene glnZ. The two-component regulatory system NtrB-NtrC and the ς54 factor (RpoN) in A. brasilense have been characterized (40, 48). NtrC is involved in nitrate utilization (40) and (methyl)ammonium uptake (69). Notably, the rpoN mutant has a pleiotropic effect: nitrogen fixation, nitrate assimilation, ammonium uptake, and flagellar biosynthesis are impaired (48).
We report here the isolation and characterization of a nitrogen-regulated (methyl)ammonium transporter gene from A. brasilense.
MATERIALS AND METHODS
Bacterial strains, plasmids, media, and growth conditions.
The E. coli and A. brasilense strains used are listed in Table 1. Plasmids mentioned in the text are also described in this table. Sequencing constructs and intermediate constructs are not given. A genomic library of A. brasilense Sp7 was constructed by ligation of fragments generated by partial EcoRI digestion of total DNA into cosmid pLAFR1. These constructs were packed into phage particles, transferred to E. coli HB101, and selected for isolation of tetracycline-resistant colonies. E. coli strains were grown in Luria-Bertani (LB) medium (57) at 37°C. A. brasilense was grown in LB medium supplemented with 2.5 mM CaCl2 and 2.5 mM MgSO4 (LB* medium) at 30°C. For solid media, 15 g of agar per liter was added. Conjugal transfers of recombinant plasmids, derived either from pLAFR1 or pLAFR3, from E. coli to A. brasilense were performed on D plates (containing 8 g of Bacto nutrient broth [Difco], 0.25 g of MgSO4 · 7H2O, 1.0 g of KCl, and 0.01 g of MnCl2 per liter). After conjugation, MMAB minimal medium (71) with 0.5% malate as the C source was used for selection of A. brasilense transconjugants. MMAB medium was also used in growth experiments and in [14C]methylammonium uptake, nitrogenase activity, and β-glucuronidase assays. Growth rates in liquid minimal medium supplemented with 20 mM NH4+, 2 mM NH4+, 8 mM nitrate, or 10 mM aspartate as the nitrogen source were measured by monitoring the optical density at 595 nm (OD595). Solid medium was used for growth experiments involving low NH4+ concentrations (0.7 to 0.1 mM). For RNA work, minimal K medium (18) supplemented with sodium lactate (5 g per liter) was used.
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant properties | Source or reference |
|---|---|---|
| A. brasilense | ||
| Sp7 (ATCC29145) | Wild-type strain, isolated from Digitaria decumbens rhizosphere soil, Brazil | 66 |
| FAJ301 | rpoN::kan Kmr | 48 |
| FAJ310 | amtB::kan Kmr | This work |
| 7148 | ntrC::Tn5-148 Kmr | 40 |
| 7194 | ntrB::Tn5-194 Kmr | 40 |
| 7606 | glnB::kan Kmr | 15 |
| E. coli | ||
| DH5 α | hsdR17 endA1 thi-1 gyrA96 relA1 recA1 supE44 ΔlacU169 (φ80lacZΔM15) | Gibco BRL |
| HB101 | recA hsdR hsdM pro leu lacZ supE F− Strr | 5 |
| Plasmids | ||
| pUC18 | Cloning vector; ColE1 replicon, Apr | 77 |
| pUC18-2 | pUC18 with 1.8-kb Kmr cassette from Tn5, Apr Kmr | 13 |
| pSUP202 | Mobilizable plasmid, suicide vector for A. brasilense; ColE1 replicon, Cmr Tcr Apr | 62 |
| pRK2013 | Tra+ helper plasmid | 20 |
| pKW117 | pUC8 + gusA + trpA terminator, Apr | 74 |
| pLAFR1 | IncP1 broad-host-range cosmid | 21 |
| pLAFR3 | pLAFR1 derivative containing the pUC8 multiple cloning site | 64 |
| pFAJ302 | pLAFR3 containing an amtB::gusA translational fusion | This work |
| pFAJ304 | pUC19 containing an amtB::gusA translational fusion | This work |
| pFAJ308 | pUC18 containing the A. brasilense amtB gene as a 1.9-kb EcoRI-NarI fragment cloned in EcoRI-AccI | This work |
| pFAJ309 | pLAFR3 containing the A. brasilense amtB gene as a 1.9-kb EcoRI-HindIII fragment from pFAJ308, amtB+ | This work |
| pFAJ312 | pUC19 containing the 1.6-kb EcoRI-SalI fragment of amtB | This work |
| pFAJ314 | pUC18 containing the 14-kb EcoRI fragment with the A. brasilense amtB gene | This work |
DNA methods.
Standard methods, as described by Sambrook et al. (57), were used for plasmid isolation, chromosomal DNA preparation, restriction analysis, ligations, transformations, Southern blotting, and hybridization. DNA fragments were recovered from agarose gels with the Nucleotrap kit (Macherey-Nagel Filter Service, Eupen, Belgium). For Southern hybridization, DNA was transferred to a Hybond-N membrane (Amersham, Ghent, Belgium). Hybridization was done overnight at 68°C with a DNA probe labeled with digoxigenin-dUTP by a random-primed labeling kit (Boehringer Mannheim, Brussels, Belgium). The signal was detected with a chemiluminescence detection kit (Boehringer Mannheim).
PCR.
To identify A. brasilense genes encoding NH4+ transporter proteins, degenerate PCR primers were designed based on conserved amino acid sequences of reported NH4+ transporter homologs (43, 45, 52, 76). Two of them (Fig. 1) (5′-GTGAATTCGGNGCNTTYGCNGARCGNATG-3′ and 5′-TCGAATTCRTTRAANCCRAACCANCCRAACCA-3′) yielded a single amplification product of the expected size (344 bases), which was cloned and sequenced (data not shown). The deduced amino acid sequence corresponding to this amplification product showed significant homology with those of reported NH4+ transporter proteins.
FIG. 1.
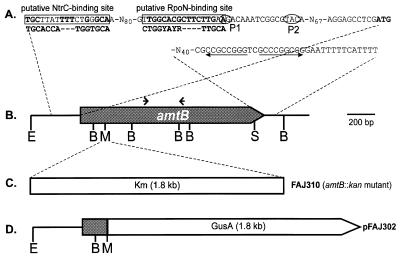
Schematic representation of the A. brasilense Sp7 amtB gene and constructs used. (A) Promoter and terminator regions. A putative NtrC-binding sequence and a RpoN-binding sequence are boxed, and the corresponding consensus sequences (3, 41) are given below. The two transcription start sites, P1 and P2, are circled. The start codon (ATG) is in boldface. Arrows indicate the dyad symmetry of a putative Rho-independent terminator. (B) Physical map of the amtB gene. Arrows indicate the locations of the PCR primers used to amplify a 344-bp internal sequence. E, EcoRI; B, BglI; M, SmaI; S, SalI. (C) Insertion site of the kanamycin resistance cartridge used to construct an amtB::kan mutant (FAJ310). (D) Construction of the amtB::gusA fusion (pFAJ302) in broad-host-range vector pLAFR3. Abbreviations are as defined for panel B.
An EcoRI recognition site had been added to the 5′ end of each primer to facilitate the cloning of the amplified PCR product into a pUC18 vector. PCRs were performed in a thermocycler (TRIO-Thermoblock; Biometra, Göttingen, Germany) in a reaction volume of 25 μl containing 0.5 U of Taq DNA polymerase (Boehringer Mannheim) and each of the primers at a 1 μM concentration. A PCR cycle consisting of 1 min of denaturation at 94°C, 1 min of primer annealing at 50°C, and 1 min of primer extension at 72°C was applied 30 times. These 30 cycles were preceded by 5 min of denaturation (94°C) and were followed by 7 min of primer extension (72°C).
DNA sequencing and analysis.
The chain-terminating dideoxynucleotide triphosphate method (58) was performed by using an automated sequencer (ALF; Pharmacia Biotech Benelux, Roosendaal, The Netherlands) with the Autoread sequencing kit (Pharmacia Biotech Benelux). The PALIGN, CLUSTAL, PROSITE, SOAP, NOVOTNY, BETATURN, HELIXMEM, and PSIGNAL programs of the P.C. Gene software package (Intelligenetics) were used to process and analyze the sequencing data. Potential coding regions were identified with the GCWIND program (60). The Blast program (1) was used to search for related sequences.
RNA preparation and Northern blot analysis.
RNA was prepared from wild-type and glnB::kan mutant cells grown under nitrogen fixation conditions (0.5% O2) in minimal medium with 10 mM sodium aspartate or 20 mM NH4Cl as the nitrogen source to an OD600 of 0.5. Total RNA was extracted with hot phenol according to the method of Gubler and Hennecke (23). Fifteen micrograms of RNA was separated on a 1.2% formaldehyde-agarose gel and transferred to a Hybond-N membrane (Amersham). The 1.6-kb EcoRI-SalI DNA fragment containing the first 1,260 bp of amtB was, after purification from plasmid pFAJ312, labeled with [α-32P]dCTP by using a random primer labeling kit (Amersham). Hybridization with the radiolabeled probe was performed for 6 h at 68°C in the presence of Rapid-hyb buffer from Amersham.
Primer extension.
Primer extension was performed as described by Ausubel et al. (2) with two primers labeled with [γ-32P]dATP: AMTB-15 (5′-CAGACCGGCCAAGCCCAGAATCGCCGCCAT-3′) and AMTB-17 (5′-GGCGGCGCTTTCCTGGGCGAGGGCGGC-3′). Ten picomoles of each primer added to 20 μg of total RNA was heated in the hybridization buffer with 80% formamide for 10 min at 85°C, and the mixture was subsequently incubated overnight at 30°C. The primer extension was performed for 90 min at 42°C with 50 U of avian myeloblastosis virus reverse transcriptase (Boehringer Mannheim). The extension products were run on a sequencing gel adjacent to the DNA sequence obtained by using the same oligonucleotide as the primer.
Construction of an amtB::gusA fusion and β-glucuronidase assay.
The 1.8-kb SmaI-HindIII cassette from pKW117 (74) containing the gusA gene was ligated behind the 525-bp EcoRI-SmaI fragment (Fig. 1) containing the 5′ region of the amtB gene. This was achieved by replacing the 1.1-kb SmaI-HindIII fragment of pFAJ312 with the 1.8-kb SmaI-HindIII cassette from pKW117, yielding plasmid pFAJ304. The construction was tested with the 22-bp oligonucleotide complementary to the gusA coding strand (29). The 2.3-kb EcoRI-HindIII fragment from pFAJ304, containing the amtB promoter region fused to the promoterless gusA gene, was cloned in broad-host-range vector pLAFR3 yielding plasmid pFAJ302.
β-Glucuronidase activity was measured as described by Vande Broek et al. (67). Cells were grown overnight in minimal MMAB medium containing the indicated nitrogen source. Nitrogen-fixing conditions were created in airtight 80-ml tubes with 3 ml of nitrogen-free MMAB medium by replacing the 77-ml air volume with N2 plus 0.5% O2 or in semisolid (0.07% agarose) nitrogen-free MMAB medium. Units of activity were calculated as described by Miller (49).
Uptake of [14C]methylammonium.
Cells were pregrown in minimal MMAB medium with 10 mM aspartate as the nitrogen source. When the cultures had reached a concentration of approximately 109 cells per ml they were centrifuged and resuspended in a double volume of the same medium without a nitrogen source. After 15 min of incubation at room temperature, [14C]methylammonium (Amersham; 2.11 GBq/mmol) was added to a final concentration of 8.75 μM. Samples (100 μl) were filtered through a MultiScreen Durapore (type DV; 0.65-μm-pore-size) filtration plate, which was placed on a multiscreen filtration manifold. Filters were dried, and radioactivity was measured with a liquid scintillation counter (Wallac 1410; Pharmacia Biotech Benelux). Protein concentrations were determined by the bicinchoninic acid assay (63) after lysis of the cells in 1 N NaOH at 65°C.
Uptake of NH4+.
Cells were pregrown as described for the [14C]methylammonium uptake assay. Cell suspensions were centrifuged, washed once with the assay buffer containing 10 mM MgSO4, 20 mM HEPES (pH 7) and 5 g of malate per liter, and resuspended in the same buffer. The original culture was concentrated 10 times in the assay buffer. After 15 min of incubation at room temperature, ammonium was added to a final concentration of 0.1 mM. The NH4+ concentration was measured with a selective ammonium electrode (F2322NH4 AMMONIUM SELECTRODE; Radiometer, Copenhagen, Denmark). Values were stored every 30 s. The detection limit of the electrode was 10 μM; this value was reached after approximately 25 min.
Ammonium excretion.
Ammonium excretion was measured in liquid MMAB medium containing 8 mM KNO3 as the nitrogen source. At different growth stages, the presence of ammonium in the cell supernatants was assayed by the indophenol blue method as described by Chaney and Marbach (9).
Construction of an amtB::kan insertion mutant.
A 1.8-kb BamHI fragment containing the kanamycin resistance cassette from pUC18-2 was made blunt with T4 DNA polymerase and was ligated into the SmaI site of the 1.6-kb EcoRI-SalI fragment (Fig. 1) containing the amtB gene. This 1.6-kb EcoRI-SalI fragment had previously been ligated into suicide vector pSUP202 as an EcoRI-PstI fragment. The resulting plasmid was conjugated to A. brasilense Sp7, with helper plasmid pRK2013 providing the tra genes. Transconjugants were selected on the basis of Kmr (for the presence of the cassette) and Tcs (loss of the pSUP202 plasmid). Two PCR primers designed to amplify a 627-bp internal part of the kan gene were used to confirm the presence of the Kmr cassette, and the insertion of the Kmr cassette in the amtB gene was also checked by hybridization of mutant and wild-type total DNA with the 1.6-kb EcoRI-SalI DNA fragment containing the first 1,260 bp of amtB (data not shown).
Nitrogenase assay.
Strains were pregrown overnight in rich LB* medium. After being washed, cell suspensions were brought to equal OD595 values and 25 μl of these exponential-phase cells was inoculated into 5 ml of semisolid nitrogen-free MMAB medium (0.07% agarose). After overnight incubation at 30°C, acetylene was added (10% of the air volume), and ethylene production was measured after 5, 9, 12, and 25 h with a Hewlett-Packard 5890A gas chromatograph with a PLOT (porous layer open tubular) fused silica column. Propane was used as the internal standard.
Nucleotide sequence accession number.
Sequence data has been submitted to the DDBJ/EMBL/GenBank database under accession no. AF005275.
RESULTS
Cloning of an A. brasilense Sp7 gene encoding a putative NH4+ transporter.
Degenerate PCR primers were used to amplify a 344-bp internal fragment of the putative NH4+ transporter gene (see Materials and Methods). This amplification product was used to screen a pLAFR1-derived genomic library of A. brasilense Sp7, made by cloning partial EcoRI-digested genomic DNA (see Materials and Methods). The 14-kb EcoRI fragment of a positively hybridizing cosmid clone was subcloned as a 1.6-kb EcoRI-SalI fragment, and sequence analysis identified the first 1,249 bp of a putative A. brasilense NH4+ transporter gene. Cloning the original 14-kb EcoRI fragment in vector pUC18 (pFAJ314) permitted the sequencing of the remainder of the gene by using primer walking from the 3′-terminal part of the known sequence. Figure 1 shows the organization of the sequenced DNA region.
The G+C content of the entire open reading frame (ORF) is 67.2%. This is consistent with the high G+C content of A. brasilense DNA. The G+C content in the third position of the codons is 91.9%. The potential ATG start codon is preceded by a putative ribosome-binding site. Immediately downstream of the ORF there is a G+C-rich sequence with interrupted dyad symmetry (ΔG[25°] = −21.6 kcal) followed by a T-rich region. This suggests the presence of a Rho-independent transcription terminator (56) (Fig. 1).
DNA sequence analysis did not reveal other ORFs in the 330-bp region immediately downstream of the putative NH4+ transporter gene or in the 350 bp preceding the ORF.
Analysis of the deduced amino acid sequence.
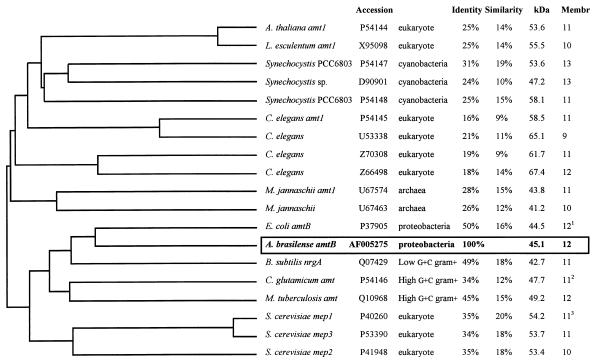
A comparison of the deduced amino acid sequence of the product of the A. brasilense amtB gene with those of reported putative NH4+ transporter proteins revealed that the sequence of this protein showed the highest level of similarity (approximately 50%) to the deduced amino acid sequences of the products of the E. coli amtB gene and the B. subtilis nrgA gene (data not shown). In Fig. 2 a phylogenetic tree built by multiple sequence alignment of reported putative NH4+ transporter proteins is shown. Besides the proteins included in this figure, proteins belonging to the Mep/Amt family (44) have been reported for Mycobacterium leprae (accession no. L78818), Rhodobacter capsulatus (accession no., X12359), and Methanobacterium thermautotrophicum (accession no. AE000846). However, no NH4+ transporter homolog was found by examining the complete genomic sequences of Haemophilus influenzae and Mycobacterium genitalium, two bacteria whose natural environment is human tissues (44).
FIG. 2.
Dendrogram of the multiple sequence alignment of putative NH4+ transporter proteins according to the methods of Higgins and Sharp (25, 26). Genome sequencing of the archaebacterium Methanococcus jannaschii (7), the cyanobacterium Synechocystis (30, 31), and the nematode Caenorhabditis elegans (75) predicted several NH4+ transporter-coding genes. Percentages of similarity and identity to the A. brasilense protein sequence were calculated by the method of Myers and Miller (50). Predicted molecular masses are given in kilodaltons. The Membr column gives the predicted numbers of membrane-associated helices. Unless cited in the literature, these numbers were calculated by the method of Eisenberg et al. (17). Superscripts 1, 2, and 3 (Membr column) indicate that the data came from references 70, 61, and 43, respectively.
Analysis of the promoter region and the transcription of the amtB gene.
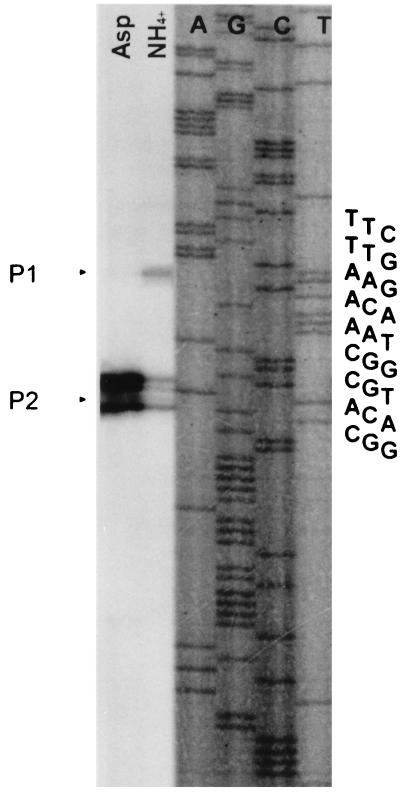
To localize the transcription start site of amtB, primer extension analysis was performed. RNA was isolated from wild-type (Sp7) cells grown in the presence of different nitrogen sources. The complementary sequence of nucleotides between positions 379 and 408 was used as a primer (AMTB-15). Figure 3 shows that the P2 transcription start site, located 80 to 82 nucleotides upstream of the translation start site of amtB, is preferentially used when 10 mM aspartate is the nitrogen source. In conditions of ammonium excess, a weak but significant signal was obtained at the P2 transcription start site. Another weak signal also appeared on the autoradiogram at the P1 transcription start site (94 to 95 nucleotides upstream of the translational start site of amtB). Under conditions of nitrogen fixation the transcription of amtB starts at the P2 transcription start site, but less efficiently than in the presence of aspartate (data not shown). Using another oligonucleotide (AMTB-17; see Materials and Methods) allowed the detection of the same transcription start sites (P1 and P2) (data not shown). The results obtained under conditions of nitrogen fixation or with 10 mM aspartate as the nitrogen source were the same for a glnB-null mutant strain (7606) (data not shown).
FIG. 3.
Primer extension analysis of the amtB promoter region to identify the transcription initiation sites under different nitrogen conditions. RNA was prepared from wild-type Sp7 grown on minimal medium with 10 mM aspartate (Asp) or 20 mM NH4+ (NH4+) as the nitrogen source. The DNA sequence in the transcription start region is indicated next to the sequencing gel. Two transcription start sites, P1 and P2, are indicated by arrowheads, and the corresponding sequences are shown circled in Fig. 1.
An examination of the DNA region upstream from these transcription start sites reveals a putative RpoN-binding consensus sequence at positions −26 to −14 relative to the P2 transcription start site (Fig. 1). This sequence differs by two nucleotides from consensus sequence CTGGYAYR-N4-TTGCA (four positions previously defined as invariable are underlined) (3) for RpoN-dependent promoters. The occurrence of A instead of C at position −14 is also found in the Rhizobium leguminosarum nifH promoter sequence (55) and in the R. leguminosarum biovar phaseoli nifH1, nifH2, and nifH3 promoter regions (47). A partially conserved NtrC-binding consensus sequence is present at positions −128 through −112 (relative to the P2 transcription start site; the consensus sequence is TGCACCA-N3-TGGTGCA) (41) (Fig. 1).
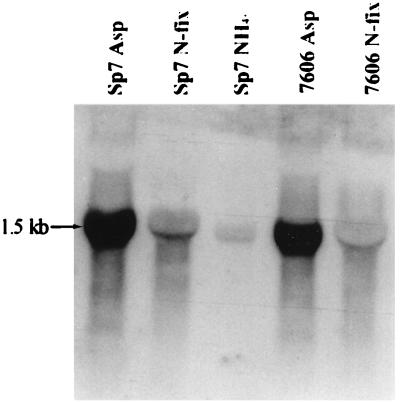
Northern blot analyses were performed with RNA extracted from wild-type Sp7 and glnB::kan mutant (7606) cells grown under different nitrogen conditions. Figure 4 shows the hybridization of total RNA with an amtB probe. A single transcript of 1.5 kb is detected under all physiological conditions, indicating that amtB is transcribed as a monocistronic unit. This is consistent with the absence of ORFs in the vicinity of amtB and the presence of a putative downstream Rho-independent terminator close to the 3′ end of amtB (Fig. 1).
FIG. 4.
Northern blot analysis of total RNA hybridized with the 1.6-kb EcoRI-SalI amtB probe. Total RNA was prepared from Sp7 (wild-type) and 7606 (glnB::kan mutant) cells grown in minimal medium with 10 mM aspartate (Asp) or 20 mM ammonium (NH4+) as the nitrogen source or grown under nitrogen-fixing conditions (N-fix).
As was also observed in the primer extension experiment, the level of the amtB mRNA signal is high in cells grown in aspartate-containing minimal medium, reduced in cells grown under conditions of nitrogen fixation, and very low in cells grown in the presence of 20 mM NH4+. This is in agreement with negative regulation by ammonium.
Northern blot analysis of a glnB::kan mutant strain shows that the absence of PII does not affect the transcription of the amtB gene under conditions of nitrogen fixation or when 10 mM aspartate is the nitrogen source. Thus, in contrast to what is found for the regulation of nif gene expression (14), the transduction of the nitrogen signal for amtB gene expression does not require PII under the conditions tested.
Expression of an amtB-gusA fusion in wild-type and regulatory mutant strains.
To investigate the regulation of amtB transcription in A. brasilense, an amtB::gusA translational fusion was constructed in vector pLAFR3, resulting in pFAJ302 (Fig. 1D). As NH4+ transport is reported to be regulated by the Ntr system in E. coli (27, 59) and Klebsiella pneumoniae (35), the pFAJ302 vector was conjugated into wild-type A. brasilense and three different A. brasilense ntr mutants: 7194 (ntrB::Tn5-194), 7148 (ntrC::Tn5-148), and FAJ301 (rpoN::kan). Expression of the amtB::gusA fusion is maximal in the presence of 10 mM aspartate or 2 mM NH4+ and is reduced eightfold in the presence of 20 mM NH4+ (Table 2), in agreement with the results of the Northern blot analysis. This confirms that the nitrogen status of the cell influences amtB transcription. amtB::gusA expression levels were significantly lowered in the ntr mutants under all physiological conditions tested. This suggests that the Ntr system is involved in the nitrogen regulation of amtB transcription. The slightly higher amtB::gusA expression levels observed in the ntr mutants grown on 20 mM NH4+ could be explained by assuming that in these circumstances the first promoter, located upstream of the P2 transcription start site (Fig. 1 and 3), participates in the transcription of amtB.
TABLE 2.
Expression of the amtB::gusA fusion in different genetic backgrounds
| Strainb | β-Glucuronidase activity (U)a with indicated N source
|
|||
|---|---|---|---|---|
| 10 mM Asp | 2 mM NH4+ | 20 mM NH4+ | N2 | |
| FAJ301 | 3 | 2 | 9 | 0 |
| 7194 | 2 | 5 | 8 | 2 |
| 7148 | 1 | 4 | 6 | 0 |
| Sp7 | 123 | 95 | 16 | 75 |
β-Glucuronidase activity was measured after overnight incubation in minimal medium containing one of the four nitrogen sources. Units of activity were calculated as described by Miller (49). The values shown are the means of at least three independent replicates.
For a description of the strains, see Table 1.
Ammonium uptake in ntr mutants.
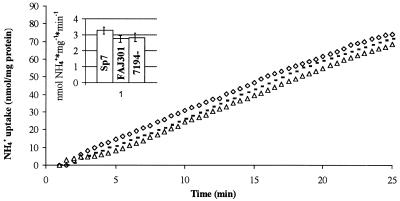
In line with the nearly absent expression of the amtB-gusA fusion in ntr mutants, these mutants do not show any [14C]methylammonium uptake (48). In order to measure the transport of the natural substrate for the AmtB transporter protein, a selective ammonium electrode was used to measure the uptake of NH4+ added to a cell suspension of wild-type Sp7 cells, rpoN::kan mutant cells (FAJ301), and ntrB::Tn5 mutant cells (7194). In contrast to the results of the [14C]methylammonium uptake studies, no significant difference between wild-type cells and ntr mutant cells was observed (Fig. 5).
FIG. 5.
Uptake of NH4+ in the wild-type A. brasilense Sp7 strain (◊), rpoN::kan mutant strain FAJ301 (▵), and ntrB::Tn5-194 mutant strain 7194 (-). Uptake was measured as the disappearance of NH4+ in the assay buffer after the addition of ammonium to a final concentration of 0.1 mM at time zero. The detection limit of the selective ammonium electrode used was reached after 25 min. The inset gives a representation of the mean slope values for the wild-type A. brasilense Sp7 strain, rpoN::kan mutant strain FAJ301, and ntrB::Tn5-194 mutant strain 7194. The values shown are the means of two independent replicates. Standard deviations are indicated as vertical bars.
Construction and phenotypic characterization of an A. brasilense amtB::kan insertion mutant.
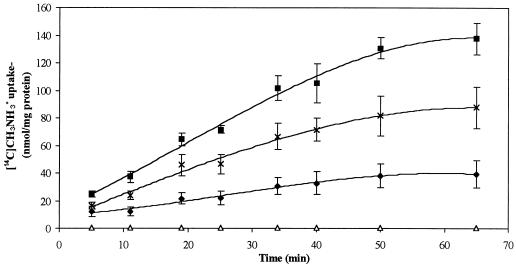
An Sp7 mutant strain was constructed by inserting a kanamycin resistance gene (kan) into the structural amtB gene (Fig. 1C). This mutant strain is unable to take up [14C]methylammonium (Fig. 6). It should be noted that although the activity of the AmtB carrier is measured with radioactively labeled [14C]methylammonium, the affinity of this carrier for CH3NH3+ is considerably less than its affinity for NH4+, and methylammonium cannot serve as a carbon or nitrogen source for A. brasilense (24).
FIG. 6.
Uptake of [14C]methylammonium in the wild-type A. brasilense Sp7 strain (⧫), the amtB::kan mutant (▵), the amtB::kan mutant complemented with the A. brasilense amtB gene (pFAJ309) (×), and the wild-type Sp7 strain containing extra copies of the A. brasilense amtB gene (pFAJ309) (▪). Values shown are the means of at least three independent replicates. Standard deviations are indicated as vertical bars. The standard deviation for the amtB::kan mutant (▵) was less than 0.6 nmol/mg.
Growth rates on rich medium or on minimal medium with 20 mM NH4+, 2 mM NH4+, 8 mM nitrate, or 10 mM aspartate were similar for both the wild-type and mutant strains. As with the wild-type strain, the amtB::kan mutant fixes nitrogen in nitrogen-free minimal medium and at low oxygen concentration. No ammonium excretion exceeding the minimum detection level of the assay method (30 mM) could be measured.
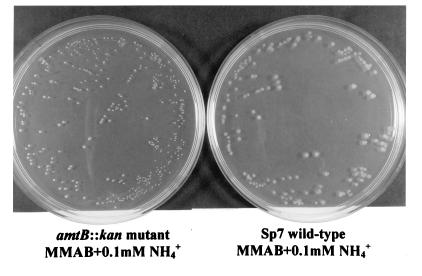
At low ammonium concentrations (0.1 mM) growth of the amtB::kan mutant cells was reduced compared to that of wild-type cells (Fig. 7).
FIG. 7.
Growth of the wild type and the amtB mutant on minimal medium with 0.1 mM NH4+ at pH 7. Cells were grown for 2 days at 30°C.
Complementation of an A. brasilense amtB::kan insertion mutant and overexpression of amtB in wild-type cells.
The amtB gene, expressed from its own promoter, was inserted in the broad-host-range vector pLAFR3 (resulting in pFAJ309). This construct was transferred to both wild-type cells and amtB::kan mutant cells. [14C]methylammonium uptake was restored in the mutant and occurred at even a higher rate than that in wild-type cells (Fig. 6). This could be due to the presence of extra copies of the amtB gene carried on the low-copy-number pLAFR3 vector (4 to 7 copies per cell) (33). Another possibility is that the lacZ promoter adjacent to the multiple-cloning site in pLAFR3 is highly expressed in A. brasilense. Since the amtB gene is oriented in the direction of expression of the lacZ promoter, this could result in enhanced amtB expression. The enhancement of [14C]methylammonium uptake in wild-type strain Sp7 when pFAJ309 was introduced is in line with these hypotheses.
DISCUSSION
When Azospirillum cells are grown under conditions of nitrogen fixation, no ammonium can be detected in the growth medium. It was postulated that bacteria have an active ammonium uptake system to compensate for the loss of NH3 by diffusion through the plasma membrane (36). Here we report on the genetic and biochemical characterization of AmtB, a (methyl)ammonium transport protein of A. brasilense.
In contrast to that of E. coli, the A. brasilense amtB gene is not part of an operon with a glnB-like gene. The coexistence of two PII-like proteins, encoded by glnB and glnZ, has been established in A. brasilense (15). Comparing physical maps of the DNA fragments containing the A. brasilense glnB, glnZ, and amtB genes confirms that they are located on different restriction fragments. In E. coli, amtB is cotranscribed with glnK, which encodes a nitrogen regulatory PII-like protein (70). In B. subtilis the nrgA gene is also part of an operon with nrgB, encoding a PII-like protein, but nrgA is located upstream of nrgB. In M. jannaschii two genes which encode putative NH4+ transporter proteins are located just next to a gene homologous to PII (7). C. glutamicum is the only prokaryote for which it has also been reported that amt is not located in an operon structure with a gene encoding a PII-like protein (61).
The synthesis of most prokaryotic NH4+ carriers is repressed by NH4+ (37). This is also the case for the yeast S. cerevisiae (16, 43), but Arabidopsis thaliana AMT1 activity is not lowered in cells grown in the presence of NH4+ (52). Transcription of the E. coli glnKamtB operon and the C. glutamicum amt gene is under nitrogen control in both organisms (61, 70). This is also observed for the A. brasilense amtB gene. The level of expression of an amtB::gusA fusion is significantly lowered in a ntrC mutant (Table 2). This indicates that the Ntr system is involved in the nitrogen-regulated transcription of amtB.
The NH4+ uptake rates of ntr mutant strains, as well as the observation that an amtB::kan mutant is still able to grow on ammonium concentrations as low as 0.1 mM (Fig. 7), suggest the existence of a second NH4+ transport mechanism. The existence of multiple NH4+ transporter proteins has been reported for other organisms. In yeast three genes encoding an NH4+ transporter (MEP1, MEP2, and MEP3) (43, 44) were isolated. Evidence also exists for the presence of two transporter systems in Nostoc muscorum (32), Rhodobacter sphaeroides (12), and Anacystis nidulans (4). In these three bacteria, one of the transport systems is constitutive and cannot transport methylammonium, while the other system is repressed in high NH4+ concentrations and is capable of transporting methylammonium.
The ammonium uptake profiles of ntr mutants, in K+-free assay buffer (Fig. 5), indicate that in these conditions the second ammonium transport mechanism does not correspond to simple diffusion. Indeed, in this assay, the external ammonium concentration, as measured by the ammonium electrode, drops below 42 μM, which is the apparent Km (Michaelis constant) of the GS enzyme of A. brasilense (54). The Km value of the main ammonium-assimilating enzyme, which is the GS enzyme in A. brasilense (73), can be used to estimate the steady-state internal ammonium concentration (11). K+ uptake systems have been reported to support NH4+ uptake in E. coli (8), and in Rhodobacter capsulatus (22). Since NH4+ ions are similar both in charge and size to K+ ions, the possibility that a K+ transporter can mediate NH4+ uptake in K+-free conditions cannot be excluded. Further investigation will be needed to establish the mode of ammonium transport in mutants lacking the nitrogen-regulated (methyl)ammonium transporter.
An analysis of the distribution of hydrophobic and hydrophilic amino acid residues shows that the AmtB amino acid sequence has highly hydrophobic stretches (38). The algorithm of Klein et al. (34) predicted 12 transmembrane domains and classified the protein as an integral membrane protein. The stretches separating the putative transmembrane regions are rich in charged amino acid residues and correspond to potential beta-turns (10). According to the positive-inside rule of von Heijne (72), the protein is oriented with its charged C and N termini exposed to the cytoplasm. These predictions are in agreement with the situation found for most bacterial carrier proteins (42).
In conclusion, we have characterized an A. brasilense (methyl)ammonium transporter. The corresponding gene, amtB, is the first A. brasilense gene known to require NtrC for expression. Indeed, this transcriptional activator is not required for the transcription of nifA, glnA, or glnB in A. brasilense. The AmtB NH4+ transporter is necessary for growth on low NH4+ concentrations. However, it appears not to be the sole mechanism for NH4+ uptake in A. brasilense.
ACKNOWLEDGMENTS
We thank C. Elmerich and A. Milcamps for kindly providing us the A. brasilense ntrB and ntrC mutants and the A. brasilense rpoN mutant, respectively. We thank K. Marchal for the PCR primers used to amplify part of the kan gene. We are also very grateful to C. Kennedy and P. Rudnick for sharing the amino acid sequence of the A. vinelandii NH4+ transporter sequence prior to publication and for interesting and fruitful exchanges. R. De Mot helped us a lot to identify the amino-terminal signal sequence of the AmtB protein. D. Corkery was so kind to revise the manuscript.
A.V.D. is a recipient of the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen. Part of this work was supported by funds of the Fonds voor Wetenschappelijk Onderzoek-Vlaanderen and the Flemish Government (GOA) to J.V.
REFERENCES
- 1.Altschul S F, Gish W, Milles W, Myersand E W, Lipman D J. Basic local alignment tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: Wiley Interscience; 1991. pp. 4.8.1–4.8.5. [Google Scholar]
- 3.Ausubel F M. Regulation of nitrogen fixation genes. Cell. 1984;37:5–6. doi: 10.1016/0092-8674(84)90294-0. [DOI] [PubMed] [Google Scholar]
- 4.Boussiba S, Dilling W, Gibson J. Methylammonium transport in Anacystis nidulans R-2. J Bacteriol. 1984;160:204–210. doi: 10.1128/jb.160.1.204-210.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 6.Bueno R, Pahel G, Magasanik B. Role of glnB and glnD gene products in regulation of the glnALG operon of Escherichia coli. J Bacteriol. 1985;164:816–822. doi: 10.1128/jb.164.2.816-822.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bult C J, White O, Olsen G J, Zhou L, Fleischmann R D, Sutton G G, Blake J A, FitzGerald L M, Clayton R A, Gocayne J D, Kerlavage A R, Dougherty B A, Tomb J-F, Adams M D, Reich C I, Overbeek R, Kirkness E F, Weinstock K G, Merrick J M, Glodek A, Scott J L, Geoghagen N S M, Weidman J F, Fuhrmann J L, Presley E A, Nguyen D, Utterback T R, Kelley J M, Peterson J D, Sadow P W, Hanna M C, Cotton M D, Hurst M A, Roberts K M, Kaine B P, Borodovsky M, Klenk H-P, Fraser C M, Smith H O, Woese C R, Venter J C. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- 8.Buurman E T, Teixeira de Mattos M J, Neijssel O M. Futile cycling of ammonium ions via the high affinity potassium uptake system (Kdp) of Escherichia coli. Arch Microbiol. 1991;155:391–395. doi: 10.1007/BF00243460. [DOI] [PubMed] [Google Scholar]
- 9.Chaney A L, Marbach E P. Modified reagents for determination of urea and ammonia. Clin Chem. 1962;8:130–132. [PubMed] [Google Scholar]
- 10.Chou P Y, Fasman G D. Prediction of beta-turns. Biophys J. 1979;26:367–384. doi: 10.1016/S0006-3495(79)85259-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cleland W W. Steady state kinetics. In: Boyer P D, editor. The enzymes. Vol. 2. New York, N.Y: Academic Press; 1970. pp. 1–65. [Google Scholar]
- 12.Cordts M L, Gibson J. Ammonium and methylammonium transport in Rhodobacter sphaeroides. J Bacteriol. 1987;169:1632–1638. doi: 10.1128/jb.169.4.1632-1638.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Croes C, Van Bastelaere E, DeClercq E, Eyers M, Vanderleyden J, Michiels K. Identification and mapping of loci involved in motility, adsorption to wheat roots, colony morphology, and growth in minimal medium on the Azospirillum brasilense Sp7 90-Mda Plasmid. Plasmid. 1991;26:83–93. doi: 10.1016/0147-619x(91)90048-2. [DOI] [PubMed] [Google Scholar]
- 14.de Zamaroczy M, Paquelin A, Elmerich C. Functional organization of the glnB-glnA cluster of Azospirillum brasilense. J Bacteriol. 1993;175:2507–2515. doi: 10.1128/jb.175.9.2507-2515.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Zamaroczy M, Paquelin A, Peltre G, Forchhammer K, Elmerich C. Coexistence of two structurally similar but functionally different PII proteins in Azospirillum brasilense. J Bacteriol. 1996;178:4143–4149. doi: 10.1128/jb.178.14.4143-4149.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dubois E, Grenson M. Methylamine/ammonia uptake systems in Saccharomyces cerevisiae: multiplicity and regulation. Mol Gen Genet. 1979;175:67–76. doi: 10.1007/BF00267857. [DOI] [PubMed] [Google Scholar]
- 17.Eisenberg D, Schwarz E, Komaromy M, Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984;179:125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- 18.Elmerich C. Le cycle du glutamate, point de départ du métabolisme de l’azote, chez Bacillus megaterium. Eur J Biochem. 1972;27:216–224. doi: 10.1111/j.1432-1033.1972.tb01829.x. [DOI] [PubMed] [Google Scholar]
- 19.Fabiny J M, Jayakumar A, Chinault A C, Barnes E M., Jr Ammonium transport in Escherichia coli: localization and nucleotide sequence of the amtA gene. J Gen Microbiol. 1991;137:983–989. doi: 10.1099/00221287-137-4-983. [DOI] [PubMed] [Google Scholar]
- 20.Figurski D A, Helinski D R. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Natl Acad Sci USA. 1979;76:1648–1652. doi: 10.1073/pnas.76.4.1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Friedman A M, Long S R, Brown S E, Buikema W J, Ausubel F M. Construction of a broad host range cosmid cloning vector and its use in the genetic analysis of Rhizobium mutants. Gene. 1982;18:289–296. doi: 10.1016/0378-1119(82)90167-6. [DOI] [PubMed] [Google Scholar]
- 22.Golby P, Carver M, Jackson J B. Membrane ionic currents in Rhodobacter capsulatus. Evidence for electrophoretic transport of K+, Rb+ and NH4+ Eur J Biochem. 1990;187:589–597. doi: 10.1111/j.1432-1033.1990.tb15341.x. [DOI] [PubMed] [Google Scholar]
- 23.Gubler M, Hennecke H. Regulation of the fixA gene and fixBC operon in Bradyrhizobium japonicum. J Bacteriol. 1988;170:1205–1214. doi: 10.1128/jb.170.3.1205-1214.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartmann A, Kleiner D. Ammonium (methylammonium) transport by Azospirillum spp. FEMS Microbiol Lett. 1982;15:65–67. [Google Scholar]
- 25.Higgins D G, Sharp P M. CLUSTAL: a package for performing multiple sequence alignment on a microcomputer. Gene. 1988;73:237–244. doi: 10.1016/0378-1119(88)90330-7. [DOI] [PubMed] [Google Scholar]
- 26.Higgins D G, Sharp P M. Fast and sensitive multiple sequence alignments on a microcomputer. Comput Appl Biosci. 1989;5:151–153. doi: 10.1093/bioinformatics/5.2.151. [DOI] [PubMed] [Google Scholar]
- 27.Jayakumar A, Schulman I, MacNeil D, Barnes E M., Jr Role of the Escherichia coli glnALG operon in regulation of ammonium transport. J Bacteriol. 1986;166:281–284. doi: 10.1128/jb.166.1.281-284.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jayakumar A, Hwang S J, Fabiny J M, Chinault A C, Barnes E M., Jr Isolation of an ammonium or methylammonium ion transport mutant of Escherichia coli and complementation by the cloned gene. J Bacteriol. 1989;171:996–1001. doi: 10.1128/jb.171.2.996-1001.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jefferson R A. Assaying chimeric genes in plants: the gus gene fusion system. Plant Mol Biol. 1987;5:387–405. [Google Scholar]
- 30.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, Kimura T, Hosouchi T, Matsuno A, Muraki A, Nakazaki N, Naruo K, Okumura S, Shimpo S, Takeuchi C, Wada T, Watanabe A, Yamada M, Yasuda M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 31.Kaneko T, Tanaka A, Sato S, Kotani H, Sazuka T, Miyajima N, Sugiura M, Tabata S. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. I. Sequence features in the 1 Mb region from map positions 64% to 92% of the genome. DNA Res. 1995;2:153–166. doi: 10.1093/dnares/2.4.153. [DOI] [PubMed] [Google Scholar]
- 32.Kashyap A K, Johar G. Genetic control of ammonium transport in nitrogen-fixing cyanobacterium Nostoc muscorum. Mol Gen Genet. 1984;197:509–512. [Google Scholar]
- 33.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 34.Klein P, Kanehisa M, DeLisi C. The detection and classification of membrane-spanning proteins. Biochim Biophys Acta. 1985;815:468–476. doi: 10.1016/0005-2736(85)90375-x. [DOI] [PubMed] [Google Scholar]
- 35.Kleiner D. Ammonium (methylammonium) transport by Klebsiella pneumoniae. Biochim Biophys Acta. 1982;688:702–708. doi: 10.1016/0005-2736(82)90282-6. [DOI] [PubMed] [Google Scholar]
- 36.Kleiner D. Bacterial ammonium transport. FEMS Microbiol Rev. 1985;32:87–100. [Google Scholar]
- 37.Kleiner D. NH4+ transport systems. In: Bakker E P, editor. Alkali cation transport systems in prokaryotes. Boca Raton, Fla: CRC Press; 1993. pp. 379–396. [Google Scholar]
- 38.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 39.Lauter F R, Ninnemann O, Bucher M, Riesmeyer J W, Frommer W B. Preferential expression of an ammonium transporter and of two putative nitrate transporters in root hairs of tomato. Proc Natl Acad Sci USA. 1996;93:8139–8144. doi: 10.1073/pnas.93.15.8139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang Y Y, Arsène F, Elmerich C. Characterization of the ntrBC genes of Azospirillum brasilense Sp7: their involvement in the regulation of nitrogenase synthesis and activity. Mol Gen Genet. 1993;240:188–196. doi: 10.1007/BF00277056. [DOI] [PubMed] [Google Scholar]
- 41.Magasanik B. Regulation of nitrogen utilization. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 1344–1356. [Google Scholar]
- 42.Maloney P C, Wilson T H. Ion-coupled transport and transporters. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: ASM Press; 1996. pp. 1130–1148. [Google Scholar]
- 43.Marini A-M, Vissers S, Urrestarazu A, André B. Cloning and expression of the MEP1 gene encoding an ammonium transporter in Saccharomyces cerevisiae. EMBO J. 1994;13:3456–3463. doi: 10.1002/j.1460-2075.1994.tb06651.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Marini A-M, Soussi-Boudekou S, Vissers S, André B. A family of ammonium transporters in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:4282–4293. doi: 10.1128/mcb.17.8.4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Meletzus D, Doetsch N, Green A, He L, Rudnick P, Yan D, Kennedy C. Genetic characterization of ammonium sensing and signal transduction in Azotobacter vinelandii. In: Tikhonovich I A, et al., editors. Nitrogen fixation: fundamentals and applications. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1995. p. 220. [Google Scholar]
- 46.Merrick M J, Edwards R A. Nitrogen control in bacteria. Microbiol Rev. 1995;59:604–622. doi: 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Michiels J, D’hooghe I, Verreth C, Pelemans H, Vanderleyden J. Characterization of the Rhizobium leguminosarum biovar phaseoli nifA gene, a positive regulator of nif gene expression. Arch Microbiol. 1994;161:404–408. doi: 10.1007/BF00288950. [DOI] [PubMed] [Google Scholar]
- 48.Milcamps A, Van Dommelen A, Stigter J, Vanderleyden J, de Bruijn F. The Azospirillum brasilense rpoN gene is involved in nitrogen fixation, nitrate assimilation, ammonium uptake and flagellar biosynthesis. Can J Microbiol. 1996;42:467–478. doi: 10.1139/m96-064. [DOI] [PubMed] [Google Scholar]
- 49.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 50.Myers E W, Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988;4:11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- 51.Neuwald A F, Krishnan B R, Brikun I, Kulakauskas S, Suziedelis K, Tomcsanyi T, Leyh T S, Berg D E. cysQ, a gene needed for cysteine synthesis in Escherichia coli K-12 only during aerobic growth. J Bacteriol. 1992;174:415–425. doi: 10.1128/jb.174.2.415-425.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ninneman O, Jauniaux J-C, Frommer W B. Identification of a high affinity NH4+ transporter from plants. EMBO J. 1994;13:3464–3471. doi: 10.1002/j.1460-2075.1994.tb06652.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Okon Y. Azospirillum-plant root associations. Boca Raton, Fla: CRC Press; 1994. [Google Scholar]
- 54.Pirola M C, Monopoli R, Aliverti A, Zanetti G. Isolation and characterization of glutamine synthetase from the diazotroph Azospirillum brasilense. Int J Biochem. 1992;24:1749–1754. [Google Scholar]
- 55.Roelvink P W, Harmsen M, van Kammen A, van den Bos R C. The nifH promoter region of Rhizobium leguminosarum: nucleotide sequence and promoter elements controlling activation by NifA protein. Gene. 1990;87:31–36. doi: 10.1016/0378-1119(90)90492-a. [DOI] [PubMed] [Google Scholar]
- 56.Rosenberg M, Court D. Regulatory sequences involved in the promotion and termination of RNA transcription. Annu Rev Genet. 1979;13:319–353. doi: 10.1146/annurev.ge.13.120179.001535. [DOI] [PubMed] [Google Scholar]
- 57.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 58.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain termination inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Servin-Gonzales L, Bastarrachea F. Nitrogen regulation of the synthesis of the high affinity methylammonium transport system of Escherichia coli. J Gen Microbiol. 1984;130:3071–3077. doi: 10.1099/00221287-130-12-3071. [DOI] [PubMed] [Google Scholar]
- 60.Shields D C, Higgins D G, Sharp P M. GCWIND: a microcomputer program for identifying open reading frames according to codon positional G + C content. Comput Appl Biosci. 1992;8:521–523. doi: 10.1093/bioinformatics/8.5.521. [DOI] [PubMed] [Google Scholar]
- 61.Siewe R M, Weil B, Burkovski A, Eikmanns B J, Eikmanns M, Kramer R. Functional and genetic characterization of the (methyl)ammonium uptake carrier of Corynebacterium glutamicum. J Biol Chem. 1996;271:5398–5403. doi: 10.1074/jbc.271.10.5398. [DOI] [PubMed] [Google Scholar]
- 62.Simon R, Priefer U, Pühler A. A broad host range mobilisation system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 63.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujitomo E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 64.Staskawicz B, Dahlbeck D, Keen N, Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987;169:5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stock J B, Ninfa A J, Stock A M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989;53:450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tarrand J J, Krieg N R, Döbereiner J. A taxonomic study of the Spirillum lipoferum group, with the description of a new genus, Azospirillum gen. nov. and two species, Azospirillum lipoferum (Beijerinck) comb. nov. and Azospirillum brasilense sp. nov. Can J Microbiol. 1978;24:967–980. doi: 10.1139/m78-160. [DOI] [PubMed] [Google Scholar]
- 67.Vande Broek A, Michiels J, Van Gool A, Vanderleyden J. Spatial-temporal colonization patterns of Azospirillum brasilense on wheat root surface and expression of bacterial nifH during the association. Mol Plant-Microbe Interact. 1993;6:592–600. [Google Scholar]
- 68.Vande Broek A, Vanderleyden J. Review: genetics of the Azospirillum-plant root association. Crit Rev Plant Sci. 1995;14:445–466. [Google Scholar]
- 69.Van Dommelen A, Van Bastelaere E, Keijers V, Vanderleyden J. Genetics of Azospirillum brasilense with respect to ammonium transport, sugar uptake and chemotaxis. Plant Soil. 1997;194:155–160. [Google Scholar]
- 70.Van Heeswijk W C, Hoving S, Molenaar D, Stegeman B, Kahn D, Westerhoff H V. An alternative PII protein in the regulation of glutamine synthetase in Escherichia coli. Mol Microbiol. 1996;21:133–146. doi: 10.1046/j.1365-2958.1996.6281349.x. [DOI] [PubMed] [Google Scholar]
- 71.Vanstockem M, Michiels K, Vanderleyden J, Van Gool A P. Transposon mutagenesis of Azospirillum brasilense and Azospirillum lipoferum: physical analysis of Tn5 and Tn5-Mob insertion mutants. Appl Environ Microbiol. 1987;53:410–415. doi: 10.1128/aem.53.2.410-415.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.von Heijne G. Membrane protein structure prediction. Hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]
- 73.Westby C A, Enderlin C S, Steinberg N A, Joseph C M, Meeks J C. Assimilation of 13NH4+ by Azospirillum brasilense grown under nitrogen limitation and excess. J Bacteriol. 1987;169:4211–4214. doi: 10.1128/jb.169.9.4211-4214.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Wilson K J, Sessitsch A, Corbo J C, Giller K E, Akkermans A D L, Jefferson R A. β-Glucuronidase (gus) transposons for ecological and genetic studies of rhizobia and other gram-negative bacteria. Microbiology. 1995;141:1691–1705. doi: 10.1099/13500872-141-7-1691. [DOI] [PubMed] [Google Scholar]
- 75.Wilson R, Ainscough R, Anderson K, Baynes C, Berks M, Bonfield J, Burton J, Connell M, Copsey T, Cooper J, Coulson A, Craxton M, Dear S, Du Z, Durbin R, Favello A, Fulton L, Gardner A, Green P, Hawkins T, Hillier L, Jier M, Johnston L, Jones M, Kershaw J, Kirsten J, Laister N, Latreille P, Lightning J, Lloyd C, McMurray A, Mortimore B, O’Callaghan M, Parsons J, Percy C, Rifken L, Roopra A, Saunders D, Shownkeen R, Smaldon N, Smith A, Sonnhammer E, Staden R, Sulston J, Thierry-Mieg J, Thomas K, Vaudin M, Vaughan K, Waterston R, Watson A, Weinstock L, Wilkinson-Sproat J, Wohldman P. 2.2 Mb of contiguous nucleotide sequence from chromosome III of C. elegans. Nature. 1994;368:32–38. doi: 10.1038/368032a0. [DOI] [PubMed] [Google Scholar]
- 76.Wray L V, Jr, Atkinson M R, Fisher S H. The nitrogen-regulated Bacillus subtilis nrgAB operon encodes a membrane protein and a protein highly similar to the Escherichia coli glnB-encoded PII protein. J Bacteriol. 1994;176:108–114. doi: 10.1128/jb.176.1.108-114.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]