Abstract
The dctA gene, coding for the dicarboxylate transport protein, has an inducible promoter dependent on activation by the two-component sensor-regulator pair DctB and DctD. LacZ fusion analysis indicates that there is a single promoter for dctB and dctD. The dctA promoter is also induced by nitrogen limitation, an effect that requires DctB-DctD and NtrC. DctB alone is able to detect dicarboxylates in the absence of DctA and initiate transcription via DctD. However, DctA modifies signal detection by DctB such that in the absence of DctA, the ligand specificity of DctB is broader. dctAp also responds to heterologous induction by osmotic stress in the absence of DctA. This effect requires both DctB and DctD. A transposon insertion in the dctA-dctB intergenic region (dctA101) which locks transcription of dctA at a constitutive level independent of DctB-DctD results in improper signalling by DctB-DctD. Strain RU150, which carries this insertion, is defective in nitrogen fixation (Fix−) and grows very poorly on ammonia as a nitrogen source whenever the DctB-DctD signalling circuit is activated by the presence of a dicarboxylate ligand. Mutation of dctB or dctD in strain RU150 reinstates normal growth on dicarboxylates. This suggests that DctD-P improperly regulates a heterologous nitrogen-sensing operon. Increased expression of DctA, either via a plasmid or by chromosomal duplication, restores control of DctB-DctD and allows strain RU150 to grow on ammonia in the presence of a dicarboxylate. Thus, while DctB is a sensor for dicarboxylates in its own right, it is regulated by DctA. The absence of DctA allows DctB and DctD to become promiscuous with regard to signal detection and cross talk with other operons. This indicates that DctA contributes significantly to the signalling specificity of DctB-DctD and attenuates cross talk with other operons.
Transport and catabolism of dicarboxylic acids are required to fuel nitrogen fixation by rhizobia in legume nodules. In free-living cells of Rhizobium leguminosarum and Sinorhizobium meliloti, transport of l-malate, fumarate, and succinate occurs via the C4-dicarboxylic transport (Dct) system (5, 8, 10, 39). This system consists of three genes: dctA, which codes for the putative transport protein, and two divergently transcribed genes, dctB and dctD, which activate transcription of dctA in response to the presence of dicarboxylates (6, 15, 16, 35, 38, 46). DctB and DctD are well characterized as a two-component sensor-regulator pair. In free-living cultures, mutations in any of the three dct genes result in the loss of the ability to transport and grow on C4-dicarboxylates (1, 5, 7, 16, 35, 38). Strains with mutations in dctA always display a nitrogen fixation-defective (Fix−) phenotype on plants, and isolated bacteroids do not transport C4-dicarboxylates. However, some strains with mutations in dctB or dctD fix nitrogen, and isolated bacteroids transport C4-dicarboxylates, albeit at a reduced rate (1, 5, 8).
Transcription from dctAp is dependent on Eς54, which binds to a site located 93 bp upstream from the dctA start codon in R. leguminosarum and in a similar position in S. meliloti (13). Proximal and distal upstream activator sequences which contain inverted repeats are also located approximately 75 bp upstream from the end of the putative ς54 binding site (13, 36). DctD is able to bind in a cooperative fashion to these upstream activator sites, via a C-terminal helix-turn-helix motif, to initiate transcription (20–23, 41, 42, 45). Deletion of either upstream activator sequence results in a significant reduction in transcription (20, 21). DctD has recently been shown to be able to interact with both ς54 and the β-subunit of RNA polymerase; in the process, it hydrolyzes ATP to promote open complex formation (11, 12, 22, 23).
The C terminus of DctB has homology over 200 amino acids with a large number of sensor proteins (35, 38). It is capable of both autophosphorylation, presumably at histidine 416, and phosphorylation of DctD (9). There are two putative membrane-spanning regions, between residues 25 and 42 and between residues 321 and 328, that are predicted to form a large periplasmic loop, similar to those found in the methyl-accepting chemotaxis proteins of Escherichia coli which sense chemoattractants in the periplasm (18, 26). It has therefore been suggested that in both R. leguminosarum and S. meliloti, DctB acts as a membrane-bound sensor that responds to the presence of C4-dicarboxylates and transduces this signal across the membrane to activate its cytoplasmically located C terminus. This results in autophosphorylation and phosphotransfer to DctD (38, 46).
However, a number of reports have shown that strains with mutations in dctA display constitutive transcription from dctAp, suggesting that DctA has a role in controlling its own synthesis (16, 36, 48). It has been proposed that in the absence of substrate, DctA and DctB may interact with each other in the cytoplasmic membrane, preventing activation of DctB. In the wild type, binding of a dicarboxylate would release DctB, enabling it to activate DctD, while in a dctA strain, DctB would always be in the activated mode (17, 48). A modification of this model, whereby DctB senses the C4-dicarboxylate-dependent conformational state of DctA, rather than sensing C4-dicarboxylates directly, has been proposed (48). Thus, DctA would undergo a conformational change, as a result of binding or transporting substrate, which would result in the release and/or activation of DctB.
Given the importance of the Dct system to nitrogen fixation and our detailed knowledge of the binding kinetics and activation of transcription of dctAp by DctD, the contradictions and uncertainty in our understanding of the initial signal detection by DctA and DctB are glaring. It is not even clear if DctB is a sensor of dicarboxylates in its own right. We therefore initiated a study of how the dctA, dctB, and dctD promoters are regulated by inducers and how DctA and DctB influence this process.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains, bacteriophage, and plasmids used in this study are described in Table 1. R. leguminosarum strains were grown at 28°C on either TY (3) or acid minimal salts (AMS) medium (pH 7.2) (29). All carbon and nitrogen sources were at 10 mM concentrations unless otherwise stated. Antibiotics were used at the following concentrations (in micrograms per milliliter): kanamycin, 40; streptomycin, 500; spectinomycin, 100; tetracycline, 2 (in AMS) and 5 (in TY); and gentamicin, 20. E. coli strains were grown at 37°C on Luria-Bertani medium with antibiotic concentrations as follows (in micrograms per milliliter): kanamycin, 25; tetracycline, 10; gentamicin, 5; and ampicillin, 50.
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Description | Source or reference |
|---|---|---|
| Strains | ||
| R. leguminosarum | ||
| 3841 | Strr derivative of R. leguminosarum biovar viciae strain 300 | 10, 14 |
| 3855 | Strr derivative of R. leguminosarum biovar viciae | 38 |
| CR534 | Strain 3855 dctA::Tn5 | 38 |
| CR535 | Strain 3855 dctB::Tn5 | 38 |
| CR538 | Strain 3855 dctD::Tn5 | 38 |
| RU150 | Strain 3841 dctA101 | This work |
| RU152-1 | Second-site revertant of RU150 | This work |
| RU152-14 | Second-site revertant of RU150 | This work |
| RU152-22 | Second-site revertant of RU150 | This work |
| RU436 | Strain 3841 dctA::Tn5 | 29 |
| RU437 | Strain 3841 dctA::Tn5 | 29 |
| RU711 | Strain 3841 ΔdctD::Ω Strr Spcr | 34 |
| RU714 | Strain 3841 ΔdctABD::Ω Strr Spcr | 34 |
| RU720 | Strain RU150 dctA101 ΔdctD::Ω Strr Spcr Kmr | This work |
| RU727 | Strain 3841 dctA::Ω Strr Spcr | 34 |
| RU730 | Strain 3841 dctB::Ω Strr Spcr | 33 |
| RU865 | Strain 3841 ΔdctBD::Ω Strr Spcr | 33 |
| RU873 | Strain RU150 dctA101 ΔdctBD::Ω Strr Spcr Kmr | This work |
| RU875 | Strain RU150 dctA101 dcB::Ω Strr Spcr Kmr | This work |
| RU929 | Strain 3841 ntrC::Ω Strr Spcr | 33 |
| RU938 | Strain RU150 dctAB::Tn5 dcA::Ω Strr Spcr Kmr | This work |
| E. coli S17-1 | pro hsdR recA [RP4-2(Tc::Mu) (Km::Tn7)]; RP4 integrated into its chromosome | 42 |
| Plasmids | ||
| Bluescript II SK− | Phagemid; f1− origin of replication; ColE1 replicon; SK polylinker; 2.96 kb; standard cloning vector; Ampr | Stratagene Ltd. |
| pGEMT | T overhang vector for PCR product cloning; Ampr | Promega |
| pMP220 | IncP broad-host-range mobilizable promoter probe vector employing E. coli lacZ as reporter gene; 10.5 kb; Tcr | 43 |
| pJQ200KS | Narrow host range (p15A ori); mob sacB; 5.4 kb; Gmr | 31 |
| pHP45Ω | 4.3-kb plasmid containing 2.0-kb Ω interposon encoding the gene for Spr with transcription and translation termination signals at both ends; Ampr Spcr | 30 |
| pIJ1848 | Cosmid containing dctABD from R. leguminosarum 8002; Tcr | 25 |
| pIJ1969 | pIJ1848 dctB::Tn5 | 25 |
| pIJ1970 | pIJ1848 dctA::Tn5 | 25 |
| pRU10 | 6.7-kb EcoRI chromosomal Tn5 bearing fragment from strain RU150 cloned in Bluescript SK−; Ampr | This work |
| pRU16 | 0.9-kb dctA-dctB intergenic region from pRU3001 in Bluescript SK−; Ampr | This work |
| pRU47 | 10.4-kb HindIII fragment from pIJ1848 in Bluescript SK−; Ampr | This work |
| pRU69 | 3.0-kb HindIII/EcoRI fragment from pRU150 ligated into pBC SK+; Cmr | This work |
| pRU75 | 1-kb HpaI/HindIII fragment from IS50 cloned in Bluescript SK−; Ampr | This work |
| pRU80 | 2.8-kb internal BglII fragment from Tn5 ligated in Bluescript pBC SK+; Cmr Kmr | This work |
| pRU93 | 6.0-kb EcoRI fragment containing Tn5 cloned from the chromosome of strain RU152-22 into Bluescript SK−; Ampr Kmr | This work |
| pRU103 | 0.9-kb dctA-dctB intergenic region EcoRI from pRU16 in pMP220; dctA-lacZ; Tcr | 33 |
| pRU104 | 0.9-kb dctA-dctB intergenic region EcoRI from pRU16 in pMP220; dctB-lacZ; Tcr | This work |
| pRU105 | dctA101-lacZ fusion in pMP220; Tcr Kmr | This work |
| pRU106 | dctA101B-lacZ fusion in pMP220; Tcr Kmr | This work |
| pRU108 | 3.0-kb EcoRI fragment containing dctA from pRU47 in pMP220; Tcr | This work |
| pRU110 | 0.4-kb inverse PCR fragment from strain RU152-14 cloned in pGEM-T; Ampr | This work |
| pRU123 | 1.5-kb dctA produced by PCR in pGEMT; Ampr | This work |
| pRU150 | pRU47 ClaI digested and religated (6.1 kb); contains dctABD; Ampr | This work |
| pRU155 | pRU123 dctA::Ω; Ampr Spcr | This work |
| pRU158 | ΔdctB::Ω in SK−; Ampr Spcr | This work |
| pRU168 | ΔdctD::Ω in pJQ200KS; Gmr Spcr | This work |
| pRU292 | 0.53-kb PstI fragment from pRU3001 in pMP220; dctD1-lacZ; Tcr | This work |
| pRU303 | 4.6-kb NotI/ApaI fragment in pJQ200KS; ΔdctBD::Ω; Gmr Spcr | This work |
| pRU304 | 4.5-kb NotI/ApaI fragment from pRU347 ligated into pJQ200KS; Gmr Spcr | This work |
| pRU305 | 4.0-kb NotI/ApaI fragment from pRU345 in pJQ200KS; Gmr Spcr | This work |
| pRU345 | 3.1-kb AccIII fragment from pRU10 ligated into pRU155; Ampr Spcr Kmr | This work |
| pRU347 | 6.7-kb EcoRI fragment from pRU10 ligated into pRU158; Ampr Spcr Kmr | This work |
| pRU348 | 3.6-kb BamHI/MluI fragment from pIJ1848 in Bluescript SK−; Ampr | This work |
| pRU352 | dctA101BD-lacZ fusion in pMP220; Tcr Kmr | This work |
| pRU354 | 3.6-kb BamHI/MluI fragment from pIJ1848 in pMP220; dctBD-lacZ; Tcr | This work |
| pRU392 | 2.4-kb EcoRI/XbaI fragment from pIJ1848 in pMP220; dctD2-lacZ; Tcr | This work |
| pRU402 | 0.8-kb PstI/NotI fragment of DNA immediately downstream of dctA in Bluescript pBC SK+; Cmr | This work |
| pTH2A | S. meliloti dctA::TnphoA translational fusion; Tcr | 45 |
| pRU3001 | Cosmid from strain 3841 containing the dct region; Tcr | 29 |
DNA and genetic manipulations.
All routine DNA analysis was performed essentially according to the procedures of Sambrook et al. (40). Conjugations were performed as previously described (29). Transductions were performed with bacteriophage RL38 as described by Buchanan-Wollaston (4). Linkage in the second-site suppressor strains RU152-1, RU152-14, and RU152-22 was determined by transducing the kanamycin marker of Tn5 into strain 3841. Forty kanamycin-resistant transductants of each strain were double purified and tested for growth on glucose-ammonia and succinate-ammonia. Strain 3841 was mutagenized with Tn5 by conjugation with E. coli S17-1 containing pSUP202-1::Tn5 essentially as described by Simon et al. (43). Strain RU150 was isolated due to its poor growth on succinate-ammonia. Chromosomal DNA from strain RU150 was digested with EcoRI, and a 6.7-kb fragment was cloned into Bluescript II SK+ by selecting for kanamycin resistance. The left and right arms of the transposon were isolated by BamHI and HindIII digestion followed by self-ligation. Both arms were sequenced by using a primer that binds to the ends of IS50 (P0) and SK and KS primers in the plasmid. For clarity, the mutant allele in strain RU150 has been designated dctA101.
All sequencing was performed by the cycle sequencing method by using a Promega fmol kit according to the manufacturer’s instructions. Genetics Computer Group software was used for computer-assisted sequence analysis. Custom primers used were P0 (GTTCAGGACGCTACTTG), P2 (CGCTGTCGTCCAATCTCCCAAGAC), P3 (TCTGATGGCGCAGGGGATCAAGAT), P6 (AAGGCCACAATTTCTGCGACACGG), and P7 (GTTTCTAAGGATAAGGGGATAGCG).
Inverse PCR.
Chromosomal DNA from strain RU152-14 was digested with EcoRI and electrophoresed on an agarose gel; the 1- to 2-kb bands were excised and purified to ensure that the Tn5-containing fragment was not present. These were ligated to form circular DNA, and the insert DNA was amplified by inverse PCR with primers P2 and P3, with 30 cycles of 94, 55, and 72°C, each for 1.5 min. The resulting 0.4-kb fragment was cloned into pGEM-T (pRU110) and sequenced on both strands as far as the central EcoRI site.
Northern blotting.
Total RNA was isolated according to the method of Rastogi et al. (32), and 20 μg was separated on an agarose gel and transferred by Northern blotting to an Amersham Hybond N plus membrane. A dctA-specific probe was produced by PCR amplification of pRU69 with primers P6 and P7 under the same conditions as those used for inverse PCR. The resulting 1.5-kb fragment was cloned into pGEM-T and designated pRU123. The total-RNA blot was hybridized with the dctA DNA probe (pRU123) which had been random primed with 50 μCi of [α-32P]dCTP (3,000 Ci mmol−1) by using an Amersham random priming kit according to the manufacturer’s instructions.
Transcriptional lacZ fusions.
The dctA-lacZ (pRU103) fusion consists of a 950-bp EcoRI fragment cloned in the transcriptional promoter probe vector pMP220 and has been described previously (34). The EcoRI site in dctA is present only in strain 3841 and results from a change from G to A at bp 2082 of the sequence published for R. leguminosarum (EMBL accession no. Z11529). This results in a coding change from Leu to Phe. The dctB-lacZ fusion has the same 950-bp EcoRI fragment but in the reverse orientation (Fig. 1). Two dctD-lacZ fusions and one dctBD-lacZ fusion were constructed. The first dctD-lacZ fusion, dctD1-lacZ, consists of a 0.53-kb PstI fragment cloned in pMP220. This plasmid, pRU292, contains the 339 bp preceding the translational start of dctD and extends 193 bp into dctD. The second dctD-lacZ fusion, dctD2-lacZ, consists of a 2.4-kb EcoRI/MluI fragment cloned in pMP220. This plasmid, pRU392, contains 1.3 kb of dctB preceding dctD and 1.1 kb of dctD. The dctBD-lacZ fusion consists of a 3.6-kb BamHI/MluI fragment cloned in pMP220. This plasmid, pRU354, contains all of dctB, the 5′ end of dctA, and 1.1 kb of dctD. Notably, it contains the same point of fusion to dctD as dctD2-lacZ.
FIG. 1.
Maps of the dct region. (A) Sites used in subcloning. A, AccIII; B, BamHI; E, EcoRI; M, MluI; N, NruI; P, PstI; V, EcoRV. Additional MluI, NruI, and PstI sites not used in cloning have been omitted for clarity. (B) Expanded view of the promoter region, showing the transposon insertion in strain RU150. RBS, ribosome binding site; UAS, upstream activation sequence.
The dctA101-lacZ fusion, pRU105, was made by ligating the EcoRI fragment from pRU10 into pMP220. The reverse orientation of this clone yielded the dctA101B-lacZ fusion, pRU106. The starting point for construction of a dctA101BD-lacZ fusion was pRU348, which has the 3.6-kb BamHI/MluI fragment from pRU354 cloned in Bluescript SK−. The EcoRI fragment from pRU10 was cloned into pRU348, and a partial EcoRI/XbaI digest was used to transfer the whole insert into pMP220, creating pRU352.
β-Galactosidase assays were conducted according to the method of Miller (25) with modifications as previously described (27). For calculation of substrate-dependent induction, the average background of o-nitrophenylgalactoside hydrolyzed in the presence of the vector pMP220 alone (168 nmol min−1 mg of protein−1) was used for all samples.
Construction of strain RU150 dct double mutants.
All the RU150 dct double mutants were constructed by using the sac selection system in pJQ200KS and the Ω interposon encoding spectinomycin resistance (30, 31).
The plasmid required to produce strain RU150 dctA::Ω was made by cloning a BamHI-digested Ω interposon into the BamHI site of pRU123, creating pRU155. The DNA at the 5′ end of dctA was replaced with the corresponding DNA from strain RU150 by cloning the AccIII fragment from pRU10 (EcoRI fragment containing Tn5 from strain RU150) into pRU155, creating pRU345. The entire insert from pRU345, which contains IS50L upstream of dctA, was transferred as a NotI/ApaI fragment into pJQ200KS, generating pRU305. The NotI site used is located in IS50L of Tn5. This plasmid contains 894 bp upstream of the Ω interposon and 947 bp downstream of it and is identical to the flanking DNA in strain RU150.
The plasmid required to produce RU150 dctB::Ω was made by cloning the EcoRI/EcoRV fragment (1.2 kb) from pRU47, which contains part of dctB, into Bluescript SK−. This was digested with NruI, which cuts dctB 949 bp from its translational start site, and a SmaI-digested Ω interposon was ligated in to form pRU158. The Tn5 EcoRI fragment from pRU10 was ligated into pRU158, forming pRU347, and the insert was transferred as a NotI/ApaI fragment into pJQ200KS, generating pRU304.
To produce strain RU150 ΔdctD::Ω, plasmid pRU150 was digested with EcoRV and religated. This utilizes an EcoRV site in the polylinker of the vector and removes all of dctA, dctB, and the 5′ end of dctD. The internal NruI fragment (123 bp) was deleted, and a SmaI-digested Ω interposon was ligated in. The whole insert was transferred to pJQ200KS as a NotI/ApaI fragment, forming pRU168.
To produce strain RU150 ΔdctBD::Ω, plasmid pRU150 was digested with EcoRI and religated. This utilizes an EcoRI site in the polylinker of the vector and removes dctA and the 5′ end of dctB. The internal NruI fragment (1.9 kb) was removed, and a SmaI-digested Ω interposon was ligated in. The EcoRI fragment from pRU10 was cloned in, and the whole insert was transferred as a NotI/ApaI fragment into pJQ200KS, forming pRU303.
The four plasmids pRU168, pRU303, pRU304, and pRU305 were conjugated into strain RU150 and plated onto TY containing spectinomycin and sucrose (5% [wt/vol]). This generated strains RU720 (dctA101 ΔdctD::Ω), RU873 (dctA101 ΔdctBD::Ω), RU875 (dctA101 dctB::Ω), and RU938 (dctA101 dctA::Ω). These were all confirmed as having the correct genotypes by Southern blotting (data not shown).
Transport assays.
R. leguminosarum strains were prepared, and transport assays were performed, as previously described (28); in each case, a total succinate concentration of 25 μM was used. The specific activity of [2,3-14C]succinic acid was 4 GBq mmol−1.
Plant assays.
Nodulation and acetylene reduction were determined by using plants of common vetch (Vicia sativa). Plant growth and acetylene reductions were carried out as previously described (27), except that plants were harvested 6 weeks postinoculation.
RESULTS
Transcriptional analysis of the dctA, dctB, and dctD promoters in strain 3841.
Succinate and aspartate are good inducers of dctAp in R. leguminosarum and S. meliloti (16, 20, 36, 47, 48). Consistent with this, strain 3841, containing a dctA-lacZ transcriptional fusion (pRU103), had a 19- or 14-fold increase in β-galactosidase activity after growth on succinate or aspartate (supplied as the nitrogen source), respectively, compared to that on glucose (Table 2). All fold inductions were calculated after subtraction of the average background due to the vector pMP220 (168 nmol min−1 mg of protein−1). β-Galactosidase activity increased approximately 33-fold when strain 3841 was grown in the presence of both succinate and aspartate, indicating an additive effect. Cells grown on succinate-aspartate-ammonia showed an approximately 50% reduction in β-galactosidase activity compared to succinate-aspartate-grown cells. One possibility is that cells grown on succinate-aspartate might be nitrogen limited. If this is correct, dctAp may respond to nitrogen limitation as well as to the presence of a dicarboxylate (see below).
TABLE 2.
β-Galactosidase fusion analysis of the native dctA promoter
| Strain | Genotype | Fusiona | β-Galactosidase
activity under the following growth conditionb:
|
|||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Glc-NH4 | Succ-NH4 | Glc-Asp | Succ-Asp | Glc-Succ-NH4 | Glc-Succ-Asp | Glc-Asp-NH4 | Succ-Asp-NH4 | |||
| 3841 | Wild type | dctA-lacZ | 266 ± 61 | 2,067 ± 267 | 1,533 ± 356 | 3,433 ± 101 | 2,228 ± 257 | 3,997 ± 273 | 1,944 ± 128 | 1,751 ± 253 |
| RU150 | dctA101 | dctA-lacZ | 148 ± 28 | 2,223 ± 120 | 1,251 ± 73 | |||||
| RU727 | dctA::Ω | dctA-lacZ | 297 ± 25 | 3,417 ± 248 | 2,073 ± 212 | 3,396 ± 354 | 2,558 ± 341 | |||
| RU730 | dctB::Ω | dctA-lacZ | 184 ± 27 | 273 ± 36 | 102 ± 20 | 237 ± 112 | 102 ± 33 | |||
| RU711 | dctD::Ω | dctA-lacZ | 150 ± 26 | 235 ± 6 | 91 ± 5 | 198 ± 35 | 136 ± 47 | |||
| RU865 | ΔdctBD::Ω | dctA-lacZ | 168 ± 10 | 255 ± 61 | 141 ± 53 | 460 ± 157 | 210 ± 73 | |||
| RU714 | ΔdctABD::Ω | dctA-lacZ | 179 ± 65 | 264 ± 46 | 205 ± 50 | 380 ± 122 | 161 ± 35 | |||
| RU436 | dctA::Tn5 | dctA-lacZ | 286 ± 12 | 2,945 ± 123 | 2,407 ± 122 | 4,080 ± 147 | 1,886 ± 58 | |||
| RU437 | dctA::Tn5 | dctA-lacZ | 230 ± 37 | 3,078 ± 369 | 3,057 ± 107 | 4,475 ± 143 | 2,281 ± 165 | |||
| 3855 | Wild type | dctA-lacZ | 260 ± 45 | 1,620 ± 75 | 1,960 ± 104 | |||||
| CR534 | dctA::Tn5 | dctA-lacZ | 315 ± 85 | 2,755 ± 84 | 3,119 ± 123 | |||||
| CR535 | dctB::Tn5 | dctA-lacZ | 113 ± 12 | 95 ± 3 | 97 ± 12 | |||||
| CR538 | dctD::Tn5 | dctA-lacZ | 111 ± 21 | 120 ± 12 | 111 ± 23 | |||||
| 3841 | Wild type | pMP220 | 120 ± 40 | 168 ± 21 | 180 ± 42 | 205 ± 62 | ||||
The dctA-lacZ fusion is pRU103; the vector control is pMP220.
Results are shown as o-nitrophenylgalactoside hydrolyzed (in nanomoles per minute per milligram of protein ± SEM) and are based on at least three independent cultures assayed in triplicate. Glc, glucose; Succ, succinate; Asp, aspartate. All growth substrates were used at 10 mM concentrations.
In accord with previous reports (16, 38, 48), expression from dctBp was constitutive in cells grown on all substrates tested, with a small increase evident when cells were grown on glucose-aspartate and succinate-ammonia (Table 3). Three dctD reporter fusions containing different amounts of DNA preceding the dctD translational start site were constructed (Fig. 1A). β-Galactosidase expression from both pRU292 and pRU392, which have 339 bp and 1.3 kb of DNA upstream of the DctD translational start site, respectively, was similar to that obtained from strain 3841, containing the parent pMP220 replicon (Table 3). Since pRU392 contains 1.3 kb of dctB DNA preceding the start site of dctD, it is unlikely that dctD has a significant promoter of its own. Expression of β-galactosidase from the dctBD-lacZ fusion (pRU354) displayed activity similar to that from the dctB-lacZ fusion (Table 3), indicating that dctB and dctD are a single transcriptional unit. Given that the translational stop site for DctB is only 5 bp upstream of the translational start site of DctD in R. leguminosarum, this is not an unexpected result. However, it is in marked contrast to the conclusion, in a previous report for S. meliloti, that dctD has its own promoter (16).
TABLE 3.
β-Galactosidase fusion analysis of various dct promoters
| Strain | Genotype | Fusiona | β-Galactosidase
activity under the following growth conditionb:
|
|||
|---|---|---|---|---|---|---|
| Glc-NH4 | Succ-NH4 | Glc-Asp | Succ-Asp | |||
| 3841 | Wild type | dctB-lacZ | 778 ± 90 | 1,211 ± 40 | 1,318 ± 403 | 743 ± 116 |
| RU150 | dctA101 | dctB-lacZ | 596 ± 48 | 630 ± 48 | 470 ± 47 | |
| 3841 | Wild type | dctD1-lacZ | 172 ± 5 | 214 ± 8 | 192 ± 14 | 174 ± 9 |
| 3841 | Wild type | dctD2-lacZ | 166 ± 40 | 228 ± 34 | 265 ± 79 | 230 ± 77 |
| 3841 | Wild type | dctBD-lacZ | 788 ± 18 | 1,454 ± 56 | 1,180 ± 88 | 749 ± 50 |
| RU150 | dctA101 | dctBD-lacZ | 701 ± 35 | 777 ± 43 | 741 ± 13 | |
| 3841 | Wild type | dctA101-lacZ | 406 ± 60 | 450 ± 52 | 406 ± 69 | 649 ± 117 |
| RU150 | dctA101 | dctA101-lacZ | 347 ± 30 | 348 ± 25 | 350 ± 34 | |
| RU730 | dctB::Ω | dctA101-lacZ | 405 ± 26 | 362 ± 52 | ||
| RU711 | dctD::Ω | dctA101-lacZ | 379 ± 32 | 442 ± 37 | ||
| 3841 | Wild type | dctA101B-lacZ | 1,054 ± 66 | 1,531 ± 40 | 1,149 ± 119 | 1,137 ± 127 |
| RU150 | dctA101 | dctA101B-lacZ | 1,025 ± 45 | 1,173 ± 91 | 896 ± 90 | |
| 3841 | Wild type | dctA101BD-lacZ | 975 ± 58 | 1,459 ± 66 | 1,179 ± 58 | 1,123 ± 16 |
| RU150 | dctA101 | dctA101BD-lacZ | 906 ± 75 | 1,198 ± 25 | 1,037 ± 76 | |
| 3841 | Wild type | pMP220 | 120 ± 40 | 168 ± 21 | 180 ± 42 | 205 ± 62 |
| RU150 | dctA101 | pMP220 | 142 ± 22 | 153 ± 33 | 245 ± 5 | |
The dct-lacZ fusions are as follows: dctB, pRU104; dctD1, pRU292; dctD2, pRU392; dctBD, pRU354; dctA101, pRU105; dctA101B, pRU106; dctA101BD, pRU352. pMP220 is the vector alone.
Results are shown as o-nitrophenylgalactoside hydrolyzed (in nanomoles per minute per milligram of protein ± SEM) and are based on at least three independent cultures assayed in triplicate. Glc, glucose; Succ, succinate; Asp, aspartate. All growth substrates were at 10 mM concentrations.
DctB is the sensor for dicarboxylates.
A series of models have been proposed to explain the apparent constitutive expression of dctAp in a strain lacking DctA (16, 20, 36, 47, 48). However, there has been one report that a dctA::Tn5-lacZ mutant of S. meliloti does not constitutively express dctA (2). To examine these fundamental contradictions in our understandings of how the Dct system is regulated, expression from dctAp was monitored by using β-galactosidase activity under various growth conditions. Since all these strains are defective for growth on C4-dicarboxylates, induction of the dctAp was tested by growing the strains on glucose-ammonia in the presence of succinate or aspartate. This was possible because glucose does not interfere with induction of dctAp by succinate or aspartate in the wild type (Table 2).
Strain RU727 (3841 dctA::Ω) carrying the dctA reporter fusion pRU103 had a 19- to 33-fold elevation in β-galactosidase when grown in the presence of succinate or aspartate compared to growth on glucose-ammonia (Table 2). This is similar to results for strain 3841, indicating that transcription from dctAp is still inducible in the absence of DctA. To ensure that the inducibility of dctAp is not dependent on the interposon mutant strain RU727, β-galactosidase activity from dctA-lacZ was measured for strains RU436 and RU437, which are dctA::Tn5 derivatives of strain 3841. Both mutant strains showed induction from dctAp after growth on glucose-aspartate or glucose-succinate-ammonia (Table 2). No transcription from dctAp was evident in any strain with dctB, dctD, ΔdctBD, or ΔdctABD mutations, confirming that induction of dctAp is dependent on DctB and DctD (Table 2). Similar results were obtained with R. leguminosarum 3855 (wild type), CR534 (dctA::Tn5), CR535 (dctB::Tn5), and CR538 (dctD::Tn5). These strains were chosen because they are the original R. leguminosarum strains used to characterize the Dct system (35, 37).
Since all the results in Table 2 were obtained by using pRU103, which is a transcriptional β-galactosidase fusion, we tested induction of the dct system in R. leguminosarum by using a translation phoA fusion derived from S. meliloti (48). Plasmid pTH2A was conjugated into R. leguminosarum 3841 and RU727, and the alkaline phosphatase activities were measured. After growth on glucose-NH4Cl, the activities were 13.5 ± 0.3 and 11.6 ± 0.6 nmol min−1 mg of protein−1 for strains 3841 and RU727 containing pTH2A, respectively. After growth on succinate-NH4Cl, the activities were 63.4 ± 0.5 and 59.3 ± 0.7 nmol min−1 mg of protein−1 for strains 3841 and RU727, respectively. Thus, the dctA promoter was induced by a dicarboxylic acid in both strains, indicating that the results are not dependent on the type of reporter fusion used. Furthermore, the S. meliloti and R. leguminosarum dctAp’s respond in the same way.
Induction of the Dct system by nitrogen limitation and osmotic stress.
The above data indicate that DctB acts as a sensor for dicarboxylates and that it does not simply detect the solute binding state of DctA. However, stress-dependent induction of the Dct system has been reported for a dctA strain but not for the wild type of S. meliloti (2). To examine this further, the role of each of the genes of the Dct system in induction by osmotic stress in R. leguminosarum 3841 was investigated.
Transcription from dctAp was examined in cells grown on glucose-ammonia with either no sucrose or 0.05 or 0.1 M sucrose. These concentrations of sucrose did not alter the cells’ growth rate. Strain 3841/pRU103 displayed no increase in transcription from dctAp in response to these conditions, or to higher levels of sucrose (Table 4). However, in the dctA strain (RU727), growth on 0.1 M sucrose increased transcription from dctAp 27-fold relative to growth on no sucrose (Table 4). Transcription from dctAp in strain RU727 grown in the presence of 100 mM NaCl was 1,365 nmol min−1 mg of protein−1. In contrast to this 10-fold induction, strain 3841 was unaffected. However, 100 mM NaCl slows the growth of R. leguminosarum, so most tests were conducted with sucrose. Mutations in dctB (RU730) or dctD (RU711), or deletion of dctBD (RU865) or of the whole dctABD operon (RU714), abolished osmotic induction of dctAp (Table 4).
TABLE 4.
Stress and analog induction of dctAp
| Strain | Genotype | β-Galactosidase activity
under the following growth conditiona:
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Glc-10 mM NH4 | Glc-0.5 mM NH4 | Glc-NH450 mM sucrose | Glc-NH4100 mM sucrose | Glc-NH4itaconic acid | Glc–NH4– Me-Succ | Glc–NH4– DiMe-Succ | Glc-Asn | Glc-AsnNH4 | ||
| 3841 | Wild type | 266 ± 61 | 1,040 ± 230 | 231 ± 38 | 236 ± 41 | 1,317 ± 118 | 199 ± 7 | 202 ± 3 | 290 ± 5 | 229 ± 41 |
| RU727 | dctA::Ω | 297 ± 25 | 1,362 ± 81 | 406 ± 57 | 2,011 ± 322 | 1,889 ± 233 | 1,777 ± 103 | 1,530 ± 214 | 782 ± 88 | 505 ± 71 |
| RU730 | dctB::Ω | 184 ± 27 | 182 ± 43 | 266 ± 14 | 210 ± 8 | 75 ± 21 | 143 ± 27 | 65 ± 28 | 87 ± 63 | |
| RU711 | dctD::Ω | 150 ± 26 | 252 ± 62 | 191 ± 14 | 171 ± 25 | 211 ± 81 | 164 ± 37 | 85 ± 25 | 94 ± 5 | |
| RU865 | ΔdctBD::Ω | 168 ± 10 | 204 ± 44 | 117 ± 12 | 78 ± 21 | |||||
| RU714 | ΔdctABD::Ω | 179 ± 65 | 205 ± 35 | 123 ± 41 | 168 ± 51 | |||||
| RU929 | ntrC::Ω | 139 ± 10 | 93 ± 15 | |||||||
Results are shown as o-nitrophenylgalactoside hydrolyzed (in nanomoles per minute per milligram of protein ± SEM) and are based on at least three independent cultures assayed in triplicate. In all cases, the lacZ fusion is carried by pRU103. Unless otherwise stated, all compounds are at 10 mM concentrations. Glc, glucose; Me-Succ, 2-methyl succinate; DiMe-Succ, 2,2-dimethyl succinate.
The effect of nitrogen limitation on the induction of the Dct system was also examined because of the possible interaction between succinate and aspartate shown above. Furthermore, it has been demonstrated that a mutated allele of ntrC causes constitutive transcription from dctAp (19), and it has been suggested that wild-type NtrC may be capable of causing transcription from dctAp under conditions of nitrogen limitation (11a, 21). Various strains were grown on glucose with a level of ammonia (0.5 mM) shown previously to be limiting for strain 3841 after overnight growth (34). Strains 3841 and RU727 containing pRU103 grown under nitrogen-limited conditions had 9- and 12-fold increases in β-galactosidase activity, respectively (Table 4). Thus, nitrogen limitation leads to induction of dctAp regardless of the presence or absence of DctA. Induction of dctAp by nitrogen limitation is DctB-DctD dependent, since strains with dctB (RU730), dctD (RU711), ΔdctBD (RU865), or ΔdctABD (RU714) mutations did not induce transcription from dctAp in response to nitrogen limitation (Table 4). The increased induction of dctAp in cells grown on succinate-aspartate relative to succinate-aspartate-ammonia may be due to an additive effect of a substrate inducer and nitrogen limitation (see above).
Strain RU929 (ntrC::Ω) containing pRU103 did not show increased β-galactosidase activity in response to nitrogen limitation (Table 4), indicating that NtrC is necessary for induction of transcription from dctAp under these conditions. Therefore, DctB-DctD and NtrC are required for induction of dctAp under conditions of nitrogen limitation.
The ligand specificity of DctB is modified by DctA.
While the above data show that DctB is a sensor for dicarboxylates independent of DctA, the DctB-DctD-dependent osmotic stress activation of dctAp is prevented by the presence of DctA. This suggests that DctA may interact with DctB-DctD to modify its signalling ability. To test this further, we investigated the induction of dctAp by a number of nonmetabolizable analogs of succinate. In transport studies these analogs are able to compete with succinate, albeit weakly, for uptake by DctA (10). 2-Methyl succinate, itaconic acid, or 2,2-dimethyl succinic acid did not support growth of either strain 3841 (wild type) or RU727 (dctA) when present as the sole carbon source, but they did not inhibit the growth of these strains on glucose-ammonia. Therefore, strains carrying pRU103 were tested for induction of transcription from dctAp in response to these analogs when they were provided with glucose-ammonia as the growth substrates. Itaconic acid induced dctAp in strain 3841 to an extent similar to that induced by aspartate (compare Tables 2 and 4), while no induction by 2-methyl succinate, 2,2-dimethyl succinate, or asparagine was evident (Table 4). However, in strain RU727 (dctA), 2-methyl succinate and 2,2-dimethyl succinate induced transcription from dctAp to levels similar to those recorded after growth in the presence of aspartate. Thus, DctA prevents transcription from dctAp by 2-methyl succinate and 2,2-dimethyl succinate. No induction was evident in dctB or dctD strains in response to any compound, indicating that induction by succinate analogs is DctB-DctD dependent (Table 4). While 2,2-dimethyl succinate induced the dctA-lacZ fusion in strain RU727 at 5 mM, even 20 mM concentrations had no effect on strain 3841 (data not shown).
Asparagine, which supports growth of strain 3841 when supplied as a nitrogen source but not as the sole carbon source, was also tested for induction of transcription from dctAp. When cells were grown on glucose-ammonia-asparagine or on glucose-asparagine, no transcription from dctAp was evident either in strain 3841 (wild type) or in strain RU730 (dctB) or RU711 (dctD). However, in strain RU727 (dctA), a fivefold increase in transcription was evident after growth on glucose-asparagine or glucose-ammonia-asparagine. These data show that the DctB-DctD-dependent transcription of dctAp has a lower stereospecificity in the absence of DctA. The inducer profile of DctB is limited or modified by DctA so that in the absence of DctA, DctB-DctD becomes hypersensitive to induction.
Isolation of a constitutive Dct mutant.
In studies of the effect of the Dct system on the aspartate-mediated inhibition of the general l-amino acid permease of strain 3841, random Tn5 mutagenesis was carried out on strain 3841 (34, 44). One mutant (RU150) displayed an unusual phenotype in that it grew very poorly on succinate-ammonia (mean generation time, approximately 30 h) but grew well on succinate-aspartate (mean generation time, 4 h). Furthermore, aspartate does not support growth of strain 3841 as a carbon source (34). Strain RU150 also grew as well as strain 3841 when either glutamate, glutamine, asparagine, serine, alanine, proline, or histidine was supplied as the nitrogen source in conjunction with succinate. Of these amino acids, serine and asparagine did not support growth as the sole carbon/nitrogen source of strain 3841 or RU150. However, strain RU150 grew normally on glucose as the carbon source in conjunction with ammonia. Thus, strain RU150 requires an amino acid as a nitrogen source when growing on a C4-dicarboxylic acid but not on a sugar as the carbon source. This implies that either ammonia assimilation or nitrogen metabolism is disrupted specifically during growth on a C4-dicarboxylic acid.
Strains 3841 and RU150 were grown on glucose-ammonia, and the succinate transport rates were 6 ± 1 and 42 ± 3 nmol min−1 mg of protein−1 (± standard errors of the mean [SEM] from three independent cultures), respectively. After growth on glucose-aspartate, the succinate uptake rates for 3841 and RU150 were 50 ± 5 and 44 ± 4 nmol min−1 mg of protein−1, respectively. Thus, succinate transport is constitutive in strain RU150.
Strain RU150 has Tn5 inserted in the dctA-dctB intergenic region.
Chromosomal DNA from strains 3841 and RU150 was digested with EcoRI (which does not cut in Tn5), Southern blotted, and probed with the plasmid pSUP202::Tn5 and the dct-containing cosmid pIJ1848. A single band of 6.7 kb from strain RU150 hybridized with pSUP202::Tn5, but this band was absent in strain 3841 (data not shown). This indicates that Tn5 inserted once in strain RU150. When chromosomal DNA was hybridized with pIJ1848, a 0.9-kb EcoRI band present in strain 3841 was absent in strain RU150, being replaced by a band at 6.7 kb (data not shown). This 6.7-kb fragment presumably contains Tn5 (5.8 kb) and approximately 0.9 kb of flanking chromosomal DNA.
The generalized transducing phage RL38 was used to transduce Tn5 from strain RU150 into strain 3841 by selection for kanamycin resistance. One hundred kanamycin-resistant transductants had the same growth pattern as RU150 on succinate-ammonia, glucose-ammonia, glucose-aspartate, and succinate-aspartate, indicating 100% cotransduction of the mutation and transposon. This demonstrates that they are closely linked.
The transposon in strain RU150 was cloned, and the flanking DNA was found to have virtually 100% identity at the nucleotide level to the dctA-dctB intergenic region of R. leguminosarum (38). Tn5 had inserted in an AccIII site, 9 bp upstream from the ribosome binding site of dctA. The 9-bp repeats, indicative of a Tn5 insertion, were present on either side of Tn5, duplicating the AccIII site (Fig. 1B). IS50R is adjacent to dctB, while IS50L is adjacent to dctA. The direction of transcription of the genes encoding kanamycin and streptomycin resistance is towards dctB. This allele is referred to below as dctA101.
As succinate uptake is constitutive in strain RU150, the levels of transcription of dctA, dctB, and dctD were examined. Strain RU150 containing the native dctAp reporter probe (pRU103) displayed normal regulation of transcription from dctAp in response to suitable inducers (Table 2). This indicates that the sensor circuit via DctB and DctD, which detects the presence of C4-dicarboxylates and activates transcription of dctA, functions normally in strain RU150.
Reporter fusions for dctA101 (pRU105), dctA101B (pRU106), and dctA101BD (pRU352) were constructed by using the EcoRI clone of the dct intergenic region from strain RU150 carrying Tn5. Expression of β-galactosidase from dctA101 in strains RU150 and 3841 was constitutive at approximately double the level of the native dctAp under noninducing conditions (Table 3). dctA101-lacZ had approximately one-sixth the level of β-galactosidase relative to the native dctAp in cells grown in the presence of succinate or aspartate. These results are consistent with the ability of strain RU150 to transport succinate constitutively. However, they suggest that only a small increase in the level of transcription of dctA is required to obtain a high level of transport activity by DctA. This result was confirmed by Northern blotting (see below).
Expression of dctB from both its native promoter and the putative Tn5 promoter was measured in strains 3841 and RU150. In strain RU150 the native dctBp was expressed constitutively at lower levels than in strain 3841 under all growth conditions (Table 3). Strains 3841 and RU150 containing dctA101B-lacZ displayed similar levels of β-galactosidase on all growth substrates, although the levels were approximately twofold higher than those for the native dctBp (Table 3). This indicates that the level of dctB transcription in strain RU150 is increased slightly in comparison to that in strain 3841. As expected, dctBD-lacZ and dctA101BD-lacZ showed activities similar to those of dctB and dctA101B-lacZ, respectively. The dctA101-lacZ fusion was expressed constitutively in both strains RU730 (dctB::Ω) and RU711 (dctD::Ω), at a level similar to that observed in strains RU150 and 3841 (Table 3). Thus, transcription from dctA101Ap in strain RU150 is independent of DctB and DctD. In summary, the Tn5 insertion in dctA101 results in a low level of constitutive expression of dctA, an approximately twofold increase in transcription of dctB, and a slight increase in transcription of dctD. However, the DctB-DctD signalling pathway still functions in strain RU150, since there is a ligand-specific induction of the native dctAp carried by a plasmid.
The dct-containing cosmid pIJ1848 complemented strain RU150 for normal growth on succinate-ammonia, as did pIJ1969 (pIJ1848-Tn5::dctB) but not pIJ1970 (pIJ1848-Tn5::dctA). Consistent with this, pRU108, which contains dctA alone with the complete dctAB intergenic region, also complemented RU150. These results indicate that increasing the level of dctA enables strain RU150 to grow on succinate-ammonia.
Given that the above data indicate that DctB is a sensor for dicarboxylates, it appears that in strain RU150 the presence of succinate causes DctB-DctD to improperly regulate a heterologous nitrogen-sensing operon. Furthermore, it is the apparent absence of sufficient DctA that causes DctB-DctD to signal improperly when a ligand for DctB is present. The ability of DctA to regulate DctB is similar to its role in modifying the response of the dct operon to osmotic stress and succinate analogs.
The phenotype of strain RU150 is suppressed by chromosomal duplication of dctA.
Strain RU150 was plated on succinate-ammonia agar in the absence of kanamycin, and after 3 to 5 days, spontaneous revertants which grew as well as strain 3841 were obtained. Revertant strains were classified on the basis of their ability to grow in the absence or presence of kanamycin as either primary- or second-site suppressor mutants, respectively. Transduction analysis of the second-site suppressor strains indicates that the reversion in strain RU152-22 is 100% cotransducible with Tn5, while it is unlinked to Tn5 in strains RU152-1 and RU152-14.
Southern blotting of genomic DNA from strains RU152-1, RU152-14, and RU150 with either IS50L (pRU75) or an internal fragment of Tn5 (pRU80) indicates that they contain a single EcoRI-hybridizing fragment of 6.7 kb (data not shown). Strain RU152-22 has a smaller band of approximately 6.3 kb, indicating that a deletion had occurred within Tn5. In addition, strains RU152-1 and RU152-14 contain second, smaller bands of 1.8 and 2.5 kb, respectively, which are homologous to IS50 but not to the central region of Tn5 (data not shown).
The 1.8-kb DNA fragment that contains a second copy of IS50 in strain RU152-14 was amplified by inverse PCR and cloned. The 0.4-kb PCR product, which is the size of the flanking DNA minus IS50 (1.4 kb), has greater than 95% identity at the nucleotide level to the 5′ end of dctA, up to the central EcoRI site. The sequence on the other side of the EcoRI site revealed no significant homology to any published sequences. Thus, in strain RU151-14, IS50L had transposed without internal Tn5 DNA, duplicating at least the 5′ end of dctA.
Chromosomal DNA from strains 3841, RU150, RU152-1, and RU152-14 was digested with SmaI, Southern blotted, and hybridized with dctA (pRU123), IS50L (pRU75), and a DNA fragment located immediately downstream of dctA (pRU402). All three probes gave two hybridizing bands in strains RU152-1 and RU152-14 but only one band in strain RU150, demonstrating that a complete copy of dctA had transposed in the suppressor strains (data not shown).
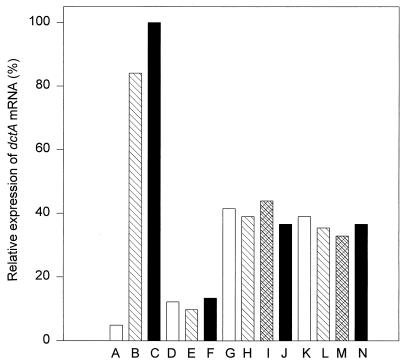
To determine if the extra copy of dctA in strains RU152-1 and RU152-14 results in an increase in transcription, the levels of dctA mRNA from several strains were measured by Northern blotting. After induction with aspartate or succinate, strain 3841 had a 20-fold increase in dctA mRNA relative to the level in glucose-grown cells (Fig. 2). Moreover, strain 3841 grown on succinate-aspartate produced the highest levels of dctA mRNA, which is similar to the result obtained from lacZ fusion analysis of dctAp. Strain RU150 produced dctA mRNA constitutively, at a level higher than that in strain 3841 grown on glucose-ammonia. However, in accord with the lacZ analysis, it was approximately one-eighth of the level in strain 3841 grown on succinate or aspartate. Strains RU152-1 and RU152-14 also produced dctA mRNA constitutively, but the level was significantly elevated in comparison to that in strain RU150. Extra dctA mRNA is presumably made in these strains due to the presence of an extra copy of dctA. The levels of dctA mRNA produced in strains RU152-1 and RU152-14 were slightly different from each other and were 25 to 50% of that observed in strain 3841 grown on succinate-aspartate or glucose-aspartate. Therefore, strain RU150 is complemented for growth on succinate-ammonia either by the presence of dctA on a plasmid or by its duplication in the chromosome.
FIG. 2.
Northern blot of various supressor strains. Lanes A through C, 3841; lanes D through F, RU150; lanes G through J, RU152-1; lanes K through N, RU152-14. Cultures were grown on glucose-ammonia (□), glucose-aspartate (▧), succinate-aspartate (▪), or succinate-ammonia (▩). Northern blots were scanned with a densitometer, and the values shown are relative to that for strain 3841 grown on succinate-aspartate.
Mutation of dctB or dctD in strain RU150 restores normal growth.
In order to map the location of the mutation in strain RU152-22, the 6.2-kb EcoRI fragment containing Tn5 was cloned from the chromosome into Bluescript II SK+ by selecting for kanamycin resistance (pRU93). Mapping revealed a 0.5-kb deletion, including the region between the intergenic SalI site and the first 300 bp of dctB, which removes the promoter and 5′ end of dctB. This will prevent expression of DctB and presumably DctD, since it requires dctBp. Thus, DctB and DctD play a key role in preventing strain RU150 from using ammonia as a nitrogen source when succinate is the sole carbon source for growth.
To confirm this, a series of directed dctA, dctB, and dctD double mutants were constructed in strain RU150. Strain RU938 (RU150 dctA::Ω) grew normally on glucose-ammonia and glucose-aspartate but did not grow on succinate-ammonia or succinate-aspartate due to mutation of DctA. Strains RU150, RU875 (RU150 dctB::Ω), RU720 (RU150 dctD::Ω), and RU938 (RU150 ΔdctBD::Ω) all grew on glucose-ammonia, glucose-aspartate, and succinate-aspartate. In contrast to strain RU150, the three double mutants also grew well on succinate-ammonia. Thus, strain RU150 can be rescued for growth on succinate-ammonia either by an increase in the transcription of dctA or by mutation of dctB or dctD.
Plant properties.
Strain RU150 formed white nodules on Vicia sativa. Plants inoculated with strain RU150 and grown on nitrogen-free medium were yellow and did not reduce acetylene, while strain 3841 reduced acetylene at 0.41 μmol h−1 plant−1. These data indicate that strain RU150 is unable to fix nitrogen.
DISCUSSION
The data presented here confirm the results of previous studies showing that induction of dctAp requires DctB and DctD, while the dctB promoter is constitutive (6, 16, 35, 38, 46, 48). However, lacZ fusion analysis in this study clearly shows that dctD is transcribed from the dctB promoter. Our finding that the dctB promoter also drives expression of dctD is consistent with the translational start site of dctD being 5 bp downstream from the stop codon for dctB. This is in marked contrast to the conclusion of a previous study with S. meliloti Rm2011 that dctD has its own promoter, which is severalfold stronger than the dctB promoter (16). While this may be due to different levels of regulation in the two species, the fusion used to show that dctD has its own promoter in S. meliloti has a transposon located upstream of it in dctB. As with strain RU150 in this study, such an upstream transposon may provide an alternative promoter. In several studies, dctB mutants have been complemented with plasmids that do not contain intact copies of dctD, suggesting that dctD has its own promoter (46, 48). We have also complemented dctB mutants with plasmids lacking an intact dctD or complemented dctD mutants with plasmids containing insertions in dctB. However, these results are difficult to interpret because even weak plasmid or transposon promoters may cause sufficient transcription of a regulator protein such as DctD, which is presumably required only in very small amounts in the cell. Indeed, many of the complex allele-specific effects of chemical and transposon mutants of dctB may be caused by differential transcription of dctD, assuming that it normally uses the dctB promoter (24, 38, 46).
A key question regarding the regulation of expression of dctA concerns the relative roles of DctA and DctB. In the first study in which this was examined, R. leguminosarum CR534 (dctA::Tn5) had constitutive expression from a dctA-lacZ fusion in cells grown on glucose-ammonia (36). The level of expression of the dctA-lacZ fusion in strain CR534 was approximately one-half the level found in the wild type grown on succinate-ammonia. When this dctA strain was grown in the presence of succinate, a further twofold increase in transcription from dctAp was apparent. Thus, while expression from dctAp was deregulated in the absence of DctA, succinate increased the induction (36). In S. meliloti transcription from a plasmid-borne dctA-phoA translational fusion was constitutive in a dctA strain grown on glucose-ammonia (48). The levels were 20-fold higher than that in the wild type grown on glucose-ammonia and were double that in the wild type grown on succinate-ammonia. Transcription from dctAp in the dctA strain still increased by 25% after growth in the presence of succinate (48). It was also shown that constitutive expression from dctAp was dependent on DctB and DctD, as dctA dctB and dctA dctD double mutants had only background levels of transcription (48). In another study, a dctA-phoA fusion was constitutively expressed in a dctA strain at levels sixfold higher than those in the wild type grown on succinate-ammonia (16). This led to models in which DctB detects the presence of inducing ligands indirectly via the solute binding state of DctA. However, one study with S. meliloti showed that a chromosomal Tn5-lacZ fusion was not constitutive (2). It was also found that dctAp could be induced by osmotic stress and calcium limitation. We therefore reexamined this question with R. leguminosarum and found that dctAp remains inducible in a dctA strain. Most measurements were made by using a transcriptional lacZ fusion, but the same results were obtained with a translational phoA fusion to the S. meliloti dctA gene. Thus, the reported differences are not due to either the species or the type of fusion. Instead, dctAp becomes highly sensitive to induction in the absence of DctA. This can be seen in at least two different ways. Firstly, dctAp becomes sensitive to cross-induction by stimuli other than a dicarboxylate; an example of this is induction by osmotic stress. Secondly, the specificity for dicarboxylate ligands is much wider in a dctA strain, indicating that DctB is activated by a wider range of molecules (Table 4). It should be noted that DctB and DctD are needed for induction under all these conditions. Models requiring that DctB detect the solute binding state of DctA are not consistent with these data. Instead, DctB must act as a sensor for dicarboxylates, which is compatible with the presence of a large putative periplasmic loop having a ligand binding site. However, it is clear that DctA modifies either ligand binding or the signalling state of DctB. One possibility is that DctA interacts with DctB in the membrane and alters the conformation of DctB, changing its stereospecificity for ligand binding. An alternative is that the ligand binding stereospecificity of DctB is unaltered but that DctA alters the signalling ability of DctB. For example, DctA might interact with DctB, altering its relative kinase and phosphatase activities. There are numerous ways in which protein-protein interaction could do this, including altering the dimerization of DctB. The physiological and genetic approaches used in this study do not directly address the nature of the possible interaction between DctA and DctB. This would require a physical approach, which is made complex because both are integral membrane proteins. However, the possibility that DctA interacts with DctB and alters its proposed kinase or phosphatase activity might explain why in a dctA strain DctB and DctD cross-regulate heterologous operons (see below). It should be noted that while physical interaction between DctA and DctB is among the simplest explanations for these data, there is no direct evidence for this. The apparent contradictions in the literature regarding the inducibility of dctAp in a dctA strain may be due to the hypersensitivity of DctB-DctD in the absence of DctA, where even small differences in media and growth conditions can lead to apparent constitutive expression of dctA reporter fusions.
The isolation of strain RU150 allowed the importance of DctA in regulation of DctB-DctD to be examined because it uncoupled expression of DctA from DctB-DctD. In this strain expression of DctA is driven from a putative transposon promoter where the transcription of dctA is locked at a constitutive low level. This strain shows wild-type transport rates for dicarboxylates, which indicates that the higher levels of transcription measured in the wild type may not be needed to generate sufficient protein complex for transport. Consistent with this, it has been shown that in S. meliloti fivefold overexpression of dctA from a trp promoter does not result in an increase in the rate of succinate transport (32). One possibility is that higher levels of transcription of dctA increase the level of the DctA protein, which is needed to regulate DctB. The locking of the level of dctA transcription has a profound effect on signalling by DctB-DctD. In this mutant, DctB-DctD still shows normal transcriptional control of a plasmid copy of a dctA-lacZ fusion. This is important because it shows that the dicarboxylate-dependent induction of phosphorylation of DctB-DctD still functions. What is notable is that DctB-DctD now appears to cause improper regulation of heterologous operons. Thus, when a dicarboxylate is included in the growth medium, which will lead to phosphorylation of DctD, strain RU150 is unable to grow properly unless an organic nitrogen source is present. This effect is prevented by mutation of either DctB or DctD, which, together with the requirement for the presence of a dicarboxylate to prevent growth on ammonia, suggests that the improper regulation is caused by phosphorylated DctD. Increasing the level of transcription of dctA carried either by a plasmid or by chromosomal duplication reestablishes proper control of DctB-DctD, enabling strain RU150 to grow on succinate-ammonia. Given the conserved structure of ς54-dependent activators, such as DctD, NtrC, and NifA, it is not remarkable that DctD might be capable of interfering with the regulation of other sensing operons. Indeed, it has been shown several times before that transposon and chemical mutants of the dct system can have complex regulatory effects on other signalling systems (24, 38, 46). We do not know what system is being interfered with in strain RU150; however, the NtrC-dependent promoter for glnII was apparently regulated normally in strain RU150 (33). An important feature of this study is the demonstration that DctA has two roles in the cell; the first is as a transport protein such that DctA expressed constitutively in strain RU150 with a deletion in dctB or dctD will transport and permit growth on succinate. The second role appears to be regulation of DctB-DctD. While it is not clear how DctA regulates DctB-DctD, we suggest above that DctA may modify the kinase or phosphatase activity of DctB. If DctB-DctD becomes more sensitive to phosphorylation in the absence of DctA, then other stimuli, such as osmotic pressure, might cause activation of dctAp via cross talk. Increased sensitivity of DctB-DctD to phosphorylation could cause excessive phosphorylated DctD to improperly regulate other operons. Given the conserved structure of regulators such as DctD, NtrC, and NifA, which may have arisen by gene duplication, it is an interesting possibility that the unique product of an operon may impose signalling specificity. In this context DctA attenuates cross talk with other systems.
ACKNOWLEDGMENTS
We thank the University of Reading endowment trust for supporting this research.
We thank T. Finan for providing plasmid pTH2A.
REFERENCES
- 1.Arwas R, McKay I A, Rowney F R P, Dilworth M J, Glenn A R. Properties of organic acid utilization mutants of Rhizobium leguminosarumstrain 300. J Gen Microbiol. 1985;131:2059–2066. [Google Scholar]
- 2.Batista S, Castro S, Aguilar O M, Martinezdrets G. Induction of C4-dicarboxylate transport genes by external stimuli in Rhizobium meliloti. Can J Microbiol. 1992;38:51–55. doi: 10.1139/m92-008. [DOI] [PubMed] [Google Scholar]
- 3.Beringer J E. R factor transfer in Rhizobium leguminosarum. J Gen Microbiol. 1974;84:188–198. doi: 10.1099/00221287-84-1-188. [DOI] [PubMed] [Google Scholar]
- 4.Buchanan-Wollaston V. Generalized transduction in Rhizobium leguminosarum. J Gen Microbiol. 1979;112:135–142. [Google Scholar]
- 5.Engelke T, Jagadish M N, Pühler A. Biochemical and genetical analysis of Rhizobium meliloti mutants defective in C4-dicarboxylate transport. J Gen Microbiol. 1987;133:3019–3029. [Google Scholar]
- 6.Engelke T, Jording D, Kapp D, Pühler A. Identification and sequence analysis of the Rhizobium meliloti dctA gene encoding the C4-dicarboxylate carrier. J Bacteriol. 1989;171:5551–5560. doi: 10.1128/jb.171.10.5551-5560.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Finan T M, Oresnik I, Bottacin A. Mutants of Rhizobium melilotidefective in succinate metabolism. J Bacteriol. 1988;170:3396–3403. doi: 10.1128/jb.170.8.3396-3403.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finan T M, Wood J M, Jordan D C. Symbiotic properties of C4-dicarboxylic acid transport mutants of Rhizobium leguminosarum. J Bacteriol. 1983;154:1403–1413. doi: 10.1128/jb.154.3.1403-1413.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giblin L, Archdeacon J, Ogara F. Modular structure of the Rhizobium melilotiDctB protein. FEMS Microbiol Lett. 1996;139:19–25. doi: 10.1111/j.1574-6968.1996.tb08174.x. [DOI] [PubMed] [Google Scholar]
- 10.Glenn A R, Poole P S, Hudman J F. Succinate uptake by free-living and bacteroid forms of Rhizobium leguminosarum. J Gen Microbiol. 1980;119:267–271. [Google Scholar]
- 11.Gu B H, Lee J H, Hoover T R, Scholl D, Nixon B T. Rhizobium meliloti dctD, a ς54-dependent transcriptional activator, may be negatively controlled by a subdomain in the C-terminal end of its two-component receiver module. Mol Microbiol. 1994;13:51–66. doi: 10.1111/j.1365-2958.1994.tb00401.x. [DOI] [PubMed] [Google Scholar]
- 11a.Gu, B. H., and B. T. Nixon. Unpublished data.
- 12.Huala E, Stigter J, Ausubel F M. The central domain of Rhizobium leguminosarumDctD functions independently to activate transcription. J Bacteriol. 1992;174:1428–1431. doi: 10.1128/jb.174.4.1428-1431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang J, Gu B, Albright L M, Nixon B T. Conservation between coding and regulatory elements of Rhizobium meliloti and Rhizobium leguminosarum dctgenes. J Bacteriol. 1989;171:5244–5253. doi: 10.1128/jb.171.10.5244-5253.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnston A W B, Beringer J E. Identification of the Rhizobiumstrains in pea root nodules using genetic markers. J Gen Microbiol. 1975;87:343–350. doi: 10.1099/00221287-87-2-343. [DOI] [PubMed] [Google Scholar]
- 15.Jording D, Pühler A. The membrane topology of the Rhizobium meliloti C4-dicarboxylate permease (DctA) as derived from protein fusions with Escherichia coliK12 alkaline-phosphatase (PhoA) and beta-galactosidase (lacZ) Mol Gen Genet. 1993;241:106–114. doi: 10.1007/BF00280207. [DOI] [PubMed] [Google Scholar]
- 16.Jording D, Sharma P K, Schmidt R, Engelke T, Uhde C, Pühler A. Regulatory aspects of the C4-dicarboxylate transport in Rhizobium meliloti—transcriptional activation and dependence on effective symbiosis. J Plant Physiol. 1992;141:18–27. [Google Scholar]
- 17.Jording D, Uhde C, Schmidt R, Pühler A. The C4-dicarboxylate transport-system of Rhizobium melilotiand its role in nitrogen-fixation during symbiosis with alfalfa (medicago-sativa) Experientia. 1994;50:874–883. [Google Scholar]
- 18.Krikos A, Mutoh N, Boyd A, Simon M I. Sensory transducers of Escherichia coliare composed of discrete structural and functional domains. Cell. 1983;33:615–622. doi: 10.1016/0092-8674(83)90442-7. [DOI] [PubMed] [Google Scholar]
- 19.Labes M, Rastogi V, Watson R, Finan T M. Symbiotic nitrogen fixation by a nifA deletion mutant of Rhizobium meliloti: the role of an unusual ntrCallele. J Bacteriol. 1993;175:2662–2673. doi: 10.1128/jb.175.9.2662-2673.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ledebur H, Gu B, Sojda III J, Nixon B T. Rhizobium meliloti and Rhizobium leguminosarum dctD gene products bind to tandem sites in an activation sequence located upstream of ς54-dependent dctApromoters. J Bacteriol. 1990;172:3888–3897. doi: 10.1128/jb.172.7.3888-3897.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ledebur H, Nixon B T. Tandem DctD-binding sites of the Rhizobium meliloti dctAupstream activating sequence are essential for optimal function despite a 50-fold to 100-fold difference in affinity for DctD. Mol Microbiol. 1992;6:3479–3492. doi: 10.1111/j.1365-2958.1992.tb01783.x. [DOI] [PubMed] [Google Scholar]
- 22.Lee J H, Hoover T R. Protein cross-linking studies suggest that Rhizobium meliloti C4-dicarboxylic acid transport protein-d, a ς54-dependent transcriptional activator, interacts with sigma(54)-subunit and the beta-subunit of RNA polymerase. Proc Natl Acad Sci USA. 1995;92:9702–9706. doi: 10.1073/pnas.92.21.9702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J H, Scholl D, Nixon B T, Hoover T R. Constitutive ATP hydrolysis and transcription activation by a stable, truncated form of Rhizobium meliloti DctD, a ς54-dependent transcriptional activator. J Biol Chem. 1994;269:20401–20409. [PubMed] [Google Scholar]
- 24.Mavridou A, Barny M A, Poole P, Plaskitt K, Davies A E, Johnston A W B, Downie J A. Rhizobium leguminosarum nodulation gene (nod) expression is lowered by an allele-specific mutation in the dicarboxylate transport gene dctB. Microbiology. 1995;141:103–111. doi: 10.1099/00221287-141-1-103. [DOI] [PubMed] [Google Scholar]
- 25.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- 26.Park C, Hazelbauer G L. Mutations specifically affecting ligand interaction of the Trg chemosensory transducer. J Bacteriol. 1986;167:101–109. doi: 10.1128/jb.167.1.101-109.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poole P S, Blyth A, Reid C J, Walters K. myo-Inositol catabolism and catabolite regulation in Rhizobium leguminosarum bv viciae. Microbiology. 1994;140:2787–2795. [Google Scholar]
- 28.Poole P S, Franklin M, Glenn A R, Dilworth M J. The transport of l-glutamate by Rhizobium leguminosaruminvolves a common amino acid carrier. J Gen Microbiol. 1985;131:1441–1448. [Google Scholar]
- 29.Poole P S, Schofield N A, Reid C J, Drew E M, Walshaw D L. Identification of chromosomal genes located downstream of dctD that affect the requirement for calcium and the lipopolysaccharide layer of Rhizobium leguminosarum. Microbiology. 1994;140:2797–2809. doi: 10.1099/00221287-140-10-2797. [DOI] [PubMed] [Google Scholar]
- 30.Prentki P, Krisch H M. In vitroinsertional mutagenesis with a selectable DNA fragment. Gene. 1984;29:303–313. doi: 10.1016/0378-1119(84)90059-3. [DOI] [PubMed] [Google Scholar]
- 31.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in Gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 32.Rastogi V, Labes M, Finan T, Watson R. Overexpression of the dctA gene in Rhizobium meliloti—effect on transport of C4dicarboxylates and symbiotic nitrogen fixation. Can J Microbiol. 1992;38:555–562. doi: 10.1139/m92-091. [DOI] [PubMed] [Google Scholar]
- 33.Reid C J. Ph.D. thesis. Reading, United Kingdom: University of Reading; 1995. [Google Scholar]
- 34.Reid C J, Walshaw D L, Poole P S. Aspartate transport by the Dct system in Rhizobium leguminosarumnegatively affects nitrogen-regulated operons. Microbiology. 1996;142:2603–2612. doi: 10.1099/00221287-142-9-2603. [DOI] [PubMed] [Google Scholar]
- 35.Ronson C W. Genetic regulation of C4-dicarboxylate transport in rhizobia. In: Bothe H, Bruijn F J, Newton W E, editors. Nitrogen fixation: hundred years after. New York, N.Y: Gustav Fischer; 1988. pp. 547–551. [Google Scholar]
- 36.Ronson C W, Astwood P M. Genes involved in the carbon metabolism of bacteroids. In: Evans H J, Bottomley P J, Newton W E, editors. Nitrogen fixation research progress. Dordrecht, The Netherlands: Martinus Nijhoff; 1985. pp. 201–207. [Google Scholar]
- 37.Ronson C W, Astwood P M, Downie J A. Molecular cloning and genetic organization of C4-dicarboxylate transport genes from Rhizobium leguminosarum. J Bacteriol. 1984;160:903–909. doi: 10.1128/jb.160.3.903-909.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ronson C W, Astwood P M, Nixon B T, Ausubel F M. Deduced products of C4-dicarboxylate transport regulatory genes of Rhizobium leguminosarumare homologous to nitrogen regulatory gene products. Nucleic Acids Res. 1987;15:7921–7934. doi: 10.1093/nar/15.19.7921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ronson C W, Lyttleton P, Robertson J G. C4-dicarboxylate transport mutants of Rhizobium trifolii form ineffective nodules on Trifolium repens. Proc Natl Acad Sci USA. 1981;78:4284–4288. doi: 10.1073/pnas.78.7.4284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Scholl D, Nixon B T. Cooperative binding of DctD to the dctA upstream activation sequence of Rhizobium melilotiis enhanced in a constitutively active truncated mutant. J Biol Chem. 1996;271:26435–26442. doi: 10.1074/jbc.271.42.26435. [DOI] [PubMed] [Google Scholar]
- 42.Scholl D, Nixon T. Cooperative binding of DctD to the dctApromoter region. Protein Eng. 1995;8:75. [Google Scholar]
- 43.Simon R, Priefer U, Pühler A. A broad-host-range mobilization system for in vivogenetic engineering: transposon mutagenesis of Gram-negative bacteria. Bio/Technology. 1983;1:784–791. [Google Scholar]
- 44.Walshaw D L, Poole P S. The general l-amino acid permease of Rhizobium leguminosarumis an ABC uptake system that influences efflux of solutes. Mol Microbiol. 1996;21:1239–1252. doi: 10.1046/j.1365-2958.1996.00078.x. [DOI] [PubMed] [Google Scholar]
- 45.Wang Y-K, Hoover T R. Alterations within the activation domain of the ς54-dependent activator DctD that prevent transcriptional activation. J Bacteriol. 1997;179:5812–5819. doi: 10.1128/jb.179.18.5812-5819.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watson R J. Analysis of the C4-dicarboxylate transport genes of Rhizobium meliloti: nucleotide sequence and deduced products of dctA, dctB and dctD. Mol Plant-Microbe Interact. 1990;3:174–181. doi: 10.1094/mpmi-3-174. [DOI] [PubMed] [Google Scholar]
- 47.Watson R J, Rastogi V K, Chan Y K. Aspartate transport in Rhizobium meliloti. J Gen Microbiol. 1993;139:1315–1323. [Google Scholar]
- 48.Yarosh O K, Charles T C, Finan T M. Analysis of C4-dicarboxylate transport genes in Rhizobium meliloti. Mol Microbiol. 1989;3:813–823. doi: 10.1111/j.1365-2958.1989.tb00230.x. [DOI] [PubMed] [Google Scholar]