Abstract
Methionine residues in α/β-type small, acid-soluble spore proteins (SASP) of Bacillus species were readily oxidized to methionine sulfoxide in vitro by t-butyl hydroperoxide (tBHP) or hydrogen peroxide (H2O2). These oxidized α/β-type SASP no longer bound to DNA effectively, but DNA binding protected α/β-type SASP against methionine oxidation by peroxides in vitro. Incubation of an oxidized α/β-type SASP with peptidyl methionine sulfoxide reductase (MsrA), which can reduce methionine sulfoxide residues back to methionine, restored the α/β-type SASP’s ability to bind to DNA. Both tBHP and H2O2 caused some oxidation of the two methionine residues of an α/β-type SASP (SspC) in spores of Bacillus subtilis, although one methionine which is highly conserved in α/β-type SASP was only oxidized to a small degree. However, much more methionine sulfoxide was generated by peroxide treatment of spores carrying a mutant form of SspC which has a lower affinity for DNA. MsrA activity was present in wild-type B. subtilis spores. However, msrA mutant spores were no more sensitive to H2O2 than were wild-type spores. The major mechanism operating for dealing with oxidative damage to α/β-type SASP in spores is DNA binding, which protects the protein’s methionine residues from oxidation both in vitro and in vivo. This may be important in vivo since α/β-type SASP containing oxidized methionine residues no longer bind DNA well and α/β-type SASP-DNA binding is essential for long-term spore survival.
Methionine oxidation is a significant form of protein damage caused by endogenous or environmental oxidizing agents (35). Methionine residues may be oxidized to the sulfoxide form with t-butyl hydroperoxide (tBHP) or hydrogen peroxide (H2O2) under relatively mild conditions (11) or to the sulfone form with other oxidizing agents under harsher conditions (12). Although oxidation of methionine residues has no effect on the function of some polypeptides (8, 11), in other proteins methionine oxidation severely inhibits biological function (5, 36). Since methionine oxidation can alter protein function, it is not surprising that cells have at least two mechanisms for dealing with proteins containing oxidized methionine residues. First, it appears likely that oxidized proteins are preferentially degraded in vivo (35). Second, an enzyme termed peptidyl methionine sulfoxide reductase (MsrA), which can reduce methionine sulfoxide residues in proteins to methionine and thus restore protein function (16, 17), has been identified in both prokaryotes and eukaryotes.
Spores of various Bacillus and Clostridium species can survive for long periods in air and are very resistant to oxidizing agents (e.g., H2O2) compared to their corresponding growing cells (22, 27, 32). Since spores of Bacillus and Clostridium species are metabolically and enzymatically dormant, they are unable to deal with proteins containing oxidized methionine residues by either protein turnover or enzymatic reduction until they initiate spore germination (26, 32). Consequently, if methionine oxidation in a protein destroys the protein’s function, then dormant spores may have some mechanism(s) for preventing the initial methionine oxidation. In order to analyze methionine oxidation in a dormant spore protein, we decided to study this process in the α/β-type small, acid-soluble spore proteins (SASP) from Bacillus species.
The α/β-type SASP of Bacillus and Clostridium species are a family of highly abundant, nonspecific DNA binding proteins, which are synthesized only within the developing forespore (30). Typically, there are two major α/β-type SASP and a number of minor α/β-type SASP (30) which together saturate the spore DNA and protect it from a variety of environmental insults (30, 31); the α/β-type SASP are the major determinant of spore UV resistance, and a significant determinant of spore resistance to both heat and oxidizing agents (14, 27, 32). The primary sequence of these proteins has been highly conserved both within and between species, and nearly all α/β-type SASP identified to date (17 of 20) from species of Bacillus, Sporosarcina, and “Thermoactinomyces” contain one highly conserved methionine residue in the middle of the protein (30). In addition, some α/β-type SASP (11 of 20) contain an additional methionine residue at one of two positions near the carboxy terminus of the protein (30). In this communication, we report the results of studies on the oxidation of methionine residues within α/β-type SASP in vitro and in vivo, as well as the analysis of the phenotype of an msrA mutant of Bacillus subtilis.
MATERIALS AND METHODS
Bacteria and plasmids used and conditions for growth and peroxide killing.
The bacterial strains and plasmids used in this work are listed in Table 1; all B. subtilis strains are derivatives of strain 168. B. subtilis was transformed as previously described (1), and transformants were selected by their Cmr. B. subtilis was routinely grown and sporulated at 37°C in 2× SG medium, and spores were purified as described previously (18). All spores used were free (>98%) of sporulating cells and growing cell debris. Spore resistance to heat, H2O2, and tBHP was determined as described previously (20, 25).
TABLE 1.
Bacterial strains and plasmids used in these studies
| Bacterial strain or plasmid | Genotype and/or relevant phenotypea | Reference or source |
|---|---|---|
| Bacteria | ||
| B. subtilis | ||
| PS356 | ΔsspA ΔsspB trpC2 α− β− | 14 |
| PS832 | trp+ revertant of strain 168 (wild type) | Laboratory stock |
| PS2437 | ΔsspA ΔsspB ΔsspE trpC2 pSspCwt α− β− γ− Cmr Kmr; expresses high levels of SspC in spores | 9 |
| PS2438 | ΔsspA ΔsspB ΔsspE trpC2 pSspCG52A α− β− γ− Cmr Kmr; expresses high levels of SspCG52A in spores | 9 |
| PS2638 | msrA::pPS2519 CmrmsrA | This study |
| PS2639 | msrA::pPS2519 ΔsspA ΔsspB α− β− CmrmsrA | This study |
| E. coli | ||
| PS708 | pPS708 Ampr; expresses SspC under control of the lac promoter | 24 |
| PS837 | pPS837 Ampr; expresses SASP-C under control of the lac promoter | 24 |
| Plasmids | ||
| pJH101 | Ampr Cmr Tetr | 7 |
| pPS708 | pDG148 with a 0.6-kb fragment containing the gene encoding SspC under control of the lac promoter | 24 |
| pPS837 | pDG148 with a 0.7-kb fragment containing the gene encoding SASP-C under control of the lac promoter | 24 |
| pSspC | pUB110 with sspC under the control of the strong forespore-specific sspB promoter | 37 |
| pSspCG52A | pUB110 with the gene encoding SspCG52A under the control of the sspB promoter | 37 |
| pPS2519 | pJH101 with a 0.25-kb fragment of msrA | This study |
Abbreviations: Ampr, ampicillin (50 μg/ml) resistance; Cmr, chloramphenicol (3 μg/ml) resistance, Kmr, kanamycin (10 μg/ml) resistance; Tetr, tetracycline (10 μg/ml) resistance; α−, β−, and γ−, lacking SASP-α, β, or γ, respectively.
Large-scale peroxide treatment of spores was conducted as follows. Spores (140 to 160 mg [dry weight] at 7 to 8 mg/ml) in 20 mM sodium phosphate (pH 7.0) were routinely treated with 730 mM tBHP (Sigma) for 3 h at 47°C or 5% H2O2 for 30 min at 22°C. Both treatments resulted in ≥99.9% killing of spores. tBHP-treated spores were harvested by centrifugation, washed once with 20 ml of 50 mM dithiothreitol (DTT), and incubated in 20 ml of 50 mM DTT for 1 h on ice. H2O2 treatment of spores was halted by the addition of 7,000 U of catalase (Worthington). All peroxide-killed spores were washed once with distilled water and centrifuged, and the spore pellet was frozen and lyophilized.
Escherichia coli strains were grown at 37°C in 2× YT medium (16 g of tryptone, 10 g of yeast extract, and 5 g of NaCl per liter of H2O) with ampicillin (100 μg/ml).
α/β-type SASP purification.
Large-scale purification of α/β-type SASP was performed with E. coli strains overexpressing these proteins essentially as described previously (9, 29), but with some modifications. E. coli PS708 and PS2537 were grown to an optical density at 600 nm of 1.5 and made to 0.5 mM concentrations in isopropyl-β-d-thiogalactopyranoside to induce synthesis of α/β-type SASP (24). Cells (2 liters) were harvested by centrifugation after 1 h of further incubation and washed with 500 ml of cold 150 mM NaCl, and the cell pellet was frozen and lyophilized. Lyophilized cells (2 to 4 g) were dry ruptured (100 mg at a time) for 2 min in a dental amalgamator (Wig-L-Bug) with glass beads (150 mg). SspC and SASP-C were purified from acid extracts of dry-ruptured cells as described previously (9, 29). SspC and SspCG52A were purified from untreated and large-scale peroxide-killed spores (∼150 mg [dry weight]) essentially as described previously (9). The yields of these proteins from peroxide-killed spores were identical to those from untreated spores, and oxidized proteins copurified with unmodified protein (data not shown). All α/β-type SASP used or analyzed were >95% pure as determined by polyacrylamide gel electrophoresis (PAGE) at low pH and staining with Coomassie blue (21). All protein concentrations were determined by the Lowry method (13).
DNase protection assays.
The DNA binding activity of α/β-type SASP was determined by measuring their ability to protect linearized plasmid DNA from DNase I digestion (28). Purified α/β-type SASP (1.2 mg/ml) were incubated with EcoRI-linearized plasmid pUC19 (0.12 mg/ml) in 10 mM sodium phosphate (pH 7.5)–3 mM MgCl2 for 10 min at 22°C. This ratio of α/β-type SASP to DNA is sufficient to saturate the plasmid with protein (28). DNase I was added to a final concentration of 0.04 mg/ml, and the solution was incubated at 37°C for 10 min. The reaction was stopped by adjusting the solution to 1% sodium dodecyl sulfate (SDS)–20 mM EDTA. Plasmid DNA was precipitated with ethanol and analyzed by 1% agarose gel electrophoresis followed by staining with ethidium bromide (28).
Methionine oxidation and analysis.
In initial studies of methionine residue oxidation in α/β-type SASP, purified protein (1 mg/ml) was treated with 100 mM tBHP or H2O2 in 250 μl of 10 mM sodium phosphate (pH 7.5) for 3 h at 22°C followed by dialysis in Spectrapor 3 tubing against two changes (1 liter of each) of 10 mM sodium phosphate (pH 7.5) at 4°C. These proteins were used directly for DNase protection, enzymatic reduction with partially purified MsrA from E. coli, and reverse-phase high-performance liquid chromatography (HPLC) analysis (see below). In later oxidation protection experiments, α/β-type SASP (1 mg/ml) were incubated with or without poly(dG) · poly(dC) (0.5 mg/ml; Sigma) in 100 μl of 10 mM sodium phosphate (pH 7.5) for 20 min at 22°C prior to addition of the oxidizing agent. At this protein-to-DNA ratio, DNA is in excess and therefore all α/β-type SASP should be bound to DNA (28). Oxidizing agents (tBHP or H2O2) were added to 100 mM, and the reaction mixtures were incubated for 3 h as described above. Oxidation reactions were terminated by their injection onto a Shodex KW-800 series HPLC size exclusion column and eluted with 10 mM sodium phosphate (pH 7.5) at a flow rate of 1 ml/min. The HPLC fractions containing α/β-type SASP [with or without poly(dG) · poly(dC)] were frozen and lyophilized, and α/β-type SASP were further purified by Tris-Tricine-SDS-PAGE (23), followed by staining with Coomassie blue, and electroelution of stained proteins into 50 mM NH4HCO3–0.1% SDS by using the Elutrap (Schleicher and Schuell). Purified proteins were frozen, lyophilized, dissolved in 100 μl of Milli-Q water, and precipitated with 800 μl of cold acetone at −20°C overnight. Precipitated proteins were washed with 500 μl of cold acetone and dissolved in 60 μl of freshly prepared 8 M urea.
α/β-type SASP (∼40 μg) were digested in 100 μl of 0.8 M urea–10 mM CaCl2–200 mM NH4HCO3 for 15 h at 37°C with trypsin (5 μg; Worthington) prior to analysis of tryptic peptides by reverse-phase HPLC. Tryptic digests were injected onto a Waters Deltapak C4 (3.9 by 150 mm) column at a flow rate of 1 ml/min. After the column was washed for 5 min with 98% buffer A–2% buffer B, peptides were eluted with a linear gradient of 98% buffer A–2% buffer B to 2% buffer A–98% buffer B over 60 min. Buffer A was 0.06% aqueous trifluoroacetic acid, and buffer B was 0.052% trifluoroacetic acid in 80% acetonitrile. Peptides were detected by their UV absorbance at 214 nm. Individual tryptic peptides were identified by amino acid analyses; methionine sulfoxide-containing peptides were identified by mass spectrometry. Quantitation of methionine sulfoxide levels was by integration of HPLC peaks to determine the areas corresponding to reduced and oxidized peptides.
To reduce methionine sulfoxide residues in oxidized SspC, tBHP-treated SspC (1 mg/ml) was incubated with partially purified (∼25% pure) E. coli MsrA (0.04 mg/ml) (a generous gift from H. Weissbach) in 25 mM Tris-HCl (pH 7.4)–15 mM DTT–10 mM MgCl2 for 4 h at 24°C. The reduced SspC was then analyzed for DNase protection and digested with trypsin and peptides analyzed as described above.
Construction of a yppP (msrA) mutant.
Oligonucleotide primers were designed to PCR amplify a 253-bp fragment from within the yppP (now termed msrA) coding region. YPPP-1, a 23-mer with the sequence 5′-GGGAATTCGGGACATCGTGAAGC, and YPPP-2, a 23-mer with the sequence 5′-CCGGATCCGTCCGTTACAATCGG (nucleotides 431,735 to 431,749 and nucleotides 431,958 to 431,972 in the Subtilist B. subtilis genome project database, respectively) were designed with additional 5′ nucleotides and EcoRI and BamHI restriction sites (underlined residues), respectively, for cloning purposes. The resulting EcoRI-BamHI-digested fragment was ligated with EcoRI-BamHI-digested plasmid pJH101 (7), and the ligation mix was used to transform E. coli JM83 to Ampr. A plasmid resulting from this transformation (pPS2519) was used to transform B. subtilis PS832 and PS356 to Cmr, generating an insertional mutation within the msrA gene which truncated the coding region to 30% of the original open reading frame. Southern blot analysis (34) of chromosomal DNA from Cmr transformants confirmed that the transformation had generated the expected insertion within msrA (data not shown).
Methionine sulfoxide reductase (MsrA) enzyme assays.
N-Acetyl-l-[35S]methionine sulfoxide, a radiolabeled substrate for MsrA (3), was synthesized as follows. A 67-μl aliquot (1 mCi) of l-[35S]methionine (>1,000 Ci/mmol; New England Nuclear) was added to 933 μl of 10 mM l-methionine–3% acetic acid followed by the addition of 10 μl of 30% H2O2. After incubation at 22°C for 2 h, the solution was lyophilized and dissolved in 500 μl of glacial acetic acid, and l-[35S]methionine sulfoxide was acetylated by the addition of 500 μl of acetic anhydride. After incubation at 22°C for 2 h, the reaction was quenched with 9 ml of distilled water, lyophilized, and dissolved in 200 μl of distilled water. Conversion of l-[35S]methionine to N-acetyl-l-[35S]methionine sulfoxide was >99.9% complete as determined by analytical reverse-phase HPLC (see below) of the final compound. The specific activity of the synthesized N-acetyl-l-[35S]methionine sulfoxide was 180 cpm/pmol.
Bacterial cells were grown to late log phase (optical density at 600 nm, ∼1) in 250 ml of either 2× YT (E. coli) or 2× SG (B. subtilis) (18) medium, harvested by centrifugation, washed with 100 ml of 0.15 M NaCl, and suspended in 1 to 2 ml of cell extraction buffer (20 mM Tris-HCl [pH 7.5]–10 mM MgCl2–10 mM KCl–10 mM DTT–1 mM phenylmethylsulfonyl fluoride–10% glycerol). Cells were disrupted by 4 min of sonication at 0°C with glass beads and centrifuged at 32,000 × g for 1 h at 4°C, and the supernatant fluid was used immediately for enzyme assays. Spores (∼150 mg [dry weight]) were decoated as previously described (9), and 200 μl of 10 mM Tris-HCl (pH 8.0)–10 mM EDTA–150 mM NaCl–1 mM DTT–0.1 mM phenylmethylsulfonyl fluoride was added to the decoated spore pellet for resuspension. The decoated spore suspension was digested with lysozyme for 20 min at 37°C, sonicated twice for 20 s, centrifuged in a microcentrifuge (Fisher model 235A) for 15 min at 4°C, and adjusted to 20 mM MgCl2–10 mM DTT–10% glycerol, and the supernatant fluid was used immediately for MsrA assays. MsrA activity was routinely determined as described previously (3). Briefly, cell or spore extracts (30 and 45 μl, respectively) were incubated in cell extract or spore extract buffer with 500 μM N-acetyl-l-[35S]methionine sulfoxide in a final volume of 50 μl at 37°C for 30 min. Reactions were quenched with 250 μl of 0.5 M HCl and extracted with 700 μl of ethyl acetate, and the radioactivity of the organic extract was counted in a scintillation counter.
In some experiments a more sensitive assay was used in which any N-acetyl-l-[35S]methionine produced was purified by reverse-phase HPLC and subjected to counting. Briefly, ethyl acetate extracts of reaction mixtures were dried and then dissolved in 18 μl of 0.06% trifluoroacetic acid to which 2 μl of a solution that was 5 mM in both N-acetyl-l-methionine and N-acetyl-l-methionine sulfoxide was added, followed by resolution of N-acetyl-l-methionine and N-acetyl-l-methionine sulfoxide on a Waters μBondapak C18 reverse-phase HPLC column (3.9 by 300 mm) by isocratic elution with 0.06% trifluoroacetic acid. Peaks corresponding to N-acetyl-l-methionine and N-acetyl-l-methionine sulfoxide were collected, dried, dissolved in 100 μl of distilled water, applied to glass fiber filters, dried under an infrared lamp, and counted in a scintillation counter.
RESULTS
Oxidation of α/β-type SASP in vitro.
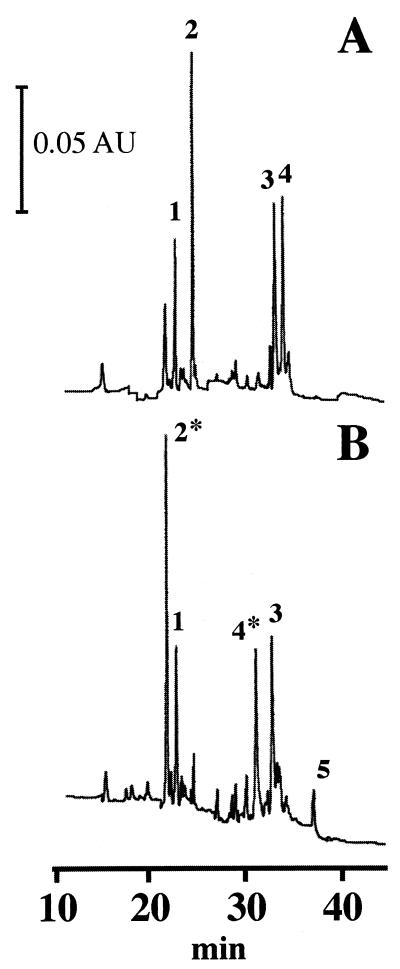
The α/β-type SASP selected for most of this study was SspC, a minor α/β-type SASP of B. subtilis. SspC and its interaction with DNA have been characterized extensively (28), and this protein has dramatic effects on spore resistance in vivo (37). In addition to containing the highly conserved methionine residue (M27) near the middle of the protein, SspC contains a second, less well conserved methionine residue near the carboxy terminus (M67) (6); the translational initiating methionine residues are removed posttranslationally from all α/β-type SASP (4, 30). Both methionine residues within SspC were substantially oxidized to methionine sulfoxide residues by treatment with tBHP or H2O2 for 3 h at room temperature, as demonstrated by large shifts in the HPLC retention times of the methionine-containing tryptic peptides of SspC (Fig. 1, peptides 2 and 4). The identities of these two peptides were confirmed by a combination of amino acid analysis and matrix-assisted laser desorption, ionization-mass spectrometry; the latter analysis revealed an increase of 16 Da in the oxidized peptides (data not shown), indicating that the methionine residues were oxidized to the sulfoxide and not the sulfone form. A small amount (∼8%) of additional oxidation also occurred at lysine 28 in SspC, which resulted in the formation of a larger tryptic peptide (S9 to R46) that contained methionine sulfoxide at M27 (Fig. 1, peptide 5). The single highly conserved methionine of SASP-C (M28), a major α/β-type SASP of Bacillus megaterium (33), was also oxidized to methionine sulfoxide by tBHP or H2O2 treatment (Table 2 and data not shown).
FIG. 1.
Analysis of tryptic peptides from untreated (A) and tBHP-oxidized (B) SspC. SspC and SspC oxidized with tBHP for 3 h were digested with trypsin, and the products were resolved by HPLC as described in Materials and Methods. Because tBHP was removed by dialysis, the actual exposure to oxidant was longer than 3 h. The SspC peptides identified are as follows: 1, A47 to K57; 2 and 2*, L62 to H72; 3, L29 to R46; 4 and 4*, S9 to K28; and 5, S9 to R46. Peptides 2*, 4*, and 5 contain methionine sulfoxide residues. Peptide 5 results from additional oxidation at lysine 28. The identity of this peptide was established by performing amino acid analysis and mass spectrometry and determining resistance to trypsin cleavage. No peptides were eluted prior to 10 min.
TABLE 2.
Methionine sulfoxide levels in α/β-type SASP oxidized in vitroa
| Protein | Treatment | % Methionine sulfoxide at:
|
||
|---|---|---|---|---|
| Conserved residueb
|
Less well conserved residue M67 | |||
| M27 | M28 | |||
| SspC | None | 3 (<1) | 21 | |
| Gel purification | 13 (<1) | 24 | ||
| tBHP | 57 (57) | 61 | ||
| tBHP plus DNA | 10 (10) | 31 | ||
| H2O2 | 89 (89) | >97 | ||
| H2O2 plus DNA | 23 (23) | 61 | ||
| SASP-C | None | 8 (<1) | ||
| Gel purification | 15 (5) | |||
| tBHP | 55 (15) | |||
| tBHP plus DNA | 17 (4) | |||
| H2O2 | 88 (23) | |||
| H2O2 plus DNA | 22 (6) | |||
SASP were oxidized, separated from the oxidizing agent, and purified by Tris-Tricine-SDS-PAGE as described in Materials and Methods, except for untreated samples, which were not electrophoretically purified. Note that in these experiments, oxidation was stopped by rapid removal of the oxidizing agent by HPLC, in contrast to the experiment shown in Fig. 1, in which the oxidizing agent was removed by dialysis.
Significant oxidation also occurred at lysines 28 and 29 of SspC and SASP-C, respectively. This resulted in larger tryptic peptides, which were fully oxidized at M27 and M28. The amounts of these larger peptides have been included in calculations of methionine oxidation, and the percentages of oxidation at lysines 28 and 29 are given in parentheses.
Since methionine residue oxidation in some cases adversely affects protein function (5, 36), we tested whether oxidized α/β-type SASP retained DNA binding activity in vitro. Strikingly, oxidation of methionine residues in SspC and SASP-C greatly reduced these proteins’ DNA binding activity as measured by the ability of the oxidized proteins to provide DNase protection to plasmid DNA (Fig. 2). Although the peroxide-treated α/β-type SASP also contained some oxidation at lysine residues, the proportions of these products were much too small (∼8 and ∼17% in SspC and SASP-C, respectively) to account for the observed decreases in DNA binding. Furthermore, DNA binding activity was restored to oxidized SspC when this protein’s methionine sulfoxide residues were reduced back to methionine by MsrA treatment as described in Materials and Methods (data not shown). Therefore, oxidation of methionine residues within α/β-type SASP, in particular the central highly conserved methionine (see below), eliminates DNA binding in vitro.
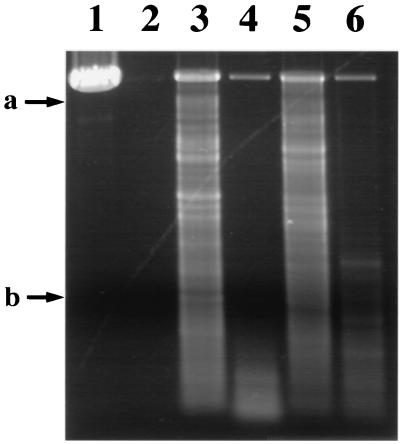
FIG. 2.
Ability of untreated and tBHP-oxidized α/β-type SASP to provide DNase protection to plasmid pUC19. Purified SspC and SASP-C were treated with tBHP for 3 h, dialyzed, complexed with linearized plasmid pCU19 DNA, and DNase treated as described in Materials and Methods. DNA was isolated and analyzed by agarose gel electrophoresis. The arrows labeled a and b indicate the positions of 2.3- and 0.56-kb DNA size markers, respectively. Samples run in the various lanes are as follows: lane 1, SspC without DNase treatment; lane 2, no α/β-type SASP; lane 3, untreated SspC; lane 4, tBHP-treated SspC; lane 5, untreated SASP-C; and lane 6, tBHP-treated SASP-C. SspC contained ∼3 and ∼21% methionine sulfoxide at M27 and M67, respectively. Oxidized SspC contained ∼79 and ∼86% methionine sulfoxide at M27 and M67, respectively. SASP-C and oxidized SASP-C contained ∼8 and ∼88% methionine sulfoxide at M28, respectively. Oxidized SspC and SASP-C also showed ∼8 and ∼17% oxidation at lysines 28 and 29, respectively. The bands at ∼2.7 kb in lanes 3 and 6 are probably microdrops of the incubation mixtures which escaped exposure to DNase.
α/β-Type SASP complexed to DNA are resistant to methionine oxidation in vitro.
A recent report demonstrated that only surface-exposed methionine residues in proteins are oxidized with tBHP, presumably due to steric constraints imposed by the bulky tert-butyl group (11). While buried methionine residues were relatively resistant to oxidation by tBHP, they were oxidized much more readily by H2O2 (11). Although α/β-type SASP are relatively unstructured in solution, they undergo a large conformational change upon binding to DNA as measured by circular dichroism spectroscopy (15). To determine whether methionine residues within α/β-type SASP become more resistant to oxidation when bound to DNA, α/β-type SASP-DNA complexes were treated with tBHP or H2O2, and the α/β-type SASP were analyzed for methionine oxidation. Untreated SspC contained only a minute amount of the sulfoxide at the conserved methionine (M27), although there was significantly more sulfoxide at the less well conserved methionine (M67) (Table 2). However, the electrophoretic purification scheme used to separate α/β-type SASP from DNA caused some methionine oxidation at M27 (Table 2). SspC that was treated with tBHP while bound to DNA showed no significant change in the levels of methionine oxidation compared to polyacrylamide gel-purified SspC and contained much less methionine sulfoxide than SspC treated with tBHP in the absence of DNA (Table 2). H2O2 treatment was more effective at oxidizing both methionine residues of SspC, but again there was significant protection afforded by DNA binding, in particular at M27 (Table 2). DNA binding also protected the single conserved methionine of SASP-C against oxidation (Table 2), indicating that the protection phenomenon is probably general to all α/β-type SASP. These experiments were also conducted with a linearized plasmid as the protective DNA, and essentially identical results were obtained (data not shown).
In the experiments described above, a significant amount of oxidation was also observed at lysines 28 and 29 of SspC and SASP-C, respectively (Table 2), although there was much less lysine oxidation when dialysis was used to remove the oxidizing agents from reactions (Fig. 1 and data not shown). Peptides containing oxidized lysine residues were identified by amino acid analysis and mass spectrometry, which measured a peptide increase of ∼16 Da. These peptides were refractory to digestion by both trypsin and cyanogen bromide, indicating that the lysine and methionine residues were both modified. The lysine residues have presumably been oxidized to α-amino-adipic semialdehyde residues (2). Although we have not studied this point further, this oxidation results in a decrease of only 1 Da in the oxidized peptide. We do not fully understand the reason for this lysine oxidation, but it appears to correlate with the lyophilization of peroxide-treated α/β-type SASP after removal of the oxidizing agent by gel filtration. Furthermore, trypsin cleavage at other lysine residues in SspC and SASP-C was unaffected by peroxide treatment, suggesting that the proximity of the oxidizable lysines 28 and 29 to the highly conserved M27 and M28 residues (respectively) plays a role in this oxidation. Accordingly, DNA binding also protected α/β-type SASP from lysine oxidation (Table 2).
Oxidation of α/β-type SASP in vivo.
With the knowledge that methionine residues in α/β-type SASP could be oxidized in vitro and that this had drastic effects on these proteins’ function, an obvious question was whether methionine residues in α/β-type SASP could be oxidized in spores. Bacterial spores are much more resistant to oxidizing agents such as tBHP and H2O2 than are vegetatively growing cells (22, 32), and the α/β-type SASP are significant determinants of this resistance to these agents; spores which lack the two major α/β-type SASP (α− β− spores) show decreased resistance to both H2O2 and tBHP compared to wild-type spores (25, 27). To determine if methionine residues in α/β-type SASP are susceptible to oxidation in vivo, B. subtilis spores which overexpress SspC as their major α/β-type SASP (PS2437) (Table 1) (9) were treated with either H2O2 or tBHP, followed by purification and analysis of SspC. The levels of methionine sulfoxide in SspC from untreated spores were similar to levels found in recombinant SspC purified from E. coli (Tables 2 and 3). However, SspC purified from peroxide-killed spores contained significantly more methionine sulfoxide at M67 than did SspC from untreated spores (Table 3). Methionine sulfoxide levels also rose slightly at M27, but this conserved methionine residue was much more resistant to oxidation than the less well conserved M67 residue (Table 3). Interestingly, SspC purified from H2O2- and tBHP-killed spores (Table 3) bound to DNA in vitro with an affinity essentially identical to that of SspC purified from untreated spores or recombinant SspC purified from E. coli (data not shown).
TABLE 3.
Methionine sulfoxide levels in SspC and SspCG52A oxidized in vivoa
| Protein | Treatment | % Methionine sulfoxide at residue:
|
|
|---|---|---|---|
| M27b | M67 | ||
| SspC | None | 3 (2) | 18 |
| tBHP | 10 (4) | 33 | |
| H2O2 | 12 (6) | 61 | |
| SspCG52A | None | 10 (5) | 25 |
| tBHP | 71 (50) | 94 | |
| H2O2 | 42 (28) | 84 | |
Spores of strains PS2437 (SspC) or PS2438 (SspCG52A) were treated and proteins were purified and analyzed as described in Materials and Methods.
Significant amounts of additional oxidation also occurred at lysine 28. This resulted in the production of a larger tryptic peptide (S9-R46), which was fully oxidized at M27. The amounts of this larger peptide have been included in calculations of M27 oxidation, and the percentage of oxidation at lysine 28 is given in parentheses.
Because DNA binding protects α/β-type SASP from methionine oxidation in vitro, we decided to assess whether it was DNA binding that was at least in part responsible for the protection of SspC against methionine oxidation in vivo. For this analysis we used spores which overexpress SspCG52A, a mutant form of SspC which does not bind to DNA in vitro with high affinity and fails to confer UV and heat resistance on spores (37). Previous work has also shown that although a labile asparagine residue in SspC is protected against deamidation by binding to DNA in vivo, in SspCG52A this residue is less well protected (9). Strikingly, sulfoxide levels at both methionine residues of SspCG52A increased dramatically as a result of peroxide treatment of PS2438 spores (Table 3); these levels were much higher than the levels measured in SspC from peroxide killed spores, particularly at M27 of SspC, which was well protected from oxidation in vivo (Table 3). Substantial oxidation also occurred at lysine 28 in SspCG52A from peroxide-killed spores (Table 3). The oxidation of lysine 28 in vivo may be due to hypohalite ions formed by peroxides and endogenous spore halides, or the enhanced reactivity of lysine 28 as mentioned above. This lysine oxidation product was not observed to any significant degree in SspC from peroxide-killed spores (Table 3).
Characterization of an msrA mutant of B. subtilis.
Although at least one critical methionine residue was relatively well protected from oxidation in spores, we did observe significant methionine residue oxidation, and it is possible that in other proteins some methionine residues may be even more readily oxidized. While it appears likely that α/β-type SASP with oxidized methionine residues are degraded during spore germination, as are all α/β-type SASP (30), it is possible that oxidized methionine residues in other proteins might be repaired by MsrA. Consequently, we wished to determine whether the MsrA protein repair pathway is present and might function in germinating spores of B. subtilis.
The B. subtilis genome sequencing project has identified an msrA homolog in B. subtilis (called yppP), which has 40% identity to MsrA of E. coli. To determine if yppP codes for MsrA and whether the enzyme plays any role in resistance of vegetative cells or dormant spores to oxidizing agents, we generated strains in which the yppP coding sequence was disrupted. MsrA activity was detected in crude extracts of wild-type B. subtilis vegetative cells at a specific activity (4.7 pmol/min/mg protein) consistent with that reported for another gram-positive bacterium, Streptococcus pneumoniae (5.7 pmol/min/mg protein) (38). However, the MsrA specific activity in the yppP disruptant was <10% of that found in wild-type cells. MsrA activity (11 pmol/min/mg protein) was also detected in extracts from wild-type spores, but again the MsrA specific activity in yppP mutant spore extracts was <5% of that in wild-type spore extracts. We therefore conclude that yppP does encode the B. subtilis MsrA homolog, and yppP is now termed msrA. In contrast to results in E. coli and Saccharomyces cerevisiae (2, 17), msrA vegetative cells were no more sensitive to H2O2 than were wild-type cells, as measured by a zone-of-inhibition assay on plates (data not shown); spores of both msrA and α− β− msrA strains (PS2638 and PS2639) were also no more sensitive to wet heat or H2O2 than were spores of their respective parental strains (PS832 and PS356) (data not shown).
DISCUSSION
Oxidative damage to proteins within bacterial spores poses a unique problem, because spores can remain dormant for long periods, during which substantial oxidative protein damage may accumulate, yet dormant spores are incapable of degrading or repairing such damaged proteins. One mechanism whereby oxidative damage to spore proteins might be dealt with is damage repair during spore germination and outgrowth. The yppP (now msrA) gene of B. subtilis is predicted to encode a homolog of the MsrA repair enzyme, and peptidyl methionine sulfoxide reductase activity is greatly diminished (>90%) within vegetative cells and spores of an msrA mutant. However, spores of the msrA mutant strains showed no increase in peroxide sensitivity compared to spores of the parental strains. Although the MsrA repair pathway may operate during spore germination, it plays no significant role in spore resistance to oxidizing agents. Similarly, growing cells of the msrA mutant were not sensitive to H2O2, presumably due to the overriding effects of other protective mechanisms. Even in E. coli, only a slight sensitivity to H2O2 was reported in an msrA mutant (17).
A second mechanism for dealing with oxidative damage to spore protein is to prevent such damage. The low permeability of spores to most chemicals is one mechanism by which spores resist oxidative damage from peroxides (32), while α/β-type SASP protect spore DNA from oxidative damage (25, 27). However, peroxides can enter the spore core; as in the absence of α/β-type SASP, spore killing by peroxides is in large part through DNA damage (25, 27). A number of spore enzymes are also inactivated within spores by H2O2 (19). While the inactivation of some spore enzymes is significantly slower than spore killing, inactivation of at least one enzyme parallels spore killing (19). However, the nature of the oxidative damage resulting in inactivation of these spore enzymes has not been established.
Because they are significant determinants of spore resistance to environmental insults, the α/β-type SASP are potentially important targets for methionine residue oxidation (30, 31). Indeed, α/β-type SASP in which the highly conserved methionine is oxidized do not bind to DNA effectively in vitro and presumably would also not confer UV and heat resistance to spores as well as their nonoxidized counterparts would. However, α/β-type SASP are resistant to peroxide-induced methionine oxidation when bound to DNA in vitro. Interestingly, the two methionine residues of SspC differ somewhat in their resistance to oxidation in the presence of DNA. In general, surface-exposed methionine residues are more susceptible to oxidation than are buried residues (11), and therefore the most obvious interpretation of the different reactivities of the M27 and M67 residues of SspC with oxidizing agents is that M27 becomes relatively inaccessible to solvent when the protein binds to DNA, while the region containing M67 is still somewhat exposed. There is evidence that protein-protein interactions occur between adjacent α/β-type SASP while bound to DNA, and some of the residues involved in these interactions (for example glutamates 30 and 34 in SASP-C) are very near the conserved methionine residue (M28 in SASP-C) (10). Therefore, α/β-type SASP packing along the DNA backbone could render the highly conserved methionine inaccessible to solvent. However, because the M27 residue of SspC is more resistant to oxidation than M67 in the absence of DNA, it is also possible that M67 is simply more reactive than M27.
As noted above, killing of wild-type B. subtilis spores with H2O2 is not due to DNA damage, indicating that α/β-type SASP continue to protect DNA even though the spore has been killed by damage to some other unknown target (27). The in vivo oxidation data presented in this study are consistent with this previous finding. Although spores (>99.9%) were killed by the peroxide treatments used, only ∼10% of the highly conserved M27 residue in SspC was oxidized to the sulfoxide form. These data indicate that in spores, the rate of oxidation of this critical methionine residue in SspC is >30-fold lower than the rate of spore killing. Comparison of the latter data with the published rates of enzyme inactivation and spore killing by H2O2 (19) indicates that the rate of oxidation of M27 in SspC in spores is at least fivefold lower than the rate of inactivation of the most stable spore enzyme analyzed. Thus, α/β-type SASP are extremely well protected against oxidative inactivation in vivo. In addition, SspC purified from H2O2-killed spores retained nearly all of its DNA binding ability, even though this protein contained approximately 60% methionine sulfoxide at the less well conserved M67 position. This result indicates that the highly conserved and well protected M27 residue of SspC is critical for DNA binding but that the less well conserved and less well protected M67 residue is less important (and perhaps not at all important).
Two of the α/β-type SASP from Bacillus species which lack the conserved central methionine residue contain phenylalanine at the position corresponding to M27 in SspC, whereas the third contains a tyrosine (30). Therefore, evolution has selected for hydrophobic residues at this position, and because the change from a hydrophobic methionine to a more hydrophilic methionine sulfoxide residue is not conservative, this change could disrupt hydrophobic interactions which may be critical to α/β-type SASP-DNA binding. Conversely, although all α/β-type SASP from Bacillus species contain hydrophobic residues (either leucine or phenylalanine) at positions corresponding to M67 in SspC, the surrounding C-terminal region of α/β-type SASP contains many sequence variations, which may indicate that this portion of the protein is not critical for function (30). Therefore, even though M67 of SspC can be substantially oxidized in vivo, the protein retains function because the methionine residue (M27) which is critical for DNA binding is largely protected from oxidation.
SspCG52A from peroxide-killed spores contained much more methionine sulfoxide at the conserved M27 position than did SspC from peroxide-killed spores. Because SspCG52A does not bind to DNA with high affinity in vitro and also fails to restore UV and heat resistance to α− β− spores in vivo (37), it appears that it is DNA binding that protects α/β-type SASP from methionine oxidation at the highly conserved M27 residue. Previous work has also shown that DNA binding protects SspC from asparagine deamidation at a very highly conserved NG sequence in vitro (9). Deamidation of this asparagine residue in SspC results in the loss of DNA binding function in vitro, and a mutant form of SspC in which the labile asparagine is replaced with an aspartate residue (mimicking the product of deamidation) fails to confer UV and heat resistance on α− β− spores of B. subtilis (9). However, SspC is well protected from deamidation in vivo due to its interaction with DNA, whereas SspCG52A is protected to a lesser degree (9). Therefore, in addition to the well-characterized effects whereby α/β-type SASP protect spore DNA from a variety of types of damage (32), DNA also protects α/β-type SASP from a variety of protein damage.
ACKNOWLEDGMENTS
We are grateful to Stacey Carrington for conducting initial experiments, to Nathan Brot for helpful discussion, and to Herbert Weissbach for generously providing MsrA.
This work was supported by a grant from the National Institutes of Health (GM19698).
REFERENCES
- 1.Anagnostopoulos C, Spizizen J. Requirements for transformation in Bacillus subtilis. J Bacteriol. 1961;81:74–76. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berlett B S, Stadtman E R. Protein oxidation in aging, disease, and oxidative stress. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 3.Brot N, Werth J, Kostar D, Weissbach H. Reduction of N-acetyl methionine sulfoxide: a simple assay for peptide methionine sulfoxide reductase. Anal Biochem. 1982;122:291–294. doi: 10.1016/0003-2697(82)90283-4. [DOI] [PubMed] [Google Scholar]
- 4.Cabrera-Martinez R M, Mason J M, Setlow B, Waites W M, Setlow P. Purification and amino acid sequence of two small, acid-soluble spore proteins from Clostridium bifermentans spores. FEMS Microbiol Lett. 1989;52:139–143. doi: 10.1016/0378-1097(89)90185-7. [DOI] [PubMed] [Google Scholar]
- 5.Chu S-T, Chu C-C, Tseng C-C, Chen Y-H. Met-8 of the β1-bungarotoxin phospholipase A2 subunit is essential for the phospholipase A2-independent neurotoxic effect. Biochem J. 1993;295:713–718. doi: 10.1042/bj2950713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Connors M J, Setlow P. Cloning of a small, acid-soluble spore protein gene from Bacillus subtilis and determination of its complete nucleotide sequence. J Bacteriol. 1985;161:333–339. doi: 10.1128/jb.161.1.333-339.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferrari F A, Nguyen A, Lang D, Hoch J A. Construction and properties of an integrable plasmid for Bacillus subtilis. J Bacteriol. 1983;154:1513–1515. doi: 10.1128/jb.154.3.1513-1515.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Glaser C B, Li C H. Reaction of bovine growth hormone with hydrogen peroxide. Biochemistry. 1974;13:1044–1047. doi: 10.1021/bi00702a033. [DOI] [PubMed] [Google Scholar]
- 9.Hayes C S, Setlow P. Analysis of deamidation of small, acid-soluble spore proteins from Bacillus subtilis in vitro and in vivo. J Bacteriol. 1997;178:6020–6027. doi: 10.1128/jb.179.19.6020-6027.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayes, C. S., and P. Setlow. 1997. Unpublished results.
- 11.Keck R G. The use of t-butyl hydroperoxide as a probe for methionine oxidation in proteins. Anal Biochem. 1996;236:52–62. doi: 10.1006/abio.1996.0131. [DOI] [PubMed] [Google Scholar]
- 12.Lischwe M A, Sung M T. Use of N-chlorosuccinimide/urea for the selective cleavage of tryptophanyl peptide bonds in proteins. Cytochrome c. J Biol Chem. 1977;252:4976–4980. [PubMed] [Google Scholar]
- 13.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 14.Mason J M, Setlow P. Essential role of small, acid-soluble spore proteins in resistance of Bacillus subtilis spores to UV light. J Bacteriol. 1986;167:174–178. doi: 10.1128/jb.167.1.174-178.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mohr, S., and P. Setlow. Unpublished results.
- 16.Moskovitz J, Jenkins N A, Gilbert D J, Copeland N G, Jursky F, Weissbach H, Brot N. Chromosomal localization of the mammalian peptide-methionine sulfoxide reductase gene and its differential expression in various tissues. Proc Natl Acad Sci USA. 1996;93:3205–3208. doi: 10.1073/pnas.93.8.3205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moskovitz J, Rahman M A, Strassman J, Yancey S O, Kushner S R, Brot N, Weissbach H. Escherichia coli peptide methionine sulfoxide reductase gene: regulation of expression and role in protecting against oxidative damage. J Bacteriol. 1995;177:502–507. doi: 10.1128/jb.177.3.502-507.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicholson W L, Setlow P. Sporulation, germination, and outgrowth. In: Harwood C R, Cutting S M, editors. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons Ltd.; 1990. pp. 391–450. [Google Scholar]
- 19.Palop A, Rutherford G C, Marquis R E. Hydroperoxide inactivation of enzymes within spores of Bacillus megaterium ATCC19213. FEMS Microbiol Lett. 1996;142:283–287. [PubMed] [Google Scholar]
- 20.Popham D L, Sengupta S, Setlow P. Heat, hydrogen peroxide, and UV resistance of Bacillus subtilis spores with increased core water content and with or without major DNA-binding proteins. Appl Environ Microbiol. 1995;61:3633–3638. doi: 10.1128/aem.61.10.3633-3638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reisfield R A, Lewis V J, Williams D E. Disk electrophoresis of basic proteins and peptides on polyacrylamide gels. Nature. 1962;195:281–283. doi: 10.1038/195281a0. [DOI] [PubMed] [Google Scholar]
- 22.Russell A D. The destruction of bacterial spores. New York, N.Y: Academic Press; 1982. [Google Scholar]
- 23.Schagger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 24.Setlow B, Hand A R, Setlow P. Synthesis of a Bacillus subtilis small, acid-soluble spore protein in Escherichia coli causes cell DNA to assume some characteristics of spore DNA. J Bacteriol. 1991;173:1642–1653. doi: 10.1128/jb.173.5.1642-1653.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Setlow B, Setlow C A, Setlow P. Killing bacterial spores by organic peroxides. J Ind Microbiol. 1997;18:384–388. [Google Scholar]
- 26.Setlow B, Setlow P. Levels of oxidized and reduced pyridine nucleotides in dormant spores and during growth, sporulation, and spore germination of Bacillus megaterium. J Bacteriol. 1977;129:857–865. doi: 10.1128/jb.129.2.857-865.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Setlow B, Setlow P. Binding of small, acid-soluble spore proteins to DNA plays a significant role in the resistance of Bacillus subtilis spores to hydrogen peroxide. Appl Environ Microbiol. 1993;59:3418–3423. doi: 10.1128/aem.59.10.3418-3423.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Setlow B, Sun D, Setlow P. Interaction between DNA and α/β-type small, acid-soluble spore proteins: a new class of DNA-binding protein. J Bacteriol. 1992;174:2312–2322. doi: 10.1128/jb.174.7.2312-2322.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Setlow P. Purification and properties of some unique low molecular weight basic proteins degraded during germination of Bacillus megaterium spores. J Biol Chem. 1975;250:8168–8173. [PubMed] [Google Scholar]
- 30.Setlow P. Small acid-soluble, spore proteins of Bacillus species: structure, synthesis, genetics, function and degradation. Annu Rev Microbiol. 1988;42:319–338. doi: 10.1146/annurev.mi.42.100188.001535. [DOI] [PubMed] [Google Scholar]
- 31.Setlow P. I will survive: protecting and repairing spore DNA. J Bacteriol. 1992;174:2737–2741. doi: 10.1128/jb.174.9.2737-2741.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Setlow P. Mechanisms for the prevention of damage to DNA in spores of Bacillus species. Annu Rev Microbiol. 1995;49:29–54. doi: 10.1146/annurev.mi.49.100195.000333. [DOI] [PubMed] [Google Scholar]
- 33.Setlow P, Ozols J. Covalent structure of protein C. A second major low molecular weight protein degraded during germination of Bacillus megaterium spores. J Biol Chem. 1980;255:8413–8416. [PubMed] [Google Scholar]
- 34.Southern E M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975;98:503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- 35.Stadtman E R. Protein oxidation and aging. Science. 1992;257:1220–1224. doi: 10.1126/science.1355616. [DOI] [PubMed] [Google Scholar]
- 36.Teh L-C, Murphy L J, Huq N L, Surus A S, Friesen H G, Lararus L, Chapman G E. Methionine oxidation in human growth hormone and human chorionic somatomammotropin. Effects on receptor binding and biological activities. J Biol Chem. 1987;262:6472–6477. [PubMed] [Google Scholar]
- 37.Tovar-Rojo F, Setlow P. Effects of mutant small, acid-soluble spore proteins from Bacillus subtilis on DNA in vivo and in vitro. J Bacteriol. 1991;173:4827–4835. doi: 10.1128/jb.173.15.4827-4835.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wizemann T M, Moskovitz J, Pearce B J, Cundell D, Arvidson C G, So M, Weissbach H, Brot N, Masure H R. Peptide methionine sulfoxide reductase contributes to the maintenance of adhesins in three major pathogens. Proc Natl Acad Sci USA. 1996;93:7985–7990. doi: 10.1073/pnas.93.15.7985. [DOI] [PMC free article] [PubMed] [Google Scholar]