Abstract
Background
Radiomics features hold significant value as quantitative imaging biomarkers for diagnosis, prognosis, and treatment response assessment. To generate radiomics features and ultimately develop signatures, various factors can be manipulated, including image discretization parameters (e.g., bin number or size), convolutional filters, segmentation perturbation, or multi-modality fusion levels. Typically, only one set of parameters is employed, resulting in a single value or “flavour” for each radiomics feature. In contrast, we propose “tensor radiomics” (TR) where tensors of features calculated using multiple parameter combinations (i.e., flavours) are utilized to optimize the creation of radiomics signatures.
Methods
We provide illustrative instances of TR implementation in positron emission tomography-computed tomography (PET-CT), magnetic resonance imaging (MRI), and CT by leveraging machine learning (ML) and deep learning (DL) methodologies, as well as reproducibility analyses: (I) to predict overall survival (OS) in lung cancer (CT) and head and neck cancer (PET-CT), TR was employed by varying bin sizes. This approach involved use of a hybrid deep neural network called ‘TR-Net’ and two ML-based techniques for combining different flavours. (II) TR was constructed by incorporating different segmentation perturbations and various bin sizes to classify the response of late-stage lung cancer to first-line immunotherapy using CT images. (III) In MRI of glioblastoma (GBM), TR was implemented to generate multi-flavour radiomics features, enabling enhanced analysis and interpretation. (IV) TR was employed via multiple PET-CT fusions in head and neck cancer. Flavours based on different fusions were created using Laplacian pyramids and wavelet transforms.
Results
Our findings demonstrated that TR outperformed conventional radiomics features in lung cancer CT and head and neck cancer PET-CT images, significantly enhancing OS prediction accuracy. TR also improved classification of lung cancer response to therapy and exhibited notable advantages in reproducibility compared to single-flavour features in MR imaging of GBM. Moreover, in head and neck cancer, TR through multiple PET-CT fusions exhibited improved performance in predicting OS.
Conclusions
We conclude that the proposed TR paradigm has significant potential to improve performance in different medical imaging tasks. By incorporating multiple flavours of radiomics features, TR overcomes limitations associated with individual features and shows promise in enhancing prognostic capabilities in clinical settings.
Keywords: Imaging biomarkers, radiomics, machine learning (ML), outcome/disease prediction, image fusion
Introduction
The term ‘radiomics’ was first introduced in 2010 by Gillies et al. (1) as “the extraction of quantitative features from radiographic images”. Radiomics features capture information about tissues and lesions (2-5). A collection or combination of radiomics features considered to be a ‘radiomics signature’ can computationally model a biological phenomenon (6). Singular radiomics features and radiomic signatures can act as imaging biomarkers and have been shown to reflect biological characteristics of lesions and can improve a range of different clinical tasks (7-9). Radiomics includes the use of single- or hybrid-imaging modalities, with the potential to identify novel imaging biomarkers for improved detection, classification, staging, prognosis, prediction and treatment planning in different cancers (9).
A radiomics feature can be generated using different parameters (e.g., pre-processing, discretized bin number or size; segmentation threshold to define region-of-interest). There have been significant efforts to establish the best values of parameters suitable for different tasks. However, determining suitable predictive features can be a difficult task. Certain features or versions of features (with specific parameters) may be non-robust to noise or can change depending on the scanner used to acquire the imaging data. This can result in a lack of reproducibility across different institutions and scanners (10-14) and radiomics features may be non-robust to noise or inter-center protocol and scanner variabilities (15,16). The vast array of feature-selection methods used in radiomics studies (17-19) attests to the need for careful pruning of features beforehand and the difficulty of such a task. Individual radiomics features may correlate with one another, hence providing no added predictive value for a radiomic signature, or they may challenge interpretability, such as many “deep features” extracted from layers of a neural network (3,20). This can result in a lack of reproducibility across radiomics trials. Although efforts have been made to standardize radiomics protocols (21) and reduce the effect of different scanners, these remain relevant issues (10,20).
This work aims to tackle the above-mentioned limitations of radiomics analyses using a different paradigm. We propose TR in which multiple flavours (i.e., versions of the same radiomics features) are generated and considered. Generally, a radiomics feature consists of a single value for an entire 2D or 3D volume. Instead of providing a single value for each feature, we compute multiple values of the feature by varying some of the parameters in the calculation. Features with different bin sizes, perturbation of segmentations, pre-processing filters [e.g., Laplacian of Gaussian (LOG) and wavelets], and fusion techniques can be considered as different flavours. We hypothesize that TR has the potential to overcome some of the shortcomings of models that use radiomics features enabling improved clinical task performances.
Methods
Patient data
In this paper, we explore application of the proposed TR paradigm to a range of data and applications. These included computed tomography (CT) images of lung cancer, positron emission tomography-CT (PET-CT) images of head and neck cancer, and magnetic resonance imaging (MRI) of glioblastoma (GBM), as listed in Table 1, and further elaborated in subsequent sections.
Table 1. Description of datasets and corresponding tasks in this work.
| Dataset | Modality | Task | Number of cases |
|---|---|---|---|
| Lung | CT | Treatment response prediction | 118 lung lesions (primary and lung metastases from 96 patients) |
| Head and neck | PET-CT | 2-year progression-free survival | 224 patients (primary tumors) |
| Brain | MRI | Repeatability analysis | 17 patients (test-retest T1- and T2-weighted images of glioblastoma) |
PET, positron emission tomography; CT, computed tomography; MRI, magnetic resonance imaging.
Study methods
The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Our original intuition to explore the TR paradigm was that features generated using multiple flavours may be more robust and reproducible (and ultimately valuable) compared to single-flavour features for outcome prediction. In TR, we build feature tensors using many flavours (Figure 1) as a method towards the optimized construction of radiomics signatures. For different clinical tasks, subsequent to the generation of the radiomics tensor, one may utilize feature selection or extraction methods, as well as machine learning (ML) or deep learning (DL) methods, for optimal construction of signatures.
Figure 1.
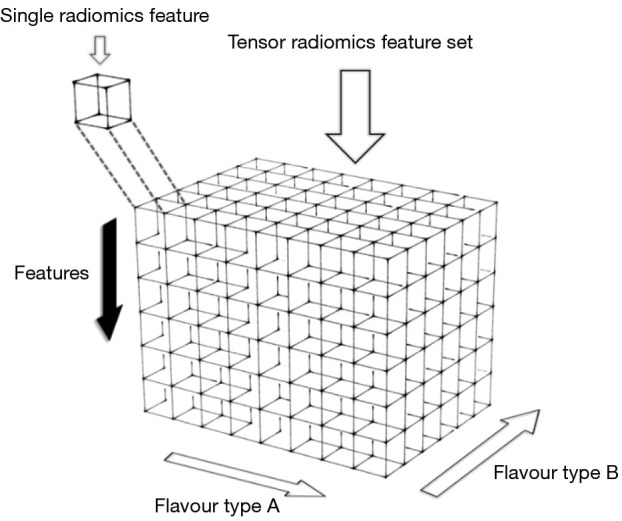
A representation of a radiomics tensor. The black arrow indicates the dimension where the different extracted radiomics features are stacked, while the flavour types (A, B, ...) encompass variations by which feature values are generated: examples include variations in discretization bin sizes, segmentations, pre-processing filters and fusion methods. Only two variants can be represented here in 3D, but even higher dimension tensors can be generated.
In what follows, we further elaborate possible approaches to TR and provide example applications in PET-CT, MRI, and CT imaging. We generate multi-flavour TR signatures by varying bin sizes in PET-CT and CT images (section Discretization flavours). Different segmentation perturbations are our next approach to build different flavours of radiomics signature in CT images (section Segmentation flavours). Multi-flavour radiomics features generated by varying hyperparameters of pre-processing filters are also considered in MRI images [section Filter (pre-processing) flavours]. Flavours built from different fusions methods on PET-CT images are also considered towards building TR signature in this effort (section Fusion flavours).
Discretization flavours
Data and task
We evaluated whether TR may enhance the prediction of outcome using PET-CT scans in patients with HNC. The 224 baseline HNC PET-CT scans were obtained from The Cancer Imaging Archive (TCIA) along with primary tumour segmentations (22). Tumours were segmented by expert radiation oncologists onto either the original PET-CT scan or a separate CT scan. In the latter case, segmentations were propagated to the PET-CT scan using intensity-based free-form deformable registration. In this work, we formulated the outcome prediction task as binary classification (2-year progression free survival). TR features were extracted from PET images using 10 different bin sizes [0.1–1.0 standard uptake value (SUV), increments of 0.1 SUV] using the PyRadiomics package v3.0.1 (23). Please see S.2 for a list of all available features. A total of 105 features were used including first order, 3D shape, gray-level co-occurrence matrix (GLCM), gray level run length matrix (GLRLM), gray level size zone matrix (GLSZM), neighboring gray tone difference matrix (NGTDM) and grey level dependence matrix (GLDM) features. From there, standard deviation, compactness 1, compactness 2, spherical disproportion, and voxel volume were excluded due to high correlation with other features.
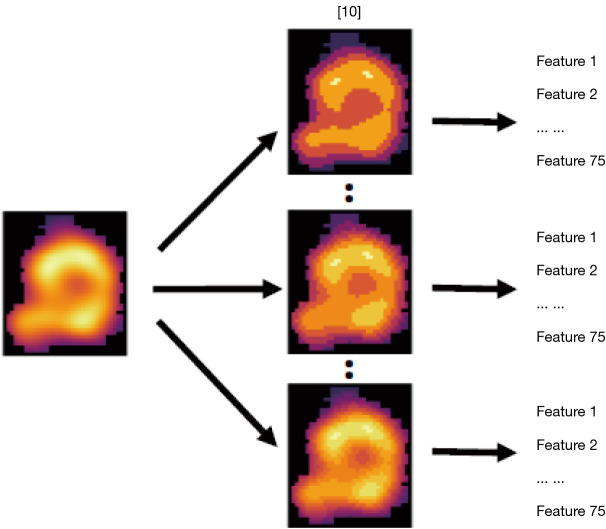
The primary realization of the proposed TR utilizes explainable, handcrafted radiomics features at varying discretization levels. Discretization is the grouping of the original range of pixel intensities into specific intervals or bins, necessary for the computational feasibility of certain features (24); e.g., fixing the bin widths (BW) or fixing the number of bins [bin counts (BC)]. Different ‘bin flavours’ calculated by different discretization strategies are shown in Figure 2. An optimal discretization would be one that consistently filters out noise while retaining the integrity of important lesion features. However, little guidance is given on achieving such an optimum in the literature, even though discretization can dramatically impact the calculated feature values. In our proposed TR paradigm, a variety of bins (flavours) can be generated, and the best ones are ultimately utilized (see Figure S1 in the supplementary material for further details).
Figure 2.
Radiomics features calculated on PET-CT images from head and neck data. 10 ‘bin flavours’ calculated by using different discretization strategies to extract 75 features. PET, positron emission tomography; CT, computed tomography.
ML-based flavour selection framework
First, we trained two ML models, support vector machines (SVM) with radial basis function (RBF) kernel and logistic regression, on concatenated features for all possible combinations of 10 flavours (1,013 different combinations with 2 or more flavours). The TR models were compared to the same ML models trained on single-flavour features.
DL-based flavour fusion framework
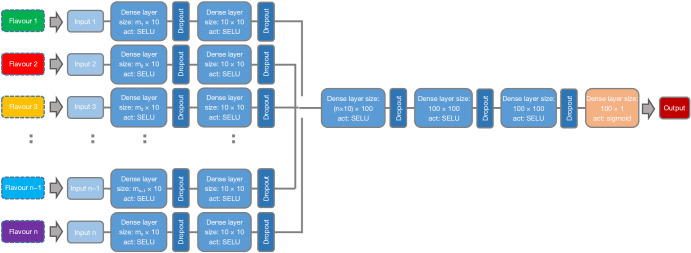
We developed a two-stage deep feed-forward neural network, named TR-Net (for TR Network), to perform end-to end-flavour fusion (see Figure 3 for further details). The input to this model were all features extracted using all flavours. TR-Net consists of legs and body: each leg is a stack of multiple dense layers separately fed with the features of each flavour in its input layer. All legs are then concatenated and connected to a few more dense layers to complete the architecture of the network. The size of the final dense layer, as well as the size of other dense layers in the architecture and the number of the layers in both the legs and the head part, were among the hyperparameters that we tuned them on the validation set.
Figure 3.
The architecture for our TR-Net. Features of a given flavour are input into the ‘legs’ of the network, which work to extract and combine the most effective features. The various flavour legs are then concatenated and put through several final layers before producing a binary prediction. SELU, scaled exponential linear unit; TR-Net, tensor radiomics network.
A sigmoid function in the last layer performs the binary outcome classification. The network was trained using a mean squared error (MSE) loss function. In binary classification tasks, the conventional approach is to employ the cross-entropy loss. In our study, we initially experimented with the cross-entropy loss during hyperparameter search. However, we encountered the issue of training loss failing to converge. To address this challenge, we empirically explored alternative options, including MSE loss. Notably, we observed loss convergence when using MSE loss, leading us to adopt it as the preferred loss function in our experiments. Random selection search using a nested 5-fold cross-validation (CV) was utilized for hyperparameter tuning. Our TR-Net is a modified version of a fusion network initially proposed in (25) for biometrics liveness detection. Intuitively, during training, TR-Net concurrently learns a transformation of features coming from its legs (different flavours in our case) into a common subspace (in the first stage) and performs classification on those features based on their fusion in that common subspace (in the second stage). To tackle class imbalance during training the network, SMOTE (26) was applied to the training set only, to up-sample the minority (positive) class.
Average balanced accuracy and F1 score metrics were computed using stratified 5-fold CV (for both ML & DL methods). Scaled exponential linear unit (SELU) activation function (27) was used in all the dense layers of the legs and the body along with dropout.
ML-based feature selection framework
Motivated by the improved performance of TR-Net, we studied the effect of applying feature selection (for any given flavour across different features) prior to performing flavour fusion on the same classification task. A range of 5 to 25 features were selected from each of the 10 flavours using sequential forward feature selection method (28) based on the mean F1 score over a nested 5-fold CV setup. These selected features were then combined for all possible combinations of 10 flavours to form our TR features, and then ranked based on mean balanced accuracy and F1 score of nested 5-fold CV using two ML classifiers, namely an RBF kernel SVM and a Logistic Regression model.
To provide clarity, our process began by extracting radiomics features for all 10 flavours. We then aggregated these features for all possible combinations of flavours, ranging from combinations of 2 to all 10 flavours together (all possible subsets of a set with 10 elements). Next, we trained two classifiers on all these combinations and reported their performance on the 5 folds of the test set. We presented a few select combinations to demonstrate the concept of TR and highlight the benefits of utilizing features extracted from multiple parameter settings in the data.
In the second part, we utilized features extracted from all flavours to train a two-stage deep neural network and reported classification metrics on 5 folds of the test set. Finally, in the third part, we performed a feature selection step to identify the most predictive single features for each flavour, and then combined those selected features to form TR features. We then aggregated these features for all possible combinations of flavours and reported the performance of classifiers trained on these combinations on the 5 folds of the test set.
Segmentation flavours
Data and task: we analyzed the viability of TR features with a Lung Volume CT dataset of late-stage non-small cell lung cancer (NSCLC) patients. This dataset is a retrospective cohort of patients with Programmed Death Li gand-1 (PDL1) scores ≥50% that received first-line single-agent pembrolizumab as the current Canadian clinical practice (29-31). Two oncology radiologists read the baseline and 1st follow-up CT to assess treatment response [e.g., disease control, including: complete/partial response (C/PR) and stable disease (SD) vs. progressive disease (PD)] using RECIST v1.1 (32). This CT dataset consists of 118 lung lesions (primary and lung metastases) from 96 patients, with up to 5 axial adjacent slices per lesion selected to provide a more supplementary descriptive CT image dataset (n=558). The ML task for this dataset was to predict patient response to pembrolizumab (i.e., Disease Control) using 2D-only TR features extracted from baseline axial CT lesion segmentations. CT volume data were from an ongoing study approved by the University of British Columbia and BC Cancer Agency review ethics board (UBC/BCCA REB #H19-01597).
We also explored TR based on segmentation flavours. Inspired by Zwanenburg et al. (33) where image perturbations to assess feature robustness were used, we incorporated combinations of translation, area adaptation, and contour randomization of the images and masks to extract a number of perturbed images per axial slice of available segmentations. This TR segmentation flavour technique was employed to leverage an established test-retest method in an effort to establish robust and stable features. For each of these perturbed images, radiomic features were extracted with PyRadiomics and used to build our radiomics tensor. Our TR feature set was used to train a ML model to execute this binary patient response task for measurement of disease control (PR + SD vs. PD). We leveraged Zwanenburg et al.’s methodology, primarily the “volume adaptation” (V) method, for our lung lesion ROIs to extract static bin size feature flavours from perturbed 2D lesion segmentations. This V method describes a process wherein the ground truth mask is either dilated or eroded using a disc-structuring element. An arbitrary and equal number of dilations and erosions are performed to capture extra parenchymal tissue either (dilation) or tissue characteristics from the core of the lesion only (erosion).
To further supplement the protocol from section Discretization flavours (bin size flavours), each vanilla feature (i.e., non-perturbed images and a static bin width) in the 2D CT feature set was recalculated using increasing bin width sizes for extraction of TR features without image perturbation. We compared the validity of both schemes of TR features, bin size changes and image perturbations respectively, with a standard rudimentary ML model, the linear discriminant analysis (LDA), for design simplicity. These models were trained on TR features chosen by Sequential Forward Feature Selection (SFS). An LDA trained on 7 vanilla radiomics features selected by SFS was chosen as the primary analysis method for comparison (5-fold nested CV and patient agnostic training-test splitting to avoid biasing the results; SMOTE resampling for class imbalance). A statistical power analysis revealed that this late-stage cancer dataset sample size (N=96), achieves 80% power for detecting a medium effect (ES =0.61) at a significance level α=0.05 for Welch’s t-test. Therefore, given our positive event rate (26:70 positive:negative patients), a 5-fold CV was selected provided that a medium effect size is maintained for each fold given the approximate number of patients in the test set.
We compared a series of TR feature flavours against a baseline LDA model trained on unperturbed images. We refer to this baseline as having “vanilla” feature flavours. We created 3 other TR feature flavour sets from images to test against this vanilla model: (I) recalculated features using increasing bin widths; (II) segmentation volume adaptation via dilating and eroding the ROI mask; and lastly (III) a combination of image translation (“T”), segmentation volume adaptation (V), and segmentation contour randomization (“C”) to produce the “TVC” TR feature flavour set.
Filter (pre-processing) flavours
Data and task: we analyzed MR images of 17 GBM patients including T1- and T2-weighted images. Test-retest scans were performed within the same imaging unit on two consecutive days (34,35). Full affine registration of test to retest images was performed with 12 degrees of freedom using mutual information cost function; three segmentations in T1 and one in T2-weighted images were performed (enhancing + necrotic, enhancing, necrotic, and edema cores) (35). N4 bias correction was performed on raw MR images. Our task in this experiment was to assess the repeatability of MR imaging radiomics features.
In this approach, tensors of radiomic features were generated after applying pre-processing filters as different flavours such as wavelets (WL, all possible combinations of applying either a high- or a low pass filter in each of the three dimensions, including HHH, HHL, HLH, HLL, LHH, LHL, LLH, and LLL), LOG (with different sigma values of 0.5 to 5 with steps 0.5), Exponential, Gradient, Logarithm, Square and Square Root scales. In addition to filter flavours, we also used discretization flavours (e.g., section Discretization flavours) of fixed bin width and fixed bin count (16, 32, 64, 128, and 256). Different features, namely first-order (FO) and textural features (GLCM, GLRLM, GLDM, GLSZM, NGTDM) were then extracted. Principal component analysis (PCA) was performed on different flavours to generate new features. One component per feature was chosen (converting multiple preprocess features to one feature). We performed this task regarding the improvement of repeatability of radiomics features across different preprocessing steps. Intraclass correlation coefficients (ICC) were calculated to assess the repeatability of features (comparing performances of conventional single-flavour features vs. newly-generated features). ICC values were classified into low, medium, high, and excellent repeatable features for ICC <50%, 50%≤ ICC <75%, 75%≤ ICC <90%, and 90%≤ ICC ≤100%, respectively.
Fusion flavours
Data and task: to evaluate fusion flavours, we used the same data and task as in section Discretization flavours (head and neck cancer PET-CT binary outcome prediction).
Fusion radiomics (36), an emerging area of investigation, has up to now meant fusing images in different ways and selecting the optimal one. In the proposed TR, we alter the paradigm and include various multi-modality fusions within the radiomics tensor for a given task, followed by subsequent model construction. We employed 15 image-level fusion techniques to combine PET-CT information (see Table S1). Detailed explanations of the fusion techniques are available in the Appendix 1. Subsequently, 211 features were extracted from each region of interest in PET-only, CT-only, and 15 fused PET-CT images through the SERA radiomics package (37). A range of optimal algorithms was pre-selected amongst various families of learner algorithms.
We considered two approaches for the prediction task: (I) we separately applied radiomics features extracted from each of the PET-only, CT-only, and 15 fused PET-CT images to three classifiers, namely logistic regression, random forest, and multilayer perceptron (MLP) classifiers. We employed ensemble voting for each classifier: specifically, we used nine different estimators for each classifier (i.e., with different optimized parameters from nine different runs/initializations/grid-search). (II) After min-max normalization, we first removed flavours with correlations over 95% to avoid redundant data and then employed a PCA algorithm to combine the remaining flavours of each TR and convert those to a single attribute. Subsequently, the polynomial feature transform was utilized to combine the attributes and generate 142,879 fusion-TR features. The ANOVA approach (38) was then utilized to select 1,000 relevant fusion-TR features among 142,879 attributes generated by the polynomial function. Finally, we applied these selected fusion-TR features to the three above-mentioned classifiers to predict survival outcome. In this work, we performed nested 5-fold CV for all datasets, including PET-only, CT-only, 15 fused PET-CT images, and fusion-TR features. In each round, we divided the dataset into a training part (four-fold) and testing part (one-fold). In the training process, we further divided the training dataset into two sub-parts, with 80% of data points for training the model and the remaining 20% for model selection. Mean accuracy in training validation was used to select the best model. Mean accuracy in nested testing was reported to validate the best model (for more information, see Figure S2).
Results
Results of using discretization flavours
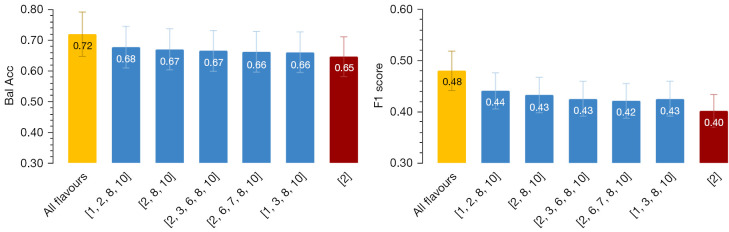
We first assessed the effectiveness of combining features from different combinations of the flavours (bin size in this part of our study) in terms of two classification score metrics: balanced accuracy, and f1 score (we also studied area under ROC curve, with similar trends; not shown). Figure 4 illustrates the enhanced metric values using 5 different flavour combinations taken from the top 20 combinations out of all possible ones (we applied the next k-fold cross validation). The rationale behind this comparison is to examine whether combining “all” features of multiple flavours improve the classification, without performing feature selection, and whether the combinations that improve the classification performance, show some meaningful combinations; e.g., evaluating if the combination of the smallest bin size with the largest bin size can improve the performance. To this end, we examined all possible combinations and reported a few of the better performers with respect to single bin size features.
Figure 4.
Balanced accuracy (left) and f1 score values (right) when using conventional radiomics (single-flavour) using ML pipeline (red), 5 different combinations of TR flavours using ML pipelines (blue), and all 10 TR-flavours of features via DL TR-Net pipeline (yellow). All features were used (no features selection methods were applied prior). ML, machine learning; TR, tensor radiomics; DL, deep learning.
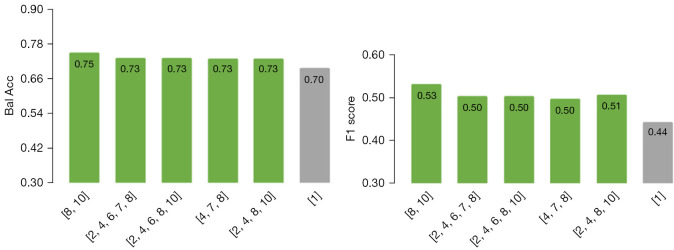
Our results show that TR features made from multiple flavours with respect to regular radiomics features improved performances from (red) conventional radiomics to (blue) TR in ML pipelines. Further improvements were obtained when utilizing (yellow) our DL TR-Net pipeline in an end-to-end fashion. We also studied the effect of feature selection prior to ML methods applied to conventional vs. TR models, with similar trends (see Figure 5). We applied a corrected t-test on the results but did not detect significant outperformance relative to the baseline.
Figure 5.
Balanced accuracy (left) and f1 score values when using feature selection (right), prior to applying conventional radiomics (single-flavour) (gray) vs. TR (multi-flavours) (green). TR, tensor radiomics.
Results of using segmentation flavours
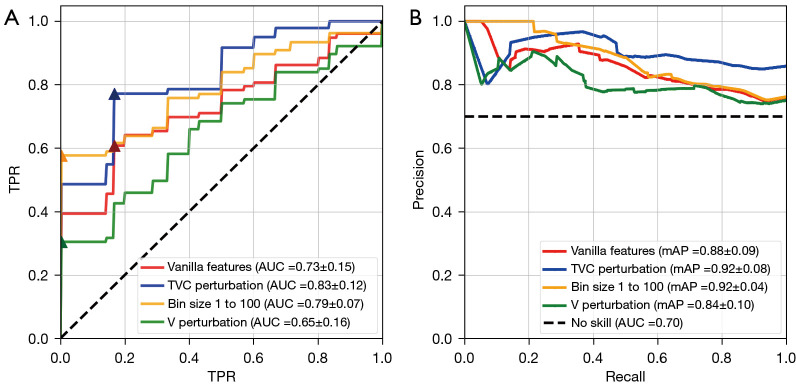
ROC and Precision-Recall analysis on vanilla feature flavour, different bin size, the combination of image translation (“T”), segmentation volume adaptation (“V”) and contour randomization (“C”) were conducted as seen in Figure 6.
Figure 6.
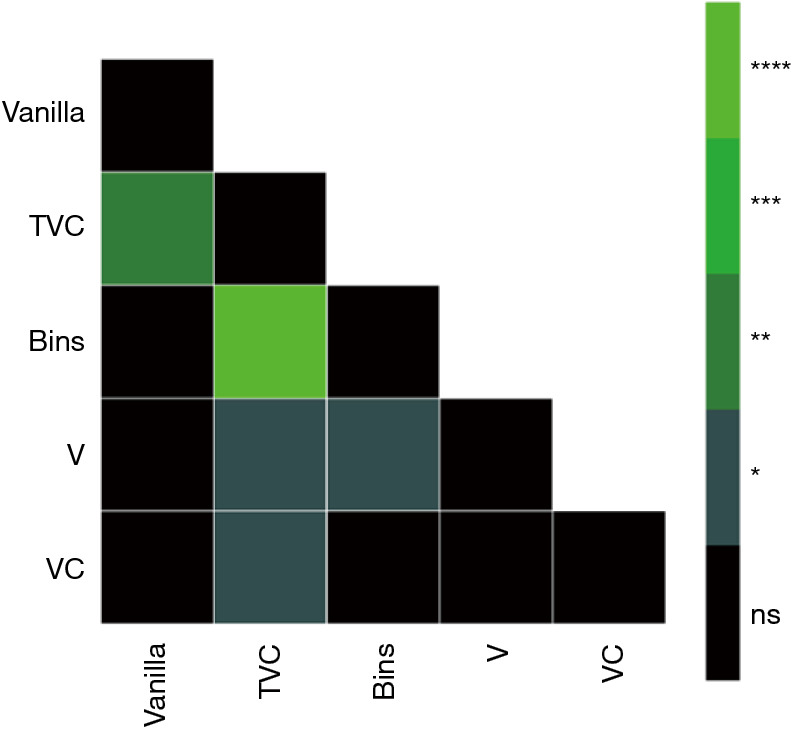
Significance matrix of five discrete LDA models in predicting treatment response to single agent pembrolizumab in late-stage NSCLC patients using radiomic features. Each model was trained with a unique feature set (image and segmentation flavours). McNemar’s test was applied to test the significance between model performances shown in the matrix (color) to the right. ns, not significant, *, 0.01<P<0.05; **, 0.001<P<0.01; ***, 0.0001<P<0.001; ****, P<0.0001. TVC, combination of image translation (T), segmentation volume adaptation (V) and contour randomization (C); LDA, linear discriminant analysis; NSCLC, non-small cell lung cancer.
As previously stated in section Segmentation flavours, a Group-k (k=5) fold CV was performed to assess the effect of our proposed segmentation TR flavours on the Lung-CT outcome prediction task. Reported metrics and uncertainties were based on the average scores of area under the ROC curve (ROC AUC), and mean average precision (mAP) and F-1 score using this CV technique. The results in terms of these classification metrics were compared to the single-flavoured (vanilla) features. Furthermore, the performance of the LDA model on the test folds by applying feature selection prior to model training is illustrated in Table 2 as well as Figures 6,7.
Table 2. The performance of the LDA model on the test folds.
| Flavour | Balanced accuracy | F1 score | ROC AUC | mAP |
|---|---|---|---|---|
| Vanilla | 0.67±0.08 | 0.79±0.08 | 0.73±0.12 | 0.88±0.09 |
| TVC | 0.77±0.06 | 0.85±0.07 | 0.83±0.09 | 0.92±0.08 |
| Bins 1-to-100 | 0.73±0.08 | 0.82±0.09 | 0.78±0.06 | 0.92±0.04 |
| V | 0.68±0.08 | 0.79±0.09 | 0.65±0.14 | 0.84±0.10 |
| VC | 0.69±0.09 | 0.87±0.02 | 0.82±0.06 | 0.93±0.03 |
Data are presented as mean ± standard deviation. ROC, receiver operator characteristic; LDA, linear discriminant analysis; AUC, area under the curve; mAP, mean average precision; TVC, combination of image translation (T), segmentation volume adaptation (V) and contour randomization (C).
Figure 7.
Model performance plots for four discrete LDA models. (A) ROC curve plot highlighting model performance with the addition of feature flavour segmentations. TVC perturbations achieved the highest ROC AUC =0.83±0.12, SN: 77.9%, SP: 83.2% at optimal threshold (triangle marker). (B) Precision-recall curve to elucidate model performance for this class imbalanced dataset. Both plots indicate a strong boost to performance of the model by implementing TR feature methods. TPR, true positive rate; FPR, false positive rate; LDA, linear discriminant analysis; ROC, receiver operator characteristic; TVC, combination of image translation (T), segmentation volume adaptation (V) and contour randomization (C); mAP, mean average precision; AUC, area under the cure; SN, sensitivity; SP, specificity; TR, tensor radiomics.
Overall, the trend indicates a strong model performance with the addition of TR feature flavours. We observe boosts to established metrics such as ROC AUC and PR AUC over the vanilla feature model as evidenced by Table 2 (and Figure 7). TVC perturbation maintained the largest F-1 score of 0.852±0.09 in comparison to the vanilla model’s 0.793±0.08. The weakest model was the solo volume adaption (V) feature flavour set in nearly all metrics. However, looking at Figure 7, one can observe a non-small model improvement in the image perturbation feature sets (TVC and V) at low false positive rates. All in all, our results suggest the TR paradigm to enable improved performance relative to conventional radiomics.
Results of using filter (pre-processing) flavours
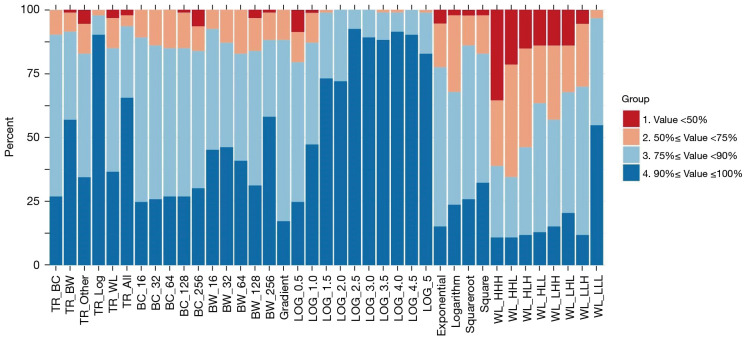
As seen in Figure 8, LOG results in the highest number of repeatable features. In LOG preprocessing with sigma 0.5 and 1 there were features with low repeatability. However, in TR_BC (bin count), there were no poorly repeatable features, and only two features showed medium repeatability, with the rest showing high and excellent repeatability. In wavelet features, different settings resulted in different numbers of non-repeatable and repeatable features; LLL results in the highest number of repeatable features. TR of wavelets showed 3 low, 11 medium, 45 high and 34 excellent repeatable features. Overall, TR increased radiomics-feature repeatability in all features and only two features Kurtosis from first order and busyness from NGTDM had low repeatability (ICC <50%). In TR, features including Complexity and Coarseness from NGTDM, CP from GLCM and Skewness from first-order had medium repeatability (50%≤ ICC <75%). Twenty-four features showed high repeatability (75%≤ ICC <90%) and 61 from 93 features showed excellent (ICC ≥90%) repeatability.
Figure 8.
The test-retest repeatability of radiomics features in MR imaging of GBM (ICC used as metric). Proposed TR-generated features (via PCA) are compared to conventional individual flavours of these features. TR, tensor radiomics; BC, bin counts; BW, bin widths; LOG, Laplacian of Gaussian; WL, wavelets; H, high; L, low; MR, magnetic resonance; GBM, glioblastoma; ICC, intra-class correlation; PCA, principal component analysis.
Results of using fusion flavours
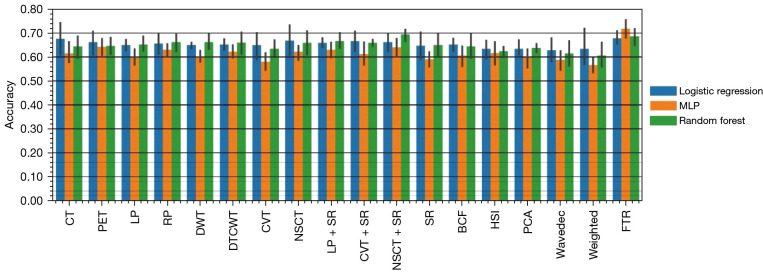
For our fusion-based TR framework, we included 211 radiomics features. After feature fusion, we selected the relevant fusion-TR features, applying them to the mentioned ensemble classifiers. The highest training validation performance of 73.6%±2.5% was obtained for fusion-TR via MLP, and nested testing of 71.8%±4.8% confirmed this finding as shown in Figure 9. For conventional non-TR scheme, the highest training validation performance of 69.7%±5.1% was obtained for Laplacian Pyramid (LP) + Sparse Representation (SR) linked with random forest, with nested testing performance of 66.6%±3.9%. Paired t-test indicated significantly improved (P<0.05) performance for the proposed TR framework compared to best non-TR performance.
Figure 9.
Bar plots of mean and standard deviation for prediction of outcome for nested testing performance. X axis: conventional radiomics (PET-only, CT-only, 15 fused PET-CT images), as well as proposed Fusion-TR, using 3 different classifiers. MLP, multi-layer perceptron; LP, Laplacian pyramid; RP, ratio of low-pass pyramid; DWT, discrete wavelet transform; DTCWT, dual-tree complex wavelet transform; CVT, curvelet transform; NSCT, nonsubsampled contourlet transform; SR, sparse representation; BCF, bilateral cross filter; HSI, hue, saturation, and intensity; PCA, principal component analysis; PET, positron emission tomography; CT, computed tomography; TR, tensor radiomics.
Discussion
Radiomics features can improve clinical task performances by capturing important pathophysiological information (7). In routine practice, radiomics features are included using a fixed set of parameters. In this study we generated TR features by varying multiple parameters such as bin sizes, exploring different segmentation perturbations, varying hyperparameters of pre-processing filters and testing different fusion techniques. We explored the tasks of survival prediction outcomes (sections Results of using discretization flavours, Results of using segmentation flavours and Results of using fusion flavours) as well as test-retest repeatability (section Results of using filter (pre-processing) flavours). Our proposed TR approach allows the learning algorithms to consider different aspects of imaging features, moving beyond conventional paradigms.
Applying bin size variations to create different flavours of TR features for the HNC study described in section Results of using discretization flavours showed its effectiveness in performance enhancement of the outcome prediction task. Compared to the radiomics features extracted in a regular manner, combining the different bin size radiomics features flavours presented more discriminative power for the classification problem. However, not all features from all flavours combined necessarily lead into a more discriminative set of information. Employing flavour combination selection and TR-Net for end-to-end flavour fusion back up this statement (Figure 4). By using SELU activation functions along with dropout regularization in every layer, TR-Net can learn a transformation of features from all flavours to a common latent space in a way that it keeps the contribution of informative features coming from different flavours while decreasing their weaknesses through suppressing the redundant features. Furthermore, motivated by the promising results obtained by TR-Net compared to non-DL results achieved by flavour combinations, we investigated the idea of performing feature selection per each flavour to make our TR feature set less redundant and more discriminative. Prior to applying ML and DL pipelines to their combinations, it is possible to use correlation between the same features from different flavours to remove the highly correlated ones. This work is basically trying to represent proof of the concept of TR and we believe that this is an important refinement that needs to be explored in future studies.
For the study of segmentation flavours in section Results of using segmentation flavours, a majority of LDA model performances saw an overall boost with the addition of TR feature flavours for segmentation perturbation as TR flavours. Notably, the more aggressive image perturbation technique (“TVC”) achieved the highest ROC AUC score and second highest PR AUC as seen in Table 2. The strong performance of the differing bin size TR feature set over the vanilla features also supports the findings of section Results of using discretization flavours vis-a-vis radiomic feature classification power improvement via changing bin size when calculating radiomic features. Continuing to look at Figure 7 the TVC and bin size methods also showed a marked improvement over the vanilla features at low false positive rates, indicating a higher sensitivity at these thresholds. In the context of this task, identifying patients who are less likely to respond to pembrolizumab with a high specificity while maintaining a sensible sensitivity is a crucial finding in patient survival. The models that are able to identify PD patients (i.e., non-responders) allow clinical decision makers to adjust from 1st-line immunotherapy with pembrolizumab (“pembro”) to combination therapy pembro and platinum-doublet chemotherapy in an effort to more aggressively combat late-stage tumour growth. The findings of the study in section Results of using segmentation flavours indicate the power of TR based features in identifying clinicopathological biomarkers that can elucidate medical utility for this homogenous patient population.
The “vanilla” baseline parameters for the first two experiments i.e., the discritization and segmentation flavours were chosen as follows. As per IBSI, image/volume preprocessing (i.e., before feature extraction) was performed using suggested default guidelines and appropriately modified depending on the requisite modality differences. Dataset exploration and analysis determined that critical parameters, such as a CT bin width of 25 HU and an intensity rescaling to the standard window [ for lung WL =−400 HU, WW =1,500 HU], resulted in a sufficient number of bins that did not generate sparse matrices for gray-level based features (e.g., GLCM, GLSZM) and therefore more robust features (as evidenced by the IBSI parameter documentation). A similar analysis pipeline was performed on the HNC PET-CT dataset. Analysis of the PET-CT scans using the Freedman-Diaconis rule indicated that the optimal discretization bin width for PET was 0.3 SUV and 2 HU for CT. Fixed bin width was chosen in lieu of a fixed bin count as it has been thought to improve feature reproducibility in PET scans (39). Other parameter considerations, such as the many possible filters that can be applied to an image with PyRadiomics v.3+ (e.g., LoG, Wavelet transforms) were also applied to the Lung CT dataset to add additional TR flavour dimensions during segmentation flavouring. We believe that this choice allows for a fairer baseline classification performance comparison between TR features and vanilla features.
In section Results of using filter (pre-processing) flavours, we considered TR based on different hyper-parameters of pre-processing filters (LOG and wavelet) increased radiomics-feature repeatability. TR made by different hyper-parameters of LOG preprocessing filter did not show any poorly-repeatable features, and only 2 features had medium repeatability. On the other hand, TR based on different flavours of wavelet filter, had some non-repeatable and repeatable features. TR based on fusion flavours also showed significant improvement over the best non-TR performer (section Results of using fusion flavours). The fusion flavour idea can be further extended to additionally include deep features from different image-fusions via deep neural networks (e.g., autoencoders), adding those to existing TR features to study potentials to further enhance performance.
Beside the effective methods employed in different studies carried out throughout the previous sections of this work, in the frame of TR, there were many other studies that were tested but not all were fruitful. However many valuable lessons were learnt that are indeed worth mentioning here. As a case in point, for bin size flavours study (section Results of using discretization flavours), we tested many other implementations involving binning flavours, but not all were led to meaningful improvements. Among them are the combination of flavours via PCA to create new, blended features. Multiple ML pipelines consisting of an unsupervised feature selection/dimensionality reduction method (univariate feature selection, PCA with a linear kernel, PCA with a RBF kernel) and an ML classifier (logistic regression, random forest, SVM, k nearest neighbors) were developed which showed no consistent increase or decrease in terms of classification scores when compared to a baseline that used only one flavour.
This approach was repeated using LDA to combine flavours with similarly inconclusive results. These results compared to that of the reported ones in section Results of using discretization flavours may indicate that a more nuanced approach is necessary to effectively include new bin flavours. Potential ideas include switching out PCA for a more sophisticated method of flavour combination, such as an autoencoder. Adding more flavour types other than bin flavours could also help to make more robust TR features. In this case LDA, PCA or any other feature extraction/dimensionality reduction method could perform an effective feature/flavour combination and as a result, a better prediction performance. These findings could back up the idea that employing domain knowledge in picking the best flavour types per each study case is crucial. In other words, beside finding the best method to combine multiple flavours, finding the most informative and relevant flavour variations for the task at hand is an important factor that needs to be taken carefully.
In this paper, we employed both machine learning and deep learning. Deep learning shows that additional features can benefit by providing more information representation of a sample, but overfitting is avoided. On the other hand, machine learning is more nuanced. We aimed to avoid the curse of dimensionality by performing feature selection and/or dimensionality reduction prior to feeding features into machine-learning models. A large pool of features was initially considered, but vetted to provide a more reasonable set of features for model training.
TR is important in the context of hand-crafted radiomics because previously published studies commonly choose some parameters a priori without necessarily reliable justification. Areas for future exploration include establishing the effectiveness of TR methods on other larger datasets, improved methods (e.g., use of methods for the analysis of longitudinal data, imagining different flavours as different time-points in feature-space), and exploring new flavours (e.g., deep features as mentioned earlier). The TR paradigm enables revisiting past efforts and re-studying them in different light. For instance, subregional intratumour radiomics were defined based on individual- and population-level clustering: combining feature flavours generated from different tumour partitions can result in a new form of TR. In addition, shell features reflecting the tumour micro environment can be extracted from different sizes of peritumoural regions and their combinations can also be explored in the proposed context of TR.
Conclusions
For radiomics analyses, fixed parameter values are commonly used to generate feature values (e.g., discretization (bin number or size), pre-processing filtering, segmentation, or multi-modality fusion). At best, results generated via different flavours are compared to one another. Our proposed paradigm is to move beyond this, to use radiomics tensors of features calculated with multiple combinations of flavours. We applied this paradigm to different modalities, tasks and algorithms (ML and DL). Our results, from different studies and modalities, revealed that TR has the potential to enable improved task performances.
Supplementary
The article’s supplementary files as
Acknowledgments
We published the arXiv version of this paper in October 2022.
Funding: This work was in part supported by BC Cancer Foundation and the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (No. RGPIN-2019-06467 to Arman Rahmim), the Canadian Institutes of Health Research (CIHR) Project Grants (Nos. PJT-162216 and PJT-173231 to Arman Rahmim), the Swiss National Science Foundation Grant (No. SNRF 320030_176052 to Habib Zaidi), and the BC Cancer Foundation (to Arman Rahmim).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). CT volume data were from an ongoing study approved by the University of British Columbia and BC Cancer Agency review ethics board (No. UBC/BCCA REB #H19-01597). Ethics approval and informed consent to participate were not needed for other studies in the paper since the study was performed on open access online dataset (PET-CT scans of head and neck cancer, and MR images of GBM).
Footnotes
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://qims.amegroups.com/article/view/10.21037/qims-23-163/coif). A.R. reports that this work was in part supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery Grant (No. RGPIN-2019-06467), the Canadian Institutes of Health Research (CIHR) Project Grants (Nos. PJT-162216 and PJT-173231), and the BC Cancer Foundation. H.Z. reports that this work was in part supported by the Swiss National Science Foundation Grant (No. SNRF 320030_176052). C.H. has received honoraria paid to self from Abbvie, Amgen, AstraZeneca, Bayer, BMS, Eisai, Jazz, Janssen, Merck, Novartis, Pfizer, Roche, Sanofi and research grants paid to the institution from AstraZeneca and Roche. A.R. and C.U. are cofounders of Ascinta technologies Inc. The other authors have no conflicts of interest to declare.
References
- 1.Gillies RJ, Anderson AR, Gatenby RA, Morse DL. The biology underlying molecular imaging in oncology: from genome to anatome and back again. Clin Radiol 2010;65:517-21. 10.1016/j.crad.2010.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Timmeren JE, Cester D, Tanadini-Lang S, Alkadhi H, Baessler B. Radiomics in medical imaging-”how-to” guide and critical reflection. Insights Imaging 2020;11:91. 10.1186/s13244-020-00887-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orlhac F, Nioche C, Klyuzhin I, Rahmim A, Buvat I. Radiomics in PET Imaging:: A Practical Guide for Newcomers. PET Clin 2021;16:597-612. 10.1016/j.cpet.2021.06.007 [DOI] [PubMed] [Google Scholar]
- 4.Koçak B, Durmaz EŞ, Ateş E, Kılıçkesmez Ö. Radiomics with artificial intelligence: a practical guide for beginners. Diagn Interv Radiol 2019;25:485-95. 10.5152/dir.2019.19321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zwanenburg A, Vallières M, Abdalah MA, Aerts HJWL, Andrearczyk V, Apte A, et al. The Image Biomarker Standardization Initiative: Standardized Quantitative Radiomics for High-Throughput Image-based Phenotyping. Radiology 2020;295:328-38. 10.1148/radiol.2020191145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang Y, Liu Z, He L, Chen X, Pan D, Ma Z, Liang C, Tian J, Liang C. Radiomics Signature: A Potential Biomarker for the Prediction of Disease-Free Survival in Early-Stage (I or II) Non-Small Cell Lung Cancer. Radiology 2016;281:947-57. 10.1148/radiol.2016152234 [DOI] [PubMed] [Google Scholar]
- 7.Tomaszewski MR, Gillies RJ. The Biological Meaning of Radiomic Features. Radiology 2021;299:E256. 10.1148/radiol.2021202553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guiot J, Vaidyanathan A, Deprez L, Zerka F, Danthine D, Frix AN, Lambin P, Bottari F, Tsoutzidis N, Miraglio B, Walsh S, Vos W, Hustinx R, Ferreira M, Lovinfosse P, Leijenaar RTH. A review in radiomics: Making personalized medicine a reality via routine imaging. Med Res Rev 2022;42:426-40. 10.1002/med.21846 [DOI] [PubMed] [Google Scholar]
- 9.Yousefirizi F, Pierre Decazes, Amyar A, Ruan S, Saboury B, Rahmim A. AI-Based Detection, Classification and Prediction/Prognosis in Medical Imaging:: Towards Radiophenomics. PET Clin 2022;17:183-212. 10.1016/j.cpet.2021.09.010 [DOI] [PubMed] [Google Scholar]
- 10.Da-Ano R, Visvikis D, Hatt M. Harmonization strategies for multicenter radiomics investigations. Phys Med Biol 2020;65:24TR02. 10.1088/1361-6560/aba798 [DOI] [PubMed] [Google Scholar]
- 11.Zwanenburg A. Radiomics in nuclear medicine: robustness, reproducibility, standardization, and how to avoid data analysis traps and replication crisis. Eur J Nucl Med Mol Imaging 2019;46:2638-55. 10.1007/s00259-019-04391-8 [DOI] [PubMed] [Google Scholar]
- 12.Ha S, Choi H, Paeng JC, Cheon GJ. Radiomics in Oncological PET/CT: a Methodological Overview. Nucl Med Mol Imaging 2019;53:14-29. 10.1007/s13139-019-00571-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li R, Xing L, Napel S, Rubin DL. Radiomics and Radiogenomics: Technical Basis and Clinical Applications. CRC Press; 2019. 442 p. Available online: https://play.google.com/store/books/details?id=cuyjDwAAQBAJ [Google Scholar]
- 14.Cook GJR, Azad G, Owczarczyk K, Siddique M, Goh V. Challenges and Promises of PET Radiomics. Int J Radiat Oncol Biol Phys 2018;102:1083-9. 10.1016/j.ijrobp.2017.12.268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galavis PE, Hollensen C, Jallow N, Paliwal B, Jeraj R. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol 2010;49:1012-6. 10.3109/0284186X.2010.498437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shiri I, Rahmim A, Ghaffarian P, Geramifar P, Abdollahi H, Bitarafan-Rajabi A. The impact of image reconstruction settings on 18F-FDG PET radiomic features: multi-scanner phantom and patient studies. Eur Radiol 2017;27:4498-509. 10.1007/s00330-017-4859-z [DOI] [PubMed] [Google Scholar]
- 17.Suter Y, Knecht U, Alão M, Valenzuela W, Hewer E, Schucht P, Wiest R, Reyes M. Radiomics for glioblastoma survival analysis in pre-operative MRI: exploring feature robustness, class boundaries, and machine learning techniques. Cancer Imaging 2020;20:55. 10.1186/s40644-020-00329-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin P, Mao N, Zhao C, Wu J, Sun C, Chen L, Hong N. Comparison of radiomics machine-learning classifiers and feature selection for differentiation of sacral chordoma and sacral giant cell tumour based on 3D computed tomography features. Eur Radiol 2019;29:1841-7. 10.1007/s00330-018-5730-6 [DOI] [PubMed] [Google Scholar]
- 19.Mayerhoefer ME, Materka A, Langs G, Häggström I, Szczypiński P, Gibbs P, Cook G. Introduction to Radiomics. J Nucl Med 2020;61:488-95. 10.2967/jnumed.118.222893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klyuzhin IS, Xu Y, Ortiz A, Ferres JML, Hamarneh G, Rahmim A. Testing the ability of convolutional neural networks to learn radiomic features. bioRxiv. medRxiv; 2020. Available online: http://medrxiv.org/lookup/doi/10.1101/2020.09.19.20198077 [DOI] [PubMed]
- 21.Hatt M, Vallieres M, Visvikis D, Zwanenburg A. IBSI: an international community radiomics standardization initiative. J Nucl Med 2018;59:287. [Google Scholar]
- 22.Vallières M, Kay-Rivest E, Perrin LJ, Liem X, Furstoss C, Aerts HJWL, Khaouam N, Nguyen-Tan PF, Wang CS, Sultanem K, Seuntjens J, El Naqa I. Radiomics strategies for risk assessment of tumour failure in head-and-neck cancer. Sci Rep 2017;7:10117. 10.1038/s41598-017-10371-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van Griethuysen JJM, Fedorov A, Parmar C, Hosny A, Aucoin N, Narayan V, Beets-Tan RGH, Fillion-Robin JC, Pieper S, Aerts HJWL. Computational Radiomics System to Decode the Radiographic Phenotype. Cancer Res 2017;77:e104-7. 10.1158/0008-5472.CAN-17-0339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yip SSF, Aerts HJW. Applications and limitations of radiomics. Phys Med Biol. 2016;61:R150. 10.1088/0031-9155/61/13/R150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Toosi A, Bottino A, Cumani S, Negri P, Sottile PL. Feature Fusion for Fingerprint Liveness Detection: a Comparative Study. IEEE Access 2017;5:23695-709.
- 26.Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP. SMOTE: Synthetic Minority Over-sampling Technique. J Artif Intell Res 2002;16:321-57. [Google Scholar]
- 27.Klambauer G, Unterthiner T, Mayr A, Hochreiter S. Self-Normalizing Neural Networks. Adv Neural Inf Process Syst 2017;30. Available online: https://proceedings.neurips.cc/paper/6698-self-normalizing-neural-networks
- 28.Aha DW, Bankert RL. A Comparative Evaluation of Sequential Feature Selection Algorithms. In: Fisher D, Lenz HJ, editors. Learning from Data: Artificial Intelligence and Statistics V. New York, NY: Springer New York; 1996. p. 199-206. [Google Scholar]
- 29.Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe S, O’Brien M, Rao S, Hotta K, Leiby MA, Lubiniecki GM, Shentu Y, Rangwala R, Brahmer JR; KEYNOTE-024 Investigators. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 2016;375:1823-33. 10.1056/NEJMoa1606774 [DOI] [PubMed] [Google Scholar]
- 30.Gandhi L, Rodríguez-Abreu D, Gadgeel S, Esteban E, Felip E, De Angelis F, et al. Pembrolizumab plus Chemotherapy in Metastatic Non-Small-Cell Lung Cancer. N Engl J Med 2018;378:2078-92. 10.1056/NEJMoa1801005 [DOI] [PubMed] [Google Scholar]
- 31.Paz-Ares L, Luft A, Vicente D, Tafreshi A, Gümüş M, Mazières J, et al. Pembrolizumab plus Chemotherapy for Squamous Non-Small-Cell Lung Cancer. N Engl J Med 2018;379:2040-51. 10.1056/NEJMoa1810865 [DOI] [PubMed] [Google Scholar]
- 32.Eisenhauer EA, Therasse P, Bogaerts J, Schwartz LH, Sargent D, Ford R, Dancey J, Arbuck S, Gwyther S, Mooney M, Rubinstein L, Shankar L, Dodd L, Kaplan R, Lacombe D, Verweij J. New response evaluation criteria in solid tumours: revised RECIST guideline (version 1.1). Eur J Cancer 2009;45:228-47. 10.1016/j.ejca.2008.10.026 [DOI] [PubMed] [Google Scholar]
- 33.Zwanenburg A, Leger S, Agolli L, Pilz K, Troost EGC, Richter C, Löck S. Assessing robustness of radiomic features by image perturbation. Sci Rep 2019;9:614. 10.1038/s41598-018-36938-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barboriak D. Data from RIDER_NEURO_MRI. Data from RIDER_NEURO_MRI. 2015. Available online: https://wiki.cancerimagingarchive.net/display/Public/RIDER+NEURO+MRI
- 35.Shiri I, Hajianfar G, Sohrabi A, Abdollahi H, P, Shayesteh S, Geramifar P, Zaidi H, Oveisi M, Rahmim A. Repeatability of radiomic features in magnetic resonance imaging of glioblastoma: Test-retest and image registration analyses. Med Phys 2020;47:4265-80. 10.1002/mp.14368 [DOI] [PubMed] [Google Scholar]
- 36.Lv W, Ashrafinia S, Ma J, Lu L, Rahmim A. Multi-Level Multi-Modality Fusion Radiomics: Application to PET and CT Imaging for Prognostication of Head and Neck Cancer. IEEE J Biomed Health Inform 2020;24:2268-77. 10.1109/JBHI.2019.2956354 [DOI] [PubMed] [Google Scholar]
- 37.Ashrafinia S. Quantitative Nuclear Medicine Imaging using Advanced Image Reconstruction and Radiomics. Johns Hopkins University, 2019; 2019. [Google Scholar]
- 38.St»hle L, Wold S. Analysis of variance (ANOVA). Chemometrics Intellig Lab Syst 1989;6:259-72. [Google Scholar]
- 39.Leijenaar RT, Nalbantov G, Carvalho S, van Elmpt WJ, Troost EG, Boellaard R, Aerts HJ, Gillies RJ, Lambin P. The effect of SUV discretization in quantitative FDG-PET Radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep 2015;5:11075. 10.1038/srep11075 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as