Abstract
The O antigen is a polymer with a repeated unit. The chain length in most Escherichia coli strains has a modal value of 10 to 18 O units, but other strains have higher or lower modal values. wzz (cld/rol) mutants have a random chain length distribution, showing that the modal distribution is determined by the Wzz protein. Cloned wzz genes from E. coli strains with short (7 to 16), intermediate (10 to 18), and long (16 to 25) modal chain lengths were transferred to a model system, and their effects on O111 antigen were studied. The O111 chain length closely resembled that of the parent strains. We present data based on the construction of chimeric wzz genes and site-directed mutagenesis of the wzz gene to show that the modal value of O-antigen chain length of E. coli O1, O2, O7, and O157 strains can be changed by specific amino acid substitutions in wzz. It is concluded that the O-antigen chain length heterogeneity in E. coli strains is the result of amino acid sequence variation of the Wzz protein.
Lipopolysaccharide (LPS), also known as endotoxin, is a major component of the outer membrane of gram-negative bacteria. It plays a very important role as a permeability barrier and protects bacteria from phagocytosis, detergents, and free fatty acids present in the mammalian colon (7, 23). LPS consists of three different regions: lipid A, core oligosaccharide, and O antigen. Lipid A and core oligosaccharide are synthesized together, while the O antigen is synthesized independently. The mechanisms for synthesis and polymerization of O antigens are relatively well known and have been reviewed many times (26, 38).
The O antigen consists of an oligosaccharide repeat unit of about two to six sugars, polymerized to give O antigen. The number of O units per molecule has a characteristic modal distribution which has been shown in several systems to be determined by the chain length determinant Wzz (previously Cld or Rol) protein, as wzz mutants have a random chain length distribution (3, 6, 12, 22). The wzz gene is located on the chromosome between the gnd and his genes. The predicted secondary structure of the Wzz protein has a large, hydrophilic loop with potential transmembrane helices at each end (3), and Morona et al. (22) demonstrated that the loop is in the periplasmic space. The precise mode of action of the Wzz protein is unknown, but it has been proposed to interact with O-antigen polymerase (Wzy, previously Rfc) in the periplasm (3) to control Wzy-mediated polymerization. It should be noted that while most O antigens are thought to be polymerized by Wzy with chain length determined by Wzz, there are a few O antigens with other modes of polymerization (39).
The O-antigen chain length of LPS for a strain is usually determined by observation of silver-stained gels of LPS, when a ladder of bands is apparent, each being of LPS with a specific number of O units. When LPSs with different O antigens are compared, one often finds that the spacing between bands varies. This is because the size of the O units varies, which affects the mass difference between LPSs in adjacent bands. Most of the LPS is in molecules with chain lengths around the modal value, but there is often a significant amount of short-chain (one to three O units) LPS. However, there is usually sufficient material, in at least some gels, in all bands up to the main group of bands to ascertain by counting bands the number of O units in the LPS of each band.
The LPS of most Escherichia coli strains has a basic O-antigen chain length of 10 to 18 O units (9, 19, 24). However, there is considerable variation, and in some cases, chain length varies among strains with the same O antigen (1, 10, 14). The LPSs of E. coli can, for convenience, be subdivided into three groups having short (7 to 16), intermediate (10 to 18), and long (16 to 25) O-antigen chains. Bastin et al. (3) have shown that wzz genes from different strains can confer different chain lengths on O111 antigen in a model system using E. coli K-12 carrying O111 genes. Use of the one O antigen for the three modes means that all tracks have the same band spacing. In this study, we used this model system to study the wzz genes from O1, O2, O7, and O157 strains, and also modification of chain length after site-directed mutagenesis of wzz, to show that substitution of one amino acid in the Wzz protein can change its modal-value specificity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and antisera.
The bacterial strains and plasmids used in this study are described in Tables 1 and 2. Bacteria were grown in nutrient broth (10 g of peptone [Amyl Media Pty., Dandenong, Victoria, Australia]), 5 g of yeast extract [Amyl Media], and 5 g of NaCl per liter of water adjusted to pH 7.2). This broth was supplemented with ampicillin at a final concentration of 25 μg ml−1 and kanamycin at a concentration of 50 μg ml−1 when necessary.
TABLE 1.
Bacterial strains used in this study
| Strain | Antigenic characteristics | Chain length (O units)a | Source or reference |
|---|---|---|---|
| M70/1-1 | O1:K1:H7 | 7–16 | 19 |
| F186 | O2:K1:H5 | 7–16 | 1 |
| E4991/76 | O2:K1:H6 | 10–18 | 1 |
| Bi7509-41 | O7:K1:H- | 7–16 | 19 |
| 79/311 | O7:K1:H7 | 10–18 | 19 |
| EDL933 | O157:H7 | 10–18 | 37 |
| C664-1992 | O157:H7 | 10–18 | Statens Seruminstitutb |
| C258-94 | O157:H39 | 16–25 | Statens Seruminstitut |
| C722-89 | O157:K52:H45 | 16–25 | Statens Seruminstitut |
| P4657c | 4 |
Chain length categories: short, 7 to 16 O units; intermediate, 10 to 18 O units; long, 16 to 25 O units.
Copenhagen, Denmark.
P4657 is K-12 strain Sφ874, which has a deletion of the O-antigen cluster, plus plasmid pPR1231 carrying the O111-antigen gene cluster.
TABLE 2.
Plasmids used in this study
| Plasmid | Characteristic(s) | Source or reference |
|---|---|---|
| pPR1231 | E. coli O111-antigen cluster; wzz | 4 |
| pPR1751 | wzz clone of EDL 933 | This work |
| pPR1752 | E4991/76-F186 wzz hybrid clone | This work |
| pPR1753 | wzz clone of E4991/76 | This work |
| pPR1754 | wzz clone of F186 | This work |
| pPR1755 | wzz clone of M70/1-1 | This work |
| pPR1756 | wzz clone of C664-1992 | This work |
| pPR1775 | Additional glycine 221 mutant of pPR1753 | This work |
| pPR1776 | V224I of pPR1753 | This work |
| pPR1793 | wzz clone of 79/311 | This work |
| pPR1794 | wzz clone of Bi7509-41 | This work |
| pPR1795 | wzz clone of C258-94 | This work |
| pPR1796 | Long-chain mode wzz clone of C722-89 | This work |
| pPR1798 | C258-94–E4991/76 wzz hybrid clone | This work |
| pPR1801 | M77I of pPR1753 | This work |
| pPR1802 | Q83S of pPR1753 | This work |
| pPR1803 | D90E of pPR1753 | This work |
| pPR1804 | D90E-L91I of pPR1753 | This work |
| pPR1815 | L91I of pPR1753 | This work |
E. coli O antisera were used to confirm the serotype of each strain. Antisera against the O157 antigen was from Denka Seiken Co. Ltd., Tokyo, Japan. Antisera against the O1, O2, and O7 antigens were from the Institute of Medical and Veterinary Science, Adelaide, South Australia, Australia.
LPS analysis.
A modified version of the proteinase K extraction method (15) was used for LPS preparation. Cultures (10 ml) were incubated overnight at 37°C with shaking. These were centrifuged at approximately 10,000 × g for 5 min, and the pellet was washed once with 10 mM Tris (pH 8) and resuspended in 10 mM Tris (pH 8) to an extinction at 625 nm of 0.6 to 0.7. Subsequently, 1.5 ml of the cell suspension was centrifuged for 5 min in an Eppendorf tube. The pellet was drained, resuspended in 40 μl of electrophoresis buffer (20), boiled for 10 min, digested with 100 μg of proteinase K (Boehringer Mannheim) at 63 to 65°C for 2 h, and subsequently boiled for 5 min to denature any traces of proteinase K. The samples were loaded on a sodium dodecyl sulfate (SDS)–12.5% polyacrylamide gel electrophoresis (PAGE) gel as described by Lugtenberg et al. (20) and run for 3 h at 50 mA. Silver staining was performed as described by Fomsgaard et al. (11), except that the developer consisted of 0.28 M sodium carbonate with 0.05% formaldehyde as described by Aucken and Pitt (2).
DNA methods.
Restriction endonucleases and ligase were obtained from Boehringer Mannheim Biochemicals. Taq polymerase was obtained from Pharmacia Biotech. The PCR was carried out as described by Saiki et al. (30), by using a DNA thermal cycler (Perkin-Elmer Cetus, Norwalk, Conn.). Oligonucleotide primers were based on the wzz gene sequence of E. coli HU1124 (5) (GenBank accession no. M89934) and synthesized by Beckman (Gladesville, New South Wales, Australia) or Auspep (Parkville, Victoria, Australia). The wzz genes from E. coli O1, O2, O7, and O157 strains were amplified by primers 465 and 466 (Fig. 1), and PCR products were purified by using the Wizard PCR purification system (Promega), digested with EcoRI and BamHI, and subsequently cloned into homologous sites of pUC18. The sequence of bases 16 to 963 of the 978-bp wzz gene was determined for the first nine strains listed in Table 1, as well as for all of the mutants described in this paper by using the dye-labelled primer technique and a Perkin-Elmer Catalyst 800 and an automated 377 DNA sequencer (Applied Biosystems) at the Sydney University and Prince Alfred Macromolecular Analysis Centre, Sydney, New South Wales, Australia.
FIG. 1.
Primers used for wzz gene amplification and site-directed mutagenesis. Primers 466 and 465 were based on the wzz gene sequence of HU1124 (5), with the addition of EcoRI and BamHI sites at the 5′ end, respectively. All of the other primers were based on the wzz gene of E4991/76. Primer 854 contains a PstI site and was used to create a chimeric wzz gene from C258-94 and E4991/76. The primers used for site-directed mutagenesis were 704, in which a glycine residue is inserted between amino acid residues 220 and 221; 705, in which isoleucine replaces valine at residue 224; 881, in which isoleucine replaces methionine at residue 77; 882, in which serine replaces glutamine at residue 83; 883, in which glutamate replaces aspartate at residue 90; 945, in which glutamate and isoleucine replace aspartate and leucine at residues 90 and 91; and 988, in which isoleucine replaces leucine at residue 91. The base substitutions are in boldface, and the PstI site in primer 854 is indicated by boldface and italics.
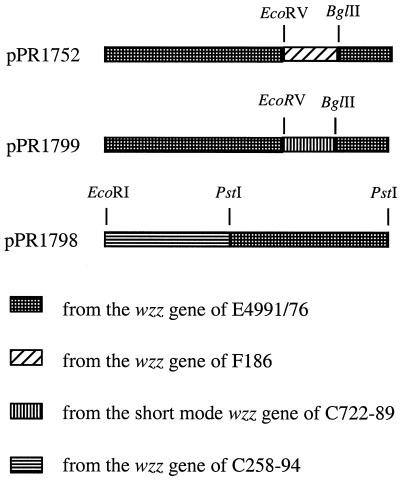
For the construction of wzz chimeric genes, DNA was subjected to restriction enzyme analysis, run on a 1.5% low-melting-point agarose gel (31), and recovered by using the Bandpure (Progen Industries Limited) protocol with the following modifications. The resin-bound DNA was decanted into a Bresaspin column unit (Bresatec Pty. Ltd., Adelaide, South Australia, Australia) and centrifuged at 10,000 × g for 30 s in a 000-MICR-190 microcentrifuge tube (Elkay, Shrewsbury, Mass.) at room temperature. The column was subsequently washed three times by centrifugation with 250 μl of the Bandpure ethanol wash solution. After the third wash, the column was centrifuged again for 3 min to remove any residue of the ethanol wash solution and the column was then transferred into another tube. The silica resin was resuspended three times with the same 60-μl aliquot of 60°C deionized water and incubated at 60°C for 5 min in a clean microcentrifuge tube. The column was centrifuged for 30 s at room temperature. An aliquot (8 to 10 μl) of the eluate was run on another gel to check the percentage recovered. The construction of chimeric wzz genes in plasmids pPR1752 and pPR1798 is described in Fig. 2.
FIG. 2.
Construction of chimeric wzz genes. The chimeric wzz gene in pPR1752 was constructed by replacing the 210-bp BglII-EcoRV fragment of the wzz gene of E4991/76 in pPR1753 with that of F186. The chimeric wzz gene in pPR1798 was made in two steps: a 424-bp EcoRI-PstI fragment, which was amplified from the wzz gene of C258-94 by using primers 466 and 854 and digested with restriction enzymes, was cloned into pUC18; subsequently, a 554-bp PstI fragment of the wzz gene of E4991/76 was cloned into the PstI site of the resulting plasmid in the right orientation for expression.
Plasmid pPR1753 carrying the wzz gene of E4991/76 was subjected to mutagenesis with a U.S.E site-directed mutagenesis kit obtained from Pharmacia Biotech. The conditions and buffers used were those recommended by the manufacturers; the primers used are listed in Fig. 1.
Phylogenetic analysis.
DNA and deduced amino acid sequences were derived and analyzed by using the Australian National Genomic Information Service (ANGIS) at Sydney University (29). The MULTICOMP program (28) was used for comparison and alignment of sequences. A phylogenetic tree was constructed by using the neighbor-joining method.
Nucleotide sequence accession numbers.
The GenBank accession numbers for the wzz gene sequences are AF011910 for E4991/76, AF011911 for F186, AF011912 for M70/1-1, AF011913 for 79/311, AF011914 for Bi7509-41, AF011915 for C664-1992, AF011916 for C258-94, AF011917 for C722-89, and AF011919 for EDL933. The GenBank accession numbers for the G7 and Bi316-41 wzz genes sequences are U39305 and U39306, respectively.
RESULTS
Cloning of wzz genes and effects of Wzz on chain length specificity.
Strain P4657 (Table 1) is a K-12 derivative which lacks the chromosomal O-antigen genes but carries those of E. coli O111 on a plasmid (3). It does not carry a wzz gene and so has a nonmodal distribution of O-antigen chain lengths (Fig. 3, track 8). This strain can be used to determine the effects of different wzz genes on chain length distribution in a common genetic background.
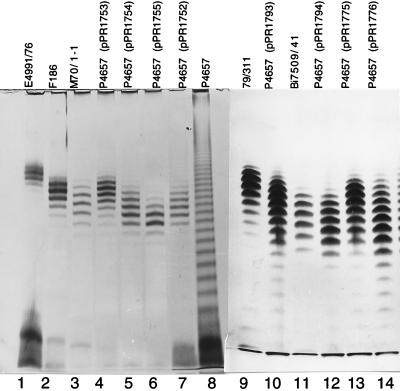
FIG. 3.
Analysis of short and intermediate chain length Wzz proteins. LPS was prepared by proteinase K digestion and analyzed by SDS–12.5% PAGE and silver staining. Tracks 1 to 8 and 9 to 14 are from two different gels. The strain used for each track is indicated above the track.
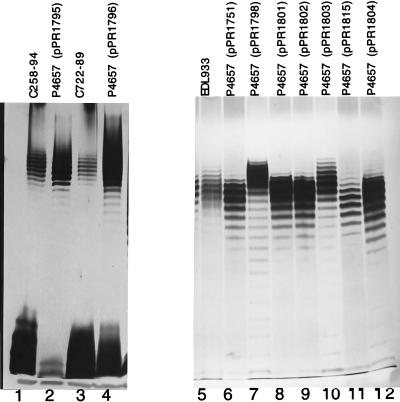
In an attempt to understand the genetic basis of O-antigen chain length heterogeneity, we transferred cloned wzz genes from E. coli O1, O2, O7, and O157 strains into strain P4657. In Fig. 3, we show short and intermediate chain length LPSs from both the parent strain and clones in the O111 model system. Tracks 1 and 2 contain LPSs from intermediate- and short-mode O2 strains, respectively. Track 3 contains the LPS of a short-mode O1 strain which has the same modal chain length as that in track 2 but different mobility due to the O-antigen difference. Tracks 4 to 6 show the LPSs of the model host strain carrying wzz genes from the same three strains. Tracks 9 and 11 show the LPSs of intermediate- and short-mode O7 strains, respectively, and tracks 10 and 12 show the LPSs of the model host strain with wzz genes from the same two strains. The LPSs of two O157 long-mode strains are shown in tracks 1 and 3 of Fig. 4, and the LPSs of the model host strain carrying their wzz genes are shown in tracks 2 and 4, respectively. We also cloned the wzz gene of intermediate-mode O157:H7 strain C664-1992 to give pPR1756, which gave results comparable to those of other intermediate-mode strains (data not shown). In summary, when the wzz genes from O1, O2, O7, and O157 strains were expressed in our model system, the length of the O111 chain, in general, corresponded well to the chain lengths of the donor strains (Fig. 3 and 4).
FIG. 4.
Analysis of long and intermediate chain length Wzz proteins. LPS was prepared by proteinase K digestion and analyzed by SDS–12.5% PAGE and silver staining. Tracks 1 to 4 and 5 to 12 are from two different gels. The strain used for each track is indicated above the track.
Sequence alignment and amino acid variation in Wzz proteins.
We sequenced the cloned wzz genes. Alignment of the deduced amino acid sequences reveals that they are highly conserved but there are minor variations which must be involved in the functional specificity of the protein (Fig. 5). The most consistent amino acid variations between intermediate- and short-mode Wzz proteins are the presence of glycine at position 221 and the change from valine to isoleucine at position 224 in the C′-terminal half of the short-mode proteins (Fig. 5). In the long-mode Wzz proteins, there is substantial difference from intermediate-mode proteins in the N′-terminal half (Fig. 5).
FIG. 5.
Comparison of amino acid sequences of wild-type and mutant Wzz proteins. Only residues which vary are shown, the consensus residues are shown at top, and for specific proteins only the amino acids which vary from the consensus are shown. Restriction enzyme names with arrows indicate the sites used for wzz chimeric gene construction. The numbers above the consensus residues indicate the positions of the amino acid residues. S, short chain; I, intermediate length chain; I-L, intermediate-to-long chain; L, long chain. Boldface letters represent the amino acids changed in pPR1753 (E4991/76) derivatives. The E. coli HU1124 wzz gene sequence is that of Batchelor et al. (5) (GenBank accession no. Z17241). The E. coli Flexneri 2a wzz gene sequence (Sfl1) is that of Morona et al. (22) (GenBank accession no. X71970). The wzz genes of E. coli G7 and Bi316-42 confer a long-chain modal distribution (data not shown). The E. coli Dysenteriae (W30864) and K-12 wzz gene sequences are from Klee et al. (18) (GenBank accession no. Y07560 and Y07559, respectively).
The new wzz sequences were combined with other E. coli sequences to construct a tree (Fig. 6), which shows that the wzz genes for the intermediate and long modes form two major clusters and the short-mode wzz genes form a third cluster derived from the intermediate-mode genes. The M92 (O111) wzz gene was used as the outgroup because of its relatively low sequence similarity with the other E. coli wzz genes (5, 12).
FIG. 6.
Phylogenetic tree generated by the neighbor-joining method on the basis of wzz gene DNA sequences. The wzz genes are represented by the strain names. The wzz genes of E. coli G7 and Bi316-42 confer a long-chain modal distribution (data not shown). The E. coli M92 wzz gene sequence was used as the outgroup (3). Abbreviations: I, intermediate-length chain mode; S, short-chain mode; L, long-chain mode.
Analysis of the genetic basis of short-mode chain length distribution.
There are a few substitutions in the region around residue 220 which distinguish short- and intermediate-mode Wzz proteins (see above). We therefore made a chimeric wzz gene with the BglII-EcoRV segment of short-mode strain F186 replacing that of intermediate-mode strain E4991/76 (Fig. 2). The resulting gene in pPR1752 conferred the short-mode O-antigen chain length distribution of 7 to 16 O units (Fig. 3, track 7), compared with 10 to 18 O units (Fig. 3, track 4) for the E4991/76 gene in pPR1753. The additional glycine at position 221 and the isoleucine at position 224 were present only in short-mode Wzz proteins (Fig. 5). We used site-directed mutagenesis of pPR1753 to make clones containing the additional glycine residue or the V224I substitution, and complementation studies were performed with P4657. Plasmid pPR1776 (V224I) conferred a chain length of the lower value of 7 to 16 O units (Fig. 3, track 14), whereas the presence of the additional glycine residue alone in pPR1775 (Fig. 3, track 13) did not cause any shift in O-antigen chain length from that conferred by pPR1753 (Fig. 3, track 4). Note that pPR1793 (Fig. 3, track 10) confers the same pattern as pPR1753 when run on the same gel (data not shown) and can replace pPR1753 for comparison with pPR1775 and pPR1776. These results suggest that the isoleucine residue alone is sufficient to cause the shift to short-mode chain length.
Analysis of the genetic basis of long-mode chain length distribution.
The long-mode Wzz proteins differ substantially from those of intermediate-mode strains in the amino-terminal half (Fig. 5). The C258-89 and E4991/76 genes differ at 103 sites in the 138-to-573 segment, of which 74 are synonymous substitutions. The flanking segments 1 to 137 and 574 to 978 contain only four and seven substitutions, respectively, of which four and six, respectively, are synonymous.
As for the analysis of short-mode chain length distribution, we started by making a chimeric gene. An EcoRI-PstI 424-bp C258-94 fragment encoding the first 140 amino acids of Wzz was cloned into pUC18, followed by insertion of an E4991/76 554-bp PstI fragment encoding the remaining 186 amino acids, to give pPR1798 (Fig. 2). The chimeric wzz gene confers a typical long-mode chain length (Fig. 4, track 7), indicating that the substitutions needed to confer a long O-antigen chain length are between residues 1 and 140. LPS from the model system with the intermediate-mode gene from EDL933 is shown for comparison. Note that it gives essentially the same LPS profile as the parent wzz gene from E4991/76.
Salmonella enterica strains generally have a long-mode chain length distribution: Wzz of S. enterica LT2 has, in the first 140 residues, only residues 77, 83, 90, and 91 in common with the long-mode E. coli strains. These residues seem most likely to be involved in determination of the long mode, and point mutations were made in pPR1753 to give substitutions M77I in pPR1801, Q83S in pPR1802, D90E in pPR1803, and L91I in pPR1815. The first two failed to change the O-antigen chain length (Fig. 4, tracks 8 and 9), but the D90E mutation in pPR1803 shifted the chain length up to approximately 13 to 21 O units (Fig. 4, track 10), midway between those of the long- and intermediate-mode parents. On the other hand, the L91I mutation shifted the chain length down to 7 to 16 O units, typical of a short-mode strain (Fig. 4, track 11). These results indicate that glutamate 90 plays an important role in determination of long-mode chain length but that the long-mode chain length of 16 to 25 O units can only be restored if one or more additional amino acid substitutions are present. We then made the D90E-L91I double mutation in pPR1804, which gave an O-antigen chain length of 7 to 16 O units, similar to that of a wild-type short-mode strain (Fig. 4, track 12). It appears that the effect of isoleucine 91 in the double mutation is dominant over that of glutamate 90.
DISCUSSION
A single amino acid substitution can change the chain length from intermediate to short.
In the present study, we looked at the genetic basis of O-antigen chain length variation in E. coli. Most strains have an intermediate chain length of 10 to 18 units, but others have short- or long-mode chain length distributions. The wzz sequences covered 948 bp, comprising all but the initial and final 15 bp. However, clones in which the 948-bp segments from long- or short-mode strains replace that segment of an intermediate-mode strain confer long- or short-mode chain length, respectively, indicating that this region is sufficient to determine the chain length mode and justifies our use of this segment for analysis. We made chimeric genes and used site-directed mutagenesis to show that substitution of one or two amino acids in these regions of wzz could determine the difference between modal values. A single amino acid change (V224I) was sufficient to change the chain length of the wzz gene of E4991/76 from the typical intermediate mode to the typical short-mode distribution. An intermediate-mode wzz gene was converted to a long-mode gene by replacement of 140 amino acids at the 5′ end, but in this case, an individual amino acid substitution (D90E) conferred only a partial shift toward the long mode.
Wide distribution of amino acids affecting chain length.
Our first experiments suggested that a specific residue was responsible for each modal value, but subsequent experiments showed the situation to be more complex.
Also, while we were completing this report, Klee et al. (18) showed that the wzz genes of E. coli K-12 and Dysenteriae type 1 confer what we call intermediate-mode distribution while E. coli Flexneri 2a (22) confers a short-mode distribution (shorter than usual at about 5 to 13 O units). We use the terms E. coli Dysenteriae and Flexneri instead of Shigella dysenteriae and S. flexneri because it is clear that E. coli and all Shigella spp. are sufficiently similar to be placed in the same species (8, 25). Klee et al. observed that only lysine 267 and phenylalanine 270 are present in strain Flexneri but not in either of K-12 or Dysenteriae and, on this basis, suggested that this region may be responsible for the different biological activities. However, E. coli Flexneri differs at six other residues from either Dysenteriae or K-12 and our data show that lysine 267 and phenylalanine 270 are not present in other intermediate-mode genes and, indeed, are unique among the sequences in Fig. 5. The wzz gene of Flexneri 2a has valine at residue 224, whereas isoleucine is characteristic of our short-mode strains and a mutation at this site can change an intermediate-mode gene to a short-mode gene. These cases all involve the same general region of the protein. Even this rule is broken by mutation L91I, which we had expected to increase the modal value of intermediate-mode strain E4991/76 but, surprisingly, shifted the O-antigen chain length to the short-mode range of 7 to 16 O units.
This does not exhaust the possibilities, as the difference between the intermediate and short modes of strains K-12 and Flexneri must involve one or more of the substitutions at residues 116, 217, 267, 270, and 285, none of which were picked up in our study.
The E. coli K-12 intermediate chain length wzz gene sequenced by Klee et al. (18) closely resembles our short-mode chain length genes, including the presence of isoleucine 224, which is characteristic of our short-mode genes. Comparison of the K-12 wzz gene with those of our short-mode strains reveals two differences: the presence of arginine 116 and methionine 285, which must, in some way, override the effect of isoleucine 224. These sites are also away from those we subjected to mutagenesis.
We conclude that while chain length distribution can be changed dramatically by substitution of a single key amino acid, it is not possible to define a specific area or amino acids for each specific mode. It appears that rather than a few specific sites determining the modal value, modal-value determination may be an overall property of the protein. All of the substitutions found to affect chain length are between the transmembrane segments in the region shown to be in the periplasm by Morona et al. (22). This indicates that the effect of Wzz on chain length occurs in the periplasm, which is perhaps to be expected, as that is where O-antigen polymerization occurs.
It is interesting that the two amino acid substitutions (V224I and L91I) directly shown to convert from the intermediate to the short mode are conservative changes in hydrophobic amino acids. It is more likely that such substitutions change the packing of residues in a protein than that they affect residues involved in some active site. We have proposed that Wzz exists in two allosteric states (3) and the substitutions may affect the ease with which Wzz moves between them, perhaps in this way influencing the modal chain length determined by Wzz. However, analysis of the tertiary structure of Wzz may be necessary to determine the conformational domains involved in the determination of O-antigen chain length specificity and the role of these residues.
Phylogeny of the wzz gene.
The short-, intermediate-, and long-mode wzz genes we sequenced form three clusters in a neighbor-joining tree (Fig. 6). The single substitutions made by mutagenesis had litte or no effect on this tree (data not shown). The most likely explanation is that the majority of nonsynonymous substitutions are not related to chain length specificity but, like the synonymous substitutions, indicate phylogenetic relationships.
The long-mode genes show substantial deviation from typical intermediate-mode genes. There are 17% synonymous substitutions in the 138- to 573-bp segment of the E. coli wzz gene, which cannot be due to selection on the protein, indicating that this segment was transferred to E. coli after a long period of divergence. For comparison, E. coli and S. enterica genes characteristically differ at 15 to 20% of the bases (32). Within that 138 to 573-bp segment of Wzz are clusters of nonsynonymous substitutions, for example, from position 231 amino acid residue 77 and position 306 amino acid residue 102. These nonsynonymous substitutions confer the difference in modal chain length, as the segment to base 420 (amino acid residue 140) in an otherwise intermediate-mode gene gives a long O-antigen chain length mode (Fig. 3 and 4). The level of synonymous substitutions is such that the segment from approximately position 138 to position 573 is most likely derived from a related species which has a long-mode wzz gene, and the lateral transfer was presumably selected because it conferred a long-mode chain length distribution.
The role of Wzz.
Wzz both confers a modal distribution and determines the modal value. There is very little information on the selective advantage of either a modal chain length or a specific average chain length, but it has been shown (36) that lack of a functional wzz gene in E. coli Flexneri 2a leads to changes in the distribution of IcsA and reduced virulence. It has been shown that the plasmid-encoded Wzz protein in E. coli Flexneri is required for serum resistance and full inflammation, while the chromosome-encoded Wzz protein is required for normal invasiveness and intercellular spread (16). There are also reports of an association of O-antigen chain length with serum resistance (13, 17, 23, 24, 33).
The effects of specific O-antigen chain lengths on serum resistance do not seem to have been studied (27), but there seems to be some association between O-antigen chain length and the pathogenicity of E. coli. Most E. coli strains have intermediate-mode chain lengths, and the few short- and long-mode strains do not seem to be randomly distributed. For example, E. coli O1:K1 strains, mostly found in urinary tract infections and newborn meningitis, all display a typical short mode (19, 21). Also, half of the E. coli O2:K1 strains found in urinary tract infections have short O-antigen chains (1). Furthermore, E. coli O83:K1 strains, which cause newborn meningitis and urinary tract infections, also show the short O-antigen chain length mode (34, 35). These facts lead us to believe that the specific chain length range of the O antigen may be an important virulence factor. However, the way in which O-antigen chain length exerts an effect is still unknown.
ACKNOWLEDGMENTS
A.V.F. was supported by a University of Sydney F. H. Loxton Postgraduate Scholarship. This work was supported by an Australian Research Council grant.
We are grateful to Michael Heuzenroeder (IMVS), Adelaide, South Australia, Australia, and Flemming Scheutz (Statens Seruminstitut, Copenhagen, Denmark) for providing strains and to Chris Murray (IMVS) for providing antisera.
REFERENCES
- 1.Achtman M, Heuzenroeder M, Kusecek B, Ochman H, Caugant D, Selander R K, Väisanen-Rhen V, Korhonen T K, Stuart S, Ørskov F, Ørskov I. Clonal analysis of Escherichia coli O2:K1 isolated from diseased humans and animals. Infect Immun. 1986;51:268–276. doi: 10.1128/iai.51.1.268-276.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aucken H, Pitt T. Serological relationships of the O antigens of Klebsiella pneumoniae O5, Escherichia coli O8 and a new serotype of Serratia marcescens. FEMS Microbiol Lett. 1991;80:93–98. doi: 10.1016/0378-1097(91)90215-v. [DOI] [PubMed] [Google Scholar]
- 3.Bastin D A, Brown P K, Haase A, Stevenson G, Reeves P R. Repeat unit polysaccharides of bacteria: a model for polymerisation resembling that of ribosomes and fatty acid synthetase, with a novel mechanism for determining chain length. Mol Microbiol. 1993;7:725–734. doi: 10.1111/j.1365-2958.1993.tb01163.x. [DOI] [PubMed] [Google Scholar]
- 4.Bastin D A, Romana L K, Reeves P R. Molecular cloning and expression in Escherichia coli K-12 of the rfb gene cluster determining the O antigen of an E. coli O111 strain. Mol Microbiol. 1991;5:2223–2231. doi: 10.1111/j.1365-2958.1991.tb02152.x. [DOI] [PubMed] [Google Scholar]
- 5.Batchelor R A, Alifano P, Biffali E, Hull S I, Hull R A. Nucleotide sequences of the genes regulating O-polysaccharide antigen chain length (rol) from Escherichia coli and Salmonella typhimurium: protein homology and functional complementation. J Bacteriol. 1992;174:5228–5236. doi: 10.1128/jb.174.16.5228-5236.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Batchelor R A, Haraguchi G E, Hull R A, Hull S I. Regulation by a novel protein of the bimodal distribution of lipopolysaccharide in the outer membrane of Escherichia coli. J Bacteriol. 1991;173:5699–5704. doi: 10.1128/jb.173.18.5699-5704.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brass J M. The cell envelope of gram-negative bacteria: new aspects of function in transport and chemotaxis. Curr Top Microbiol Immunol. 1986;129:1–92. doi: 10.1007/978-3-642-71399-6_1. [DOI] [PubMed] [Google Scholar]
- 8.Brenner D J. Enterobacteriaceae. In: Krieg N R, Holt J G, editors. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 408–420. [Google Scholar]
- 9.Brussow H, Sidoti J. Reactivity of human serum antibody with lipopolysaccharide O78 antigen from enterotoxigenic Escherichia coli. Epidemiol Infect. 1992;108:315–322. doi: 10.1017/s0950268800049785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chart H, Said B, Stokes N, Rowe B. Heterogeneity in expression of lipopolysaccharides by strains of Escherichia coli O157. J Infect. 1993;27:237–241. doi: 10.1016/0163-4453(93)91952-l. [DOI] [PubMed] [Google Scholar]
- 11.Fomsgaard A, Freudenberg M, Galanos C. Modification of the silver staining technique to detect lipopolysaccharide in polyacrylamide gels. J Clin Microbiol. 1990;28:2627–2631. doi: 10.1128/jcm.28.12.2627-2631.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Franco A V, Liu D, Reeves P R. A Wzz (Cld) protein determines the chain length of K lipopolysaccharide in Escherichia coli O8 and O9 strains. J Bacteriol. 1996;178:1903–1907. doi: 10.1128/jb.178.7.1903-1907.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldman R C, Joiner K, Leive L. Serum-resistant mutants of Escherichia coli O111 contain increased lipopolysaccharide, lack an O antigen-containing capsule, and cover more of their lipid A core with O antigen. J Bacteriol. 1984;159:877–882. doi: 10.1128/jb.159.3.877-882.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guth B, Pacheco A, von Kruger W, Ferreira L. Comparison of outer membrane protein and lipopolysaccharide profiles of enterotoxigenic Escherichia coli strains isolated in Sao Paulo, Brazil. Braz J Med Biol Res. 1995;28:545–552. [PubMed] [Google Scholar]
- 15.Hitchcock P J, Brown T M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983;154:269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hong M, Payne S M. Effect of mutations in Shigella flexneri chromosomal and plasmid-encoded lipopolysaccharide genes on invasion and serum resistance. Mol Microbiol. 1997;24:779–781. doi: 10.1046/j.1365-2958.1997.3731744.x. [DOI] [PubMed] [Google Scholar]
- 17.Jimenez-Lucho V, Foulds J. Heterogeneity of lipopolysaccharide phenotype among Salmonella typhi strains. J Infect Dis. 1990;162:763–764. doi: 10.1093/infdis/162.3.763. [DOI] [PubMed] [Google Scholar]
- 18.Klee S R, Tzschaschel B D, Timmis K N, Guzman C A. Influence of different rol gene products on the chain length of Shigella dysenteriae type 1 lipopolysaccharide O antigen expressed by Shigella flexneri carrier strains. J Bacteriol. 1997;179:2421–2425. doi: 10.1128/jb.179.7.2421-2425.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kusecek B, Wloch H, Mercer A, Vaisanen V, Pluschke G, Korhonen T, Achtman M. Lipopolysaccharide, capsule, and fimbriae as virulence factors among O1, O7, O16, O18, or O75 and K1, K5, or K100 Escherichia coli. Infect Immun. 1984;43:368–379. doi: 10.1128/iai.43.1.368-379.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lugtenberg B, Meijers J, Peters R, van der Hoek P, van Alphen L. Electrophoretic resolution of the major outer membrane proteins of Escherichia coli K12 into four bands. FEBS Lett. 1975;58:254–258. doi: 10.1016/0014-5793(75)80272-9. [DOI] [PubMed] [Google Scholar]
- 21.Moll A, Kusecek B, Pluschke G, Morelli G, Kamke M, Jann B, Jann K, Achtman M. A reexamination of the O1 lipopolysaccharide antigen group of Escherichia coli. Infect Immun. 1986;53:257–263. doi: 10.1128/iai.53.2.257-263.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Morona R, Van Den Bosch L, Manning P. Molecular, genetic, and topological characterization of O antigen chain regulation in Shigella flexneri. J Bacteriol. 1995;177:1059–1068. doi: 10.1128/jb.177.4.1059-1068.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ohno A, Isii Y, Tateda K, Matumoto T, Miyazaki S, Yokota S, Yamaguchi K. Role of LPS length in clearance rate of bacteria from the bloodstream in mice. Microbiology. 1995;141:2749–2756. doi: 10.1099/13500872-141-10-2749. [DOI] [PubMed] [Google Scholar]
- 24.Porat R, Johns M, McCabe W. Selective pressures and lipopolysaccharide subunits as determinants or resistance of clinical isolates of gram-negative bacilli to human serum. Infect Immun. 1987;55:320–328. doi: 10.1128/iai.55.2.320-328.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pupo G M, Karaolis D K R, Lan R, Reeves P R. Evolutionary relationships among pathogenic and nonpathogenic Escherichia coli strains inferred from multilocus enzyme electrophoresis and mdh sequence studies. Infect Immun. 1997;65:2685–2692. doi: 10.1128/iai.65.7.2685-2692.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reeves P R. Biosynthesis and assembly of lipopolysaccharide. New Compr Biochem. 1994;27:281–314. [Google Scholar]
- 27.Reeves P R. Role of O-antigen in the immune response. Trends Microbiol. 1995;3:381–386. doi: 10.1016/s0966-842x(00)88983-0. [DOI] [PubMed] [Google Scholar]
- 28.Reeves P R, Farnell L, Lan R. MULTICOMP: a program for preparing sequence data for phylogenetic analysis. CABIOS. 1994;10:281–284. doi: 10.1093/bioinformatics/10.3.281. [DOI] [PubMed] [Google Scholar]
- 29.Reisner A H, Bucholtz C A, Smelt J, McNeil S. Proceedings of the Twenty-Sixth Annual Hawaii International Conference on Systems Science 1. 1993. Australia’s National Genomic Information Service; pp. 595–602. [Google Scholar]
- 30.Saiki R K, Gelfand D H, Stofell S, Scharf S J, Higuchi R, Horn G T, Mullis K B, Erlich H A. Primer-directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science. 1988;239:487–491. doi: 10.1126/science.2448875. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 32.Sharp P M. Determinants of DNA sequence divergence between Escherichia coli and Salmonella typhimurium: codon usage, map position, and concerted evolution. J Mol Evol. 1991;33:23–33. doi: 10.1007/BF02100192. [DOI] [PubMed] [Google Scholar]
- 33.Stawski G, Nielsen L, Ørskov F, Ørskov I. Serum sensitivity of a diversity of Escherichia coli antigenic reference strains. Acta Pathol Microbiol Scand. 1990;98:828–838. [PubMed] [Google Scholar]
- 34.van Alphen L, van Kempen-De Troye F, Zanen H C. Characterization of cell envelope proteins and lipopolysaccharides of Escherichia coli isolates from patients with neonatal meningitis. FEMS Microbiol Lett. 1983;16:261–267. [Google Scholar]
- 35.van Alphen L, van Kempen-De Troye F, Zanen H C. Immunological detection of heterogeneous O-antigen containing lipopolysaccharides in Escherichia coli. J Gen Microbiol. 1983;129:191–198. doi: 10.1099/00221287-129-1-191. [DOI] [PubMed] [Google Scholar]
- 36.Van Den Bosch L, Manning P A, Morona R. Regulation of O-antigen chain length is required for Shigella flexneri virulence. Mol Microbiol. 1997;23:765–775. doi: 10.1046/j.1365-2958.1997.2541625.x. [DOI] [PubMed] [Google Scholar]
- 37.Wells J G, Davis B R, Wachsmuth I K, Riley L W, Remis R S, Sokolow R, Morris G K. Laboratory investigation of hemorrhagic colitis outbreaks associated with a rare Escherichia coli serotype. J Clin Microbiol. 1983;18:512–520. doi: 10.1128/jcm.18.3.512-520.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Whitfield C. Biosynthesis of lipopolysaccharide O antigens. Trends Microbiol. 1995;3:178–185. doi: 10.1016/s0966-842x(00)88917-9. [DOI] [PubMed] [Google Scholar]
- 39.Whitfield C, Amor P A, Koplin R. Modulation of the surface architecture of gram-negative bacteria by the action of surface polymer-lipid A-core ligase and determinants. Mol Microbiol. 1997;23:629–638. doi: 10.1046/j.1365-2958.1997.2571614.x. [DOI] [PubMed] [Google Scholar]