Abstract
Two membrane proteins encoded by the malonate fermentation gene cluster of Malonomonas rubra, MadL and MadM, have been synthesized in Escherichia coli. MadL and MadM were shown to function together as a malonate transport system, whereas each protein alone was unable to catalyze malonate transport. Malonate transport by MadLM is Na+ dependent, and imposition of a ΔpNa+ markedly enhanced the rate of malonate uptake. The kinetics of malonate uptake into E. coli BL21(DE3) cells synthesizing MadLM at different pH values indicated that Hmalonate− is the transported malonate species. The stimulation of malonate uptake by Na+ ions showed Michaelis-Menten kinetics, and a Km for Na+ of 1.2 mM was determined. These results suggest that MadLM is an electroneutral Na+/Hmalonate− symporter and that it is dependent on two separate genes.
Malonomonas rubra, a strictly anaerobic, gram-negative bacterium, receives its entire energy for growth from the decarboxylation of malonate to acetate and CO2 (5). As the free energy of malonate decarboxylation (ΔGo′ = −17.4 kJ/mol) is not sufficient to support the synthesis of ATP via substrate level phosphorylation, ATP synthesis must be coupled to an energy-rich ion gradient over the membrane. The decarboxylation of malonate in M. rubra was shown to be mediated by a distinct set of soluble and membrane-bound enzymes. The overall reaction was specifically activated by Na+ ions and was therefore assumed to involve Na+ ion pumping across the membrane (10, 11). Recently, the genes encoding the different proteins involved in malonate decarboxylation have been cloned and sequenced. These are located within a cluster of 14 genes. The function of most of the derived proteins could be deduced from sequence comparisons and could be related to specific steps in the malonate fermentation (1). One of the genes, madB, encodes a highly hydrophobic protein with significant sequence homologies to the membrane-bound decarboxylase subunit of other Na+-pumping decarboxylases, e.g., the oxaloacetate decarboxylase from Klebsiella pneumoniae (1, 7). Consequently, it was concluded that the malonate decarboxylase from M. rubra is also a member of the Na+ transport decarboxylase family of enzymes. The ΔμNa+ (electrochemical sodium ion gradient) generated must be used for the synthesis of ATP via an F1F0 ATPase. Part of the driving force could be consumed for the uptake of the growth substrate, malonate, into the cells. To address this possibility, we considered the gene for the putative malonate transporter to be located within the malonate fermentation gene cluster. Hydropathy analyses of the deduced gene products of this cluster indicated that along with MadB, MadL and MadM are integral membrane proteins (1). In this work, we concentrated on the madL and madM genes and their functional expression in Escherichia coli. The results indicate that the malonate carrier of M. rubra is dependent on both genes and catalyzes the ΔpNa+-driven transport of Hmalonate− over the membrane.
MATERIALS AND METHODS
Materials.
E. coli DH5α was used for general cloning purposes, and E. coli BL21(DE3) was used for expression of the madL and madM genes. pET-24a(+) (Novagen) and pACYC184 (New England Biolabs) were used as cloning and expression vectors.
General procedures.
Luria Bertani (LB) broth and LB agar were used for routine bacterial growth (19). Kanamycin was added at 50 μg ml−1, and chloramphenicol was added at 40 μg ml−1. Recombinant DNA procedures were performed as described previously (19). DNA fragments from agarose gels were isolated with QIAEX (Qiagen). Cloned PCR fragments were sequenced according to the dideoxy nucleotide chain termination method (20) by using an Amplitaq DNA sequencing kit and the model 310 Genetic Analyzer from Applied Biosystems.
Construction of the expression vectors.
The following mixture (50 μl) was used for PCR: 20 mM Tris-HCl (pH 8.0), 10 mM KCl, 6 mM (NH4)2SO4, 2 mM MgCl2, 0.5 mg of bovine serum albumin per ml, 2 mM concentrations of each deoxynucleoside triphosphate, 2.6 μM site-specific primers, and 100 ng of genomic DNA from M. rubra as template. Two units of native Pfu polymerase (Stratagene) was added. After an initial denaturation step at 94°C for 3 min, 35 cycles were carried out for 30 s at 94°C, 30 s at 60°C, and 3 min at 75°C. The forward primer madLM-for (5′-GGA GTC TCA TAT GGT CAT CTA TGG GGT A-3′) is identical to bases 9656 to 9683 of the M. rubra mad gene cluster (GenBank database accession number U89780), introducing a cleavage site for the restriction endonuclease NdeI (altered bases are underlined). The reverse primer madLM-rev2 (5′-ATT AGA AGC TTT CCG TAA ATG CCC TTC ATA A-3′) is complementary to the bases 10825 to 10855. The primer introduces a cleavage site for the restriction endonuclease HindIII (altered bases are underlined) and deletes the original stop codon of madM. The PCR product was restricted with NdeI/HindIII and ligated into NdeI/HindIII-restricted pET-24a(+), yielding pET-LM. Plasmid pET-LM carries a gene encoding, in addition to MadL, MadM comprising a C-terminal tag of six consecutive histidine residues (His tag). Subsequently, pET-LM was restricted with HindIII and BseRI, treated with T4 DNA polymerase, and religated to obtain pET-L. For construction of pET-M, pET-LM was restricted with NdeI and NheI and the recessed 3′ termini were filled in with Klenow polymerase and religated. The plasmids pET-L and pET-M were restricted with Bpu1102I, the overhanging ends were filled in a Klenow reaction, and the resulting fragments were subsequently digested with BglII. The madL- or madM-bearing fragments were ligated into BamHI- and NruI-restricted pACYC184 to obtain pACYC-L and pACYC-M, respectively. The DNA sequence of the inserts carrying the madL and madM genes and the fusion sites were confirmed by sequencing.
Expression of MadL and MadM.
E. coli BL21(DE3) cells harboring the indicated plasmid constructs were grown aerobically on LB medium (1 liter) supplemented with the appropriate antibiotics at 37°C with shaking at 180 rpm. At an optical density at 600 nm of 0.8 to 1.2, IPTG (isopropyl-β-d-thiogalactopyranoside) (final concentration, 0.2 mM) was added and the culture was grown for another 1.5 h at 37°C with shaking at 180 rpm. Cells were washed at least three times with Na+-poor 50 mM potassium phosphate, pH 7.0, unless indicated otherwise, until a Na+ concentration of about 25 μM was obtained. Cells were resuspended in the same buffer at a concentration of about 10 to 15 mg of protein/ml and stored on ice until use.
Malonate transport assays.
Transport measurements were performed at room temperature. Malonate transport was initiated by addition of 1 μl of [2-14C]malonic acid (1.8 mM, 56 mCi/mmol; ICN) to 99 μl of cells in 50 mM potassium-phosphate, pH 7.0, containing 10 mM NaCl. The transport reaction was stopped after various times by addition of 900 μl of ice-cold 0.1 M LiCl to the sample and rapid filtration through 0.45-μm-pore-size cellulose nitrate filters (Sartorius). The filters were washed once with 1 ml of ice-cold 0.1 M LiCl and placed into scintillation vials. After addition of 4 ml of scintillation fluid (Irga-Safe+; Canberra Packard), the entrapped [2-14C]malonate was determined with a liquid scintillation counter. Experimental values were corrected for zero-time controls by simultaneous addition of [2-14C]malonic acid (1 μl, 1.8 mM) and 900 μl of ice-cold 0.1 M LiCl to 99 μl of cells, followed by rapid filtration and washing of the sample as described above.
(i) Effect of alkali ions on the rate of malonate transport.
To investigate the effect of Na+ and Li+ ions on malonate transport, E. coli BL21(DE3)/pET-LM cells (97 μl) suspended in 50 mM potassium phosphate buffer, pH 7.5, were preincubated with 1 μl of 0.5 M NaCl or LiCl for 1 min at room temperature prior to the addition of 2 μl of 2.9 mM [2-14C]malonate (34,730 cpm/nmol of specific activity) to start the transport process. ΔpNa+-driven malonate uptake was initiated by addition of 3 μl of 1.67 M NaCl, containing 1.93 mM [2-14C]malonate (34,730 cpm/nmol), to 97 μl of Na+-free E. coli BL21(DE3)/pET-LM cells. After given times, the reaction was terminated by dilution and filtration as described above.
(ii) Determination of the transported malonate species.
To determine the kinetic constants (Km, Vmax) of MadLM for malonate, transport experiments were performed with E. coli BL21(DE3)/pET-LM cells (suspended in 50 mM potassium phosphate, Na+ free) at three different pH values (6.5, 7.0, and 7.5). Transport was essentially performed as described above except that the final NaCl concentration used in this assay was 50 mM. Radioactively labeled malonate was added to final concentrations ranging from 4 to 100 μM. The initial rate values were determined from 30-s measurements. The data sets were analyzed by fitting to the Michaelis-Menten or Hill equation (4) by using the computer program Sigma Plot (Jandel Scientific, SSPS). The apparent Kms (Kmapps) for the individual malonate species (H2malonate, Hmalonate−, and malonate2−) were derived by using the Henderson-Hasselbach equation.
(iii) Determination of the Km for Na+.
To determine the Na+ kinetics of Na+/malonate uptake, transport assays were performed with Na+-free washed E. coli BL21(DE3)/pET-LM cells at pH 6.5. The transport assay was started by simultaneous addition of NaCl (0.05 to 10 mM final concentration) and [2-14C]malonic acid (final concentration, 18 μM). Mixtures of NaCl and KCl were used to provide equal salt concentrations (10 mM chloride) in the separate experiments. Data sets were analyzed by fitting to the Michaelis-Menten or Hill equation as described above.
Other methods.
Protein concentration was measured by the method of Bradford (3) by using bovine serum albumin as a standard. Na+ concentrations were determined with an atomic absorption-flame emission spectrophotometer (Shimadzu, type AA-646).
RESULTS
Cloning and expression of the madL and madM genes.
The madL and madM genes were amplified from genomic M. rubra DNA. The primers used in this PCR procedure were designed to introduce restriction sites (NdeI and HindIII) flanking the genes and to delete the stop codon of the madM gene in the resulting amplification product. After restriction with NdeI and HindIII, the PCR fragment was cloned into pET24a(+), yielding pET-LM. This construct encodes (besides MadL) a MadM protein including a 6× poly-histidine extension (His tag) at its C terminus. Deletion of the madL or madM gene by restriction and religation of pET-LM resulted in the creation of pET-L and pET-M, respectively. Furthermore, pET-L and pET-M served as donors for the madL and madM genes present in the pACYC184 derivatives, pACYC-L and pACYC-M, respectively. These plasmids were used to investigate the function of the madL and madM gene products in E. coli as host. E. coli BL21(DE3) cells expressing madL and/or madM were sensitive to induction by IPTG (0.2 mM), which led to severe growth inhibition. Due to the limited synthesis of the madL and madM gene products, these proteins could not be detected by sodium dodecyl sulfate-gel electrophoresis analysis. Therefore, expression was monitored by uptake of [2-14C]malonate into cells harvested about 90 min after induction with IPTG.
Concomitant expression of madL and madM is required to evoke transport of [2-14C]malonate in E. coli.
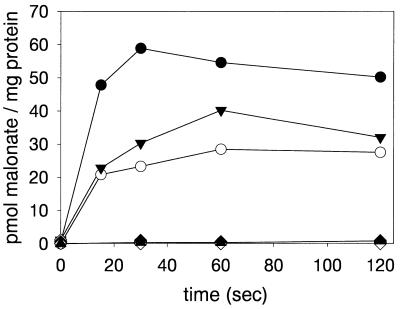
Since E. coli can neither grow nor metabolize malonate (12, 13), it provides an excellent system for studying malonate uptake transporters. Uptake of [2-14C]malonate could readily be detected in E. coli BL21(DE3)/pET-LM, whereas cells harboring the vector pET24a(+) or plasmids containing only one of the genes (pET-L, pET-M, pACYC-L, or pACYC-M) exhibited no malonate transport activity (Fig. 1). The expression of both madL and madM in trans from pET-L and pET-M in combination with the pACYC184 derivatives demonstrated that the presence of both polypeptides is imperative for malonate accumulation in the cells. Cells expressing madL and madM in trans showed about 40 to 50% of the malonate uptake activity of cells expressing both genes in cis (i.e., from plasmid pET-LM). The results clearly show the necessity for the presence of both MadL and MadM in order to render malonate uptake activity in E. coli.
FIG. 1.
Malonate uptake into E. coli cells expressing madL and/or madM. E. coli BL21(DE3) cells transformed with pET-LM (expressing madL and madM in cis) (•) or pACYC-M and pET-L (trans) (○) or pACYC-L and pET-M (trans) (▾) were compared. In controls, E. coli BL21(DE3) was transformed with pET-24a(+) (vector control) (▴), pET-L (expression of madL) (⧫), pET-M (madM) (◊), pACYC-L (madL) (▾), or pACYC-M (madM) (▿). Cells in 50 mM potassium phosphate buffer, pH 7.0, containing 10 mM NaCl (99 μl) were incubated for 1 min at room temperature, prior to the addition of 1 μl of [2-14C]malonate (1.8 mM; specific activity, 113 cpm/pmol). After the times indicated, samples were treated as described in Materials and Methods.
Reversibility of the MadLM malonate uptake system.
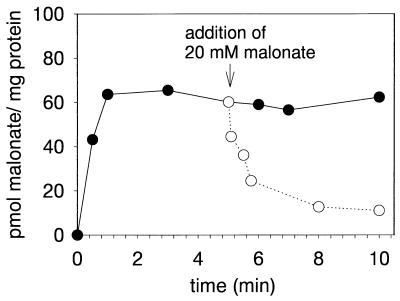
We obtained preliminary evidence for MadLM functioning as a secondary uptake system by the release of previously accumulated radioactively labeled malonate from E. coli BL21(DE3)/pET-LM cells upon addition of a 1,000-fold excess of unlabeled malonate (Fig. 2). In a period of 5 min, almost 90% of the previously accumulated [2-14C]malonate was released from the cells. This type of induced release of radioactive material is quite typical for secondary active transport systems, as has been shown, e.g., for the anaerobic dicarboxylate uptake systems in E. coli (8). Moreover, this induced release was not observed with a primary active transport system, the aerobic dicarboxylate system of E. coli (8, 14). In the case of the MadLM system, efflux of radioactive malonate could not be provoked with oxalate, succinate, fumarate, and malate, the latter two being alternative growth substrates for M. rubra (6), indicating a narrow substrate specificity of the MadLM transport system (not shown).
FIG. 2.
Release of intracellular [2-14C]malonate from E. coli BL21(DE3)/pET-LM by addition of 20 mM external malonate. E. coli BL21(DE3)/pET-LM cells suspended in 50 mM potassium phosphate, pH 7.0, containing 10 mM NaCl (99 μl) were preincubated for 1 min at room temperature. Subsequently, the transport assay was initiated by addition of 1 μl of [2-14C]malonate (1.8 mM; specific activity, 113 cpm/pmol). After 5 min, 100 μl of 40 mM potassium malonate (pH 7.0) was added (○). In the control (•), 100 μl of 50 mM potassium phosphate buffer (pH 7.0) was added. After the times indicated, samples were treated as described in Materials and Methods.
Driving forces of malonate transport and ion specificity.
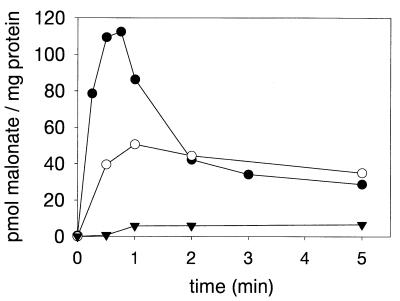
All the transport experiments described above were performed in the presence of NaCl. To obtain information about the ionic specificity of the MadLM transport system, the Na+ content of the cells was reduced as far as possible (<25 μM) by washing with potassium phosphate buffer. The resulting E. coli BL21(DE3)/pET-LM cells with less than 25 μM Na+ were unable to accumulate added [2-14C]malonate (Fig. 3). In comparison, the same cells preincubated with 50 mM NaCl (20 min) before the addition of radioactively labeled malonate showed substantial malonate transport activity (about a 13-fold stimulation of the initial rate). These results clearly indicate that malonate transport by MadLM is Na+ dependent. Preincubation of Na+-poor cells with LiCl (50 mM final concentration) resulted in a stimulation of about 3.4-fold, in comparison to cells without added salts (not shown). The stimulation of malonate uptake by Li+ ions is not surprising, since most of the Na+-dependent transport systems can use Li+ as an alternative coupling ion (7, 16). The highest malonate transport rate was obtained by simultaneous addition of NaCl (50 mM final concentration) and [2-14C]malonate to the Na+-free cells (imposition of a ΔpNa+). The initial malonate uptake rate in this experiment is about 53-fold in comparison to the uptake rate of Na+-poor washed cells and about fourfold higher than the uptake rate in cells preincubated with the same amount of NaCl. The accumulated [2-14C]malonate rapidly effluxes from the cells, suggesting a rapid breakdown of the applied Na+ gradient. The level of radioactivity remaining in the cells reached approximately the same level as observed with Na+-equilibrated cells. The results give preliminary indication of MadLM functioning as a ΔpNa+-driven, secondary active malonate transport system.
FIG. 3.
Na+ dependency of malonate transport in E. coli BL21(DE3)/pET-LM. Transport was initiated by addition of 2 μl of 2.9 mM [2-14C]malonate (34,730 cpm/nmol) to 98 μl of cells suspended in 50 mM potassium phosphate buffer, pH 7.5, without NaCl addition (<25 μM Na+) (▾) or with 50 mM NaCl (○). ΔpNa+-driven malonate uptake was initiated by addition of 3 μl of 1.67 M NaCl, containing 1.93 mM [2-14C]malonate (34,730 cpm/nmol), to 97 μl of Na+-free cells (•). After the times indicated, samples were treated as described in Materials and Methods.
Affinity of MadLM for malonate.
The Kms of MadLM for malonate were determined by ΔpNa+-driven malonate transport at three different pH values (6.5, 7.0, and 7.5). From the data obtained, Kmapps of the different malonate species were calculated. As is shown in Table 1, the Kmapps for the onefold-protonated malonate species (Hmalonate−) remained nearly constant upon variation of the pH, whereas the Kmapps for malonic acid (H2malonate) and the unprotonated malonate (malonate2−) changed by factors of 15 and 6.4, respectively, over the pH range tested. Assuming the variation of the pH in the range tested does not significantly affect the malonate binding site(s) of the malonate carrier, we conclude from these results that Hmalonate− is the actual substrate transported by MadLM.
TABLE 1.
Kinetic analysis of malonate uptakea
| pH |
Kmapp (μM)
|
Vmax | |||
|---|---|---|---|---|---|
| H2malonate | Hmalonate− | Malonate2− | Malonate total | ||
| 6.5 | 0.0003 | 1.39 | 9.81 | 11.2 | 1.52 |
| 7.0 | 0.00007 | 1.09 | 24.41 | 25.5 | 1.01 |
| 7.5 | 0.00002 | 0.88 | 62.62 | 63.5 | 0.95 |
Determination of the uptake of [2-14C]malonate was performed with E. coli BL21(DE3)/pET-LM at the pH values indicated and under conditions where a ΔpNa+ was imposed. Initial values were obtained from 30-s measurements. The concentration of the respective malonate species was calculated by using the Henderson-Hasselbach equation. The Kmapps for Hmalonate− are given in bold type. The pKas for malonate are 2.83 and 5.69. Data sets were analyzed by using Lineweaver-Burk plots. Vmax is given in nanomoles of malonate per minute per milligram of protein.
Affinity of MadLM for Na+ ions.
The experiments described provided evidence for a Na+-dependent malonate uptake system. To determine the affinity constant of MadLM for Na+ ions, we performed uptake studies with Na+-free washed E. coli(DE3)/pET-LM cells. To provide a driving force for the malonate uptake, [2-14C]malonate (18 μM final concentration) and NaCl (0.05 to 10 mM final concentration) were added simultaneously to the cells. The initial transport rates were analyzed by fitting to the Hill equation (4). The calculated K0.5 value for Na+ at pH 6.5 was 1.19 mM, and the Vmax value was 4 nmol/min/mg of protein. The calculated Hill coefficient was 0.96, pointing towards a noncooperative mechanism of Na+ transport.
DISCUSSION
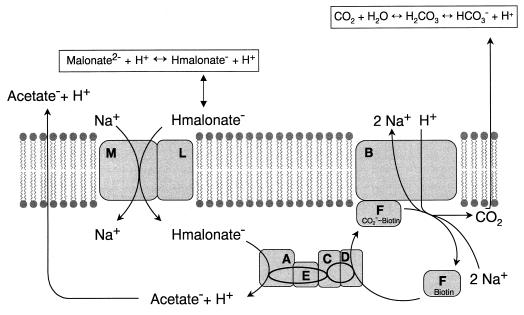
In this work, we demonstrate that the malonate carrier from M. rubra is encoded by two genes, madL and madM. Expression of both genes is imperative to render an active malonate transport system. Expression of either the madL or the madM gene in E. coli did not confer malonate transport activity on this host. To demonstrate that the lack of malonate uptake activity was not due to the lack of expression of madL or madM, we expressed both genes in trans. In this experiment, the expression of a functional malonate carrier clearly demonstrated that both genes are properly transcribed and translated (Fig. 1). Based on hydropathy plot analysis, four and seven putative transmembrane regions were predicted for MadL and MadM, respectively (1). Most secondary active carriers of bacterial origin are predicted or have been shown to consist of 10 to 12 transmembrane α helices, connected by mainly short, hydrophilic loops. The proteins usually are divided into two halves by a large hydrophilic (cytoplasmic) domain (6 + 6 symmetric arrangement) (15). Notably, if the MadLM subunit structure were heterodimeric, the transport system would theoretically contain 11 transmembrane helices in a 4 + 7 asymmetric arrangement. It is obvious from the data presented in this report that MadLM functions as a Na+-dependent malonate transporter. More difficult to interpret is the actual driving force(s) for malonate uptake into the cell. Preincubation of E. coli BL21(DE3)/pET-LM cells with 50 mM Na+ (no ΔpNa+ present) leads to accumulation of [2-14C]malonate inside these cells. More significant, however, is the accumulation of the labeled malonate in the presence of a ΔpNa+ (Fig. 3). The accumulated [2-14C]malonate rapidly effluxes from the cell, probably caused by dissipation of the ΔpNa+ by action of, e.g., Na+/H+ antiporters. This experiment furthermore demonstrated the reversibility of malonate transport by MadLM (which is further confirmed by the results of the exchange experiment shown in Fig. 2). Hence, a directed transport of malonate by action of a primary active transport system could be excluded. The identification of other driving forces, like ΔΨ (electrical potential) and ΔpH, proved to be more difficult than anticipated. We successfully performed [14C]proline uptake experiments with E. coli BL21(DE3)/pET-LM cells (in the presence of 10 mM Na+ but no ΔpNa+ imposed), and uptake of radioactively labeled proline could be forestalled by carbonyl cyanide m-chlorophenylhydrazone (CCCP) (a protonophor) (data not shown). These control experiments proved the presence of a proton-motive force across the cell membrane under the conditions tested. Analogous uptake experiments in E. coli BL21(DE3)/pET-LM using [2-14C]malonate showed that the initial uptake rate was almost unaffected by CCCP (100 μM), but the final accumulation of the substrate was about 50 to 70% that of the control without CCCP (data not shown). Moreover, investigation of [2-14C]malonate uptake with EDTA-treated cells (9) in the presence or absence of valinomycin (disrupts ΔΨ) led to similar results (not shown). Unfortunately, the interpretation of the results from these experiments was too difficult to allow a final conclusion. On the other hand, the results obtained from the kinetics of malonate and Na+ uptake experiments were essentially clear. We could determine the onefold-protonated species of malonate (Hmalonate−) as the probable substrate for MadLM (Table 1). Moreover, we could demonstrate that the Na+-dependent malonate uptake by MadLM followed Michaelis-Menten kinetics, i.e., a Hill coefficient of about 1 was found. Assuming that the presence of more than one binding site for Na+ on the MadLM system would evoke cooperative kinetics (i.e., a Hill coefficient of >1), we tend to believe that the actual stoichiometry between Na+ and Hmalonate− is one. This would then indicate an electroneutral Na+/Hmalonate− symport by MadLM, and the sole driving force would be the ΔpNa+ (and the ΔμHmalonate−). To resolve this matter, we intend to purify the MadLM complex and reconstitute it into liposomes. Elucidation of the driving force(s) will then be performed with the help of artificial ion gradients, as was successfully demonstrated in the case of the Na+-dependent citrate carrier from K. pneumoniae (17, 18). Because M. rubra growing on malonate lives at an energetical subsistence level (2 g of dry cell matter formed from 1 mol of malonate), it was surprising to find that this organism uses part of the ΔμNa+, created by conversion of the decarboxylation energy (malonate→acetate + CO2), for malonate uptake. Another possibility for malonate uptake would have been malonate/acetate antiport. Since acetate is the end product of the malonate fermentation pathway, its outwardly directed gradient could have been an excellent driving force for the uptake of malonate. Experiments were performed to address this possibility, but involvement of acetate as an antiport substrate could not be shown. Acetate is therefore more likely to leave the cell either by passive diffusion or via a specific export carrier. In analogy to the Na+ cycle operating in K. pneumoniae during citrate fermentation (2, 7), we present a model for Na+ cycling during malonate fermentation in M. rubra (Fig. 4). Inside the cell, malonate is decarboxylated to acetate by the malonate decarboxylase (encoded by the madABCDEF genes [1]), coupled to the extrusion of Na+ ions. Based on the mechanism of the well-studied oxaloacetate decarboxylase from K. pneumoniae (6), we presume that two Na+ ions are extruded per decarboxylation reaction with a concomitant consumption of one H+ originating from the extracellular environment. Another proton enters the cell by the uptake of Hmalonate−. On the other hand, protons are extruded due to the efflux of acetic acid, and by the diffusion of CO2 to the external medium and its subsequent conversion into HCO3− and H+. As the first pKa of malonic acid (2.83) is 3.5 units less than that of the carbonic acid produced (6.35), the medium turns slightly alkaline (about 0.6 pH units) during anaerobic growth of M. rubra on malonate under conditions where a 2 × 104 Pa overpressure of N2/CO2 is applied (not shown). The difference between the second pKa of malonic acid (5.69) and that of the second end product, acetic acid (4.76), cannot compensate for the observed alkanization. The uptake of malonate by the MadLM symporter proceeds electroneutrally at the expense of 1 Na+ ion. This leaves the cell with one Na+ ion (including charge) to perform other vital processes like ATP synthesis by action of an F1F0-ATP synthase. The use of an electroneutral uptake system for malonate in M. rubra in fact prevents the dissipation of the ΔΨ created by the decarboxylase and reserves this driving force for the synthesis of ATP.
FIG. 4.
Model for the Na+ cycle during malonate fermentation in M. rubra. The malonate transporter (MadLM) and the intrinsic membrane part of the malonate decarboxylase (MadB) are depicted as boxes in the membrane. The biotin-containing subunit MadF, drawn at the cytoplasmic side of the membrane, functions as a CO2 shuttle between MadB and the soluble parts of the malonate decarboxylase (MadACDE). The direction of the H+ and Na+ fluxes during malonate fermentation is indicated with arrows. Acetate is postulated to leave the cell by passive diffusion or via a specific carrier. Further details are explained in the text.
REFERENCES
- 1.Berg M, Hilbi H, Dimroth P. Sequence of a gene cluster from Malonomonas rubra encoding components of the malonate decarboxylase Na+ pump and evidence for their function. Eur J Biochem. 1997;245:103–115. doi: 10.1111/j.1432-1033.1997.00103.x. [DOI] [PubMed] [Google Scholar]
- 2.Bott M. Anaerobic citrate metabolism and its regulation in enterobacteria. Arch Microbiol. 1997;167:78–88. [PubMed] [Google Scholar]
- 3.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 4.Cornish-Bowden A. Fundamentals of enzyme kinetics. London, United Kingdom: Portland Press; 1995. [Google Scholar]
- 5.Dehning I, Schink B. Malonomonas rubra gen. nov. sp. nov., a microaerotolerant anaerobic bacterium growing by decarboxylation of malonate. Arch Microbiol. 1989;151:427–433. [Google Scholar]
- 6.Di Berardino M, Dimroth P. Aspartate 203 of the oxaloacetate decarboxylase β-subunit catalyses both the chemical and vectorial reaction of the Na+ pump. EMBO J. 1996;15:1842–1849. [PMC free article] [PubMed] [Google Scholar]
- 7.Dimroth P. Primary sodium ion translocating enzymes. Biochim Biophys Acta. 1997;1318:11–51. doi: 10.1016/s0005-2728(96)00127-2. [DOI] [PubMed] [Google Scholar]
- 8.Engel P, Krämer R, Unden G. Anaerobic fumarate transport in Escherichia coli by an fnr-dependent dicarboxylate uptake system which is different from the aerobic dicarboxylate uptake system. J Bacteriol. 1992;174:5533–5539. doi: 10.1128/jb.174.17.5533-5539.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Engel P, Krämer R, Unden G. Transport of C4-dicarboxylates by anaerobically grown Escherichia coli: energetics and mechanism of exchange, uptake and efflux. Eur J Biochem. 1994;222:605–614. doi: 10.1111/j.1432-1033.1994.tb18903.x. [DOI] [PubMed] [Google Scholar]
- 10.Hilbi H, Dehning I, Schink B, Dimroth P. Malonate decarboxylase of Malonomonas rubra, a novel type of biotin-containing acetyl enzyme. Eur J Biochem. 1992;207:117–123. doi: 10.1111/j.1432-1033.1992.tb17028.x. [DOI] [PubMed] [Google Scholar]
- 11.Hilbi H, Dimroth P. Purification and characterization of a cytoplasmic enzyme component of the Na+-activated malonate decarboxylase system of Malonomonas rubra: acetyl-S-acyl carrier protein: malonate acyl carrier protein-SH transferase. Arch Microbiol. 1994;164:48–56. [PubMed] [Google Scholar]
- 12.Hoenke S, Schmid M, Dimroth P. Sequence of a gene cluster from Klebsiella pneumoniae encoding malonate decarboxylase and expression of the enzyme in Escherichia coli. Eur J Biochem. 1997;246:530–538. doi: 10.1111/j.1432-1033.1997.00530.x. [DOI] [PubMed] [Google Scholar]
- 13.Holt J G, Krieg N R, Sneath P H A, Staley J T, Williams S T. Facultative anaerobic Gram-negative rods. 1993. pp. 175–190. , 202–252. In Bergey’s manual of determinative bacteriology, 9th ed. Williams and Wilkins, Baltimore, Md. [Google Scholar]
- 14.Lo T C Y. The molecular mechanism of dicarboxylic acid transport in Escherichia coli K 12. J Supramol Struct. 1977;7:463–480. doi: 10.1002/jss.400070316. [DOI] [PubMed] [Google Scholar]
- 15.Maloney P C. Bacterial transporters. Curr Opin Cell Biol. 1994;6:571–582. doi: 10.1016/0955-0674(94)90079-5. [DOI] [PubMed] [Google Scholar]
- 16.Poolman B, Konings W N. Secondary solute transport in bacteria. Biochim Biophys Acta. 1993;1183:5–39. doi: 10.1016/0005-2728(93)90003-x. [DOI] [PubMed] [Google Scholar]
- 17.Pos K M, Bott M, Dimroth P. Purification of two active fusion proteins of the Na+-dependent citrate carrier of Klebsiella pneumoniae. FEBS Lett. 1994;347:37–41. doi: 10.1016/0014-5793(94)00502-8. [DOI] [PubMed] [Google Scholar]
- 18.Pos K M, Dimroth P. Functional properties of the purified Na+-dependent citrate carrier of Klebsiella pneumoniae: evidence for asymmetric orientation of the carrier protein in proteoliposomes. Biochemistry. 1996;35:1018–1026. doi: 10.1021/bi951609t. [DOI] [PubMed] [Google Scholar]
- 19.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]