Abstract
Previous studies have implicated the obligatory requirement for the vir regulon (or “virulon”) of the Ti plasmid for the transfer of oncogenes from Agrobacterium tumefaciens to plant cells. The machinery used in this horizontal gene transfer has been long thought to be a transformation or conjugative delivery system. Based on recent protein sequence comparisons, the proteins encoded by the virB operon are strikingly similar to proteins involved in the synthesis and assembly of conjugative pili such as the conjugative pilus of F plasmid in Escherichia coli. The F pilus is composed of TraA pilin subunits derived from TraA propilin. In the present study, evidence is provided showing that the counterpart of TraA is VirB2, which like TraA propilin is processed into a 7.2-kDa product that comprises the pilus subunit as demonstrated by biochemical and electron microscopic analyses. The processed VirB2 protein is present exocellularly on medium on which induced A. tumefaciens had grown and appears as thin filaments of 10 nm that react specifically to VirB2 antibody. Exocellular VirB2 is produced abundantly at 19°C as compared with 28°C, an observation that parallels the effect of low temperature on the production of vir gene-specific pili observed previously (K. J. Fullner, L. C. Lara, and E. W. Nester, Science 273:1107–1109, 1996). Export of the processed VirB2 requires other virB genes since mutations in these genes cause the loss of VirB2 pilus formation and result in processed VirB2 accumulation in the cell. The presence of exocellular processed VirB2 is directly correlated with the formation of pili, and it appears as the major protein in the purified pilus preparation. The evidence provides a compelling argument for VirB2 as the propilin whose 7.2-kDa processed product is the pilin subunit of the promiscuous conjugative pilus, hereafter called the “T pilus” of A. tumefaciens.
Agrobacterium tumefaciens naturally transforms competent plant cells into tumor cells by horizontally transferring the T-DNA of the resident Ti plasmid into the nuclear genome. For many years, the nature of the mechanism of this interkingdom transfer has remained elusive since at least 24 virulence (vir) genes on the Ti plasmid were identified to play a role in this intriguing process (reviewed in reference 7). Several ideas on a potential transfer mechanism have been proposed, one of which is that Agrobacterium might be using its conjugation machinery to deliver the T-DNA into plants since the transfer system resembles interbacterial conjugative transfer mechanisms of broad-host-range plasmids (14, 20, 21, 29, 30, 32). On the basis of genetic and protein-protein interaction studies by several laboratories, the proteins encoded by the 11 virB genes of the Ti plasmid appear to be critically involved since they primarily associate with the cytoplasmic and periplasmic membranes, suggesting that they constitute part of a putative transmembrane pore or channel through which the T-DNA complex is transported (reviewed in references 1 and 7). Because the VirB proteins show amino acid sequence homologies to Tra proteins of plasmids of the IncP, IncN, and IncW groups (14, 15, 20, 21, 24a) and to some of the Tra proteins involved in the direct synthesis and assembly of the F pilus in Escherichia coli (29, 30), a VirB-specific promiscuous pilus has been proposed (15) and, indeed, has been directly observed recently (11, 16, 17, 17a).
TraA is the major structural pilin subunit of the F pilus, and VirB2 has been found to be the homolog of TraA (29). TraA is processed from a 12.7-kDa propilin into a 7.2-kDa pilin, which is the structural subunit of the F pilus (9). Likewise, VirB2 is processed from a 12.3-kDa protein into a 7.2-kDa protein (13, 29). The homology in amino acid sequence and the similarity in protein processing culminating in a product of equal size for TraA and VirB2 have led to the proposal that VirB2 is likely the propilin (15, 29). However, it was recently shown that a truncated VirB1 protein (VirB1*) is released into the medium primarily when the Agrobacterium cells are protractedly vortexed (2). The detection of exocellular VirB1* suggested that this protein might be a pilus component (2). The question is now raised as to whether or not a VirB protein constitutes the major structural component of the pilus, and if so, which VirB protein is the pilin?
In the present communication, we provide several lines of evidence in support of the hypothesis that the processed VirB2 constitutes the pilin subunit of the promiscuous conjugative pilus structure observed on vir-induced A. tumefaciens cells. We show herein that the processed 7.2-kDa VirB2 protein is consistently detectable outside the Agrobacterium cell, whereas it is not detected exocellularly when a mutation exists in virB2 and in each virB gene that was tested. In addition, the presence of exocellular VirB2 is directly correlated with the formation of pili, and it is observed as the major protein in the purified pilus fraction. These results strongly suggest that VirB2 is the major pilin subunit of the promiscuous pilus that mediates the transfer of the T-DNA from Agrobacterium to yeasts and plants (reviewed in reference 17).
MATERIALS AND METHODS
Bacteria and plasmids.
A. tumefaciens strains and plasmids used are listed in Table 1. These strains were grown on medium 523 (10 g of sucrose, 8 g of casein enzymatic hydrolysate, 4 g of yeast extract [Difco], 3 g of dibasic potassium phosphate, 0.3 g of magnesium sulfate [pH 7.0], 15 g of agar per liter) at 28°C. For the selection of specific antibiotic resistance markers, rifampin and erythromycin were used at 50 μg/ml each, and kanamycin was used at 20 μg/ml. For vir gene induction, 500 μl of overnight-cultured cells were collected by centrifugation (6,000 × g, 5 min) and resuspended in 5 ml of induction medium (I medium), which consists of medium 925 (19) plus 0.15 g of KCl, 0.01 g of CaCl2, 2.5 mg of FeSO4 · 7H2O, 50 mM 2-(N-morpholine)ethanesulfonic acid, and 20 g of glucose, pH 5.5, per liter. After growth of A. tumefaciens test strains at 28°C to mid-log phase (4 to 6 h), 500 μl of the culture was spread on 1.5% I-medium agar containing 200 μM acetosyringone (Adrich Chemical Company) and incubated for 3 days at 19°C.
TABLE 1.
Bacterial strains and plasmids
| Bacterial strain or plasmid | Relevant characteristic(s) | Reference |
|---|---|---|
| A. tumefaciens | ||
| C58 | Wild-type virulent strain containing nopaline type Ti plasmid pTiC58 | 22 |
| NT1RE | Rifampin- and erythromycin-resistant mutant of Ti plasmid-free strain A. tumefaciens C58 | 18 |
| NT1REB | Flagellum-free mutant of NT1RE | 6 |
| Plasmids | ||
| pJK270 | Tn5 insertion in the T-DNA of conjugative proficient pTiC58TraC | 18 |
| pJK502 | Tn5 insertion in virB3 of pTiC58TraC | 23 |
| pJK190 | Tn5 insertion in virB4 of pTiC58TraC | 23 |
| pJK104 | Tn5 insertion in virB5 of pTiC58TraC | 23 |
| pJK125 | Tn5 insertion in virB9 of pTiC58TraC | 23 |
| pJK210 | Tn5 insertion in virB10 of pTiC58TraC | 23 |
| pUCD4606 | virB2 nonpolar mutant of pJK270 | 13 |
| pUCD2614 | High-copy-number vir region plasmid derived from pTiC58 | 26 |
Exocellular proteins.
Exocellular secreted proteins, including surface appendages, were collected according to Roine et al. (27) with minor modifications. With the aid of an L-glass rod, the bacterial cells were scraped gently off the agar surface of each plate (150-mm diameter) into 2 ml of 10 mM sodium phosphate buffer, pH 5.3 (buffer A), and collected by centrifugation at 13,000 × g for 10 min at 4°C. The supernatant fraction (S1) contained the secreted proteins, including fragmented pili. The surface proteins and appendages were released from the cells by passing the bacterial suspension in 0.4 ml of buffer A through a hypodermic needle (25 gauge) five times. The cells were then removed by centrifugation at 13,000 × g at 4°C for 10 min, and the supernatant (S2) contained the surface proteins including substantially more lengthy pili than in S1. The pellet was resuspended and adjusted to an A600 of 10 in buffer A and represented the total cell lysate (P).
Pilus purification.
Pili of various lengths occur in approximately equal amounts in both S1 and S2 fractions originally derived from cells grown on 50 agar plates. They were collected by centrifugation at 100,000 × g for 3 h at 4°C and resuspended in 6 ml of buffer B (10 mM Tris-HCl [pH 7.5], 100 mM NaCl) containing 0.5% sodium deoxycholate and further fractionated by velocity sedimentation in a 32-ml, 25 to 70% (wt/vol) linear sucrose gradient in buffer B using a Beckman SW27 rotor at 27,400 rpm for 4.5 h at 4°C. The fractions (1.2 ml) were collected from the top of the gradient by displacing the gradient from the bottom of the centrifuge tube with fluorocarbon (Fluorinert FC-43; 3M Company). Aliquots from each fraction were examined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (see below), dialyzed overnight at 4°C against 1,000 volumes of 10 mM Tris-Cl (pH 7.5), negatively stained with 2% uranyl acetate, and viewed by transmission electron microscopy (see below).
Tricine-SDS-PAGE.
Tricine-based SDS-PAGE was performed according to Schägger and von Jagow (28) using 16.5% acrylamide and 3% bisacrylamide. Tricine as a trailing ion provides sharper resolution of small proteins than when glycine is used. Each protein sample derived from equivalent cell concentrations was mixed with an equal volume of 2× SDS-gel loading buffer (0.2 M Tris-Cl [pH 6.8], 8% SDS, 0.2% bromophenol blue, 40% glycerol, 4% β-mercaptoethanol) and incubated at 100°C for 5 min before loading. After electrophoresis, the protein bands were visualized by staining with 0.8% aqueous silver nitrate. For immunoblots, the fractionated proteins were electrotransferred onto nitrocellulose membranes (Hybond-C; Amersham, Arlington Heights, Ill.). Molecular weight marker proteins were from a commercial supplier (Amersham).
Immunoblot analysis.
Rabbit-borne polyclonal antibodies against VirB2 were prepared as described previously (13, 29). The antibody to the cytoplasmic Ros protein was kindly provided by John Archdeacon of the Davis Crown Gall Group. The VirB2-specific antibody was cross-adsorbed to sonicated cell lysates of A. tumefaciens NT1 (without Ti plasmid) at a 1:1 volume ratio and incubated for 4 h at 23°C or overnight at 4°C. The precipitated antibody-antigen complex was removed by centrifugation (13,000 × g, 10 min, 4°C). The cross-adsorbed antibodies were maintained at 4°C in the presence of 0.05% sodium azide. The nitrocellulose membranes containing the transferred proteins were incubated for 30 min at 23°C in TBST buffer (0.05 M Tris-Cl [pH 7.5], 0.85% NaCl, 0.2% Tween 20) containing 5% skim milk (Carnation) followed by treatment with specific antibodies (1:2,000 dilution in TBST containing 3% skim milk) for 1 h. After extensive washing with TBST, the membrane was incubated at 23°C with the donkey anti-rabbit horseradish peroxidase-conjugated secondary antibody (Amersham) (1:2,000 dilution in TBST containing 3% skim milk) for 30 min and then washed with TBST several times. Antibody interactions with antigen were visualized with a chemiluminescence system (ECL kit; Amersham).
Electron microscopy.
The samples were deposited on carbon-Formvar films on 300-mesh, 3-mm copper grids (Electron Microscopy Sciences, Fort Washington, Pa.). Usually 10 μl of sample was placed on each grid for 1 min and then rinsed with sterile triple-distilled water for a few seconds and stained with 2% uranyl acetate for 1 min. The samples were examined in a Phillips EM410 electron microscope at 80 kV.
RESULTS
Exocellular presence of VirB2.
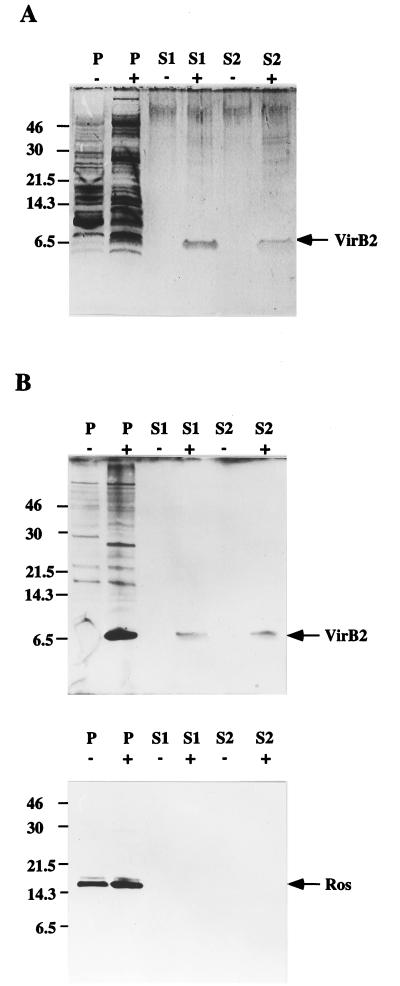
The exocellular secreted proteins in S1 and surface structure proteins in S2 prepared from A. tumefaciens C58 were fractionated by Tricine-SDS-PAGE and visualized by silver staining. A 7.2-kDa protein identical to the processed VirB2 protein was observed only in samples derived from acetosyringone-induced cells, and not in samples derived from uninduced cells (Fig. 1A). To verify that the 7.2-kDa protein was indeed VirB2, the fractionated proteins were reacted with VirB2-specific antibody. The results shown in Fig. 1B confirm that this protein is indeed the processed VirB2. To eliminate the possibility that the VirB2 protein might be derived from Agrobacterium cells that had lysed during handling, we used antibody to the cytoplasmic protein Ros, which is a repressor in the cytosol encoded by the bacterial chromosome (8), as the internal control. The immunoblot analysis of both S1 and S2 verified that the 7.2-kDa VirB2 protein did not originate from lysed cells since no Ros protein was detected in either S1 or S2 (Fig. 1B). Thus, it appears that VirB2 is exported out of the Agrobacterium cell.
FIG. 1.
Presence of the processed 7.2-kDa VirB2 protein as a surface component of acetosyringone-induced A. tumefaciens cells. Proteins prepared from total cell lysate (P), exocellular secreted proteins (S1), and the surface proteins and appendages sheared from unlysed cells (S2) of A. tumefaciens C58 induced with (+) and without (−) acetosyringone at 19°C for 3 days were fractionated by Tricine-SDS-PAGE and detected by silver staining (A) and by immunoblotting with anti-VirB2 and anti-Ros antibodies (B). The numbers on the left are the positions of the molecular mass standards in kilodaltons. The arrows indicate the positions of VirB2 and Ros.
Exocellular VirB2 production is stimulated at suboptimal growth temperature.
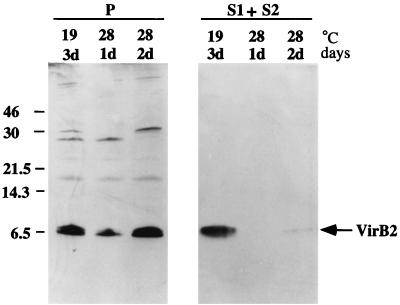
Fullner et al. (11) reported that vir gene-specific pili were produced more abundantly when A. tumefaciens cells were grown at 19 than at 28°C. We therefore examined whether the amount of exocellular VirB2 produced would be proportional to the amount of pili formed at these temperatures. Three independent experiments were performed, and the results showed consistently higher amounts of exocellular VirB2 produced at 19 than at 28°C from the same density of cells irrespective of intracellular concentrations (Fig. 2). Densitometric scanning of the exocellular VirB2 protein band obtained from cells grown at the corresponding temperatures shows that there is an average 20-fold increase at this lower temperature. The production of exocellular VirB2 protein therefore parallels that of virB-specific pilus production, suggesting that the pili might be primarily composed of exocellular VirB2.
FIG. 2.
Production of exocellular VirB2 as a function of temperature. Total cell lysate (P), exocellular secreted proteins (S1), and the sheared surface proteins and appendages (S2) were derived from acetosyringone-induced A. tumefaciens C58 cells, and the proteins derived from equivalent cell concentrations were fractionated by tricine-SDS-PAGE and analyzed by immunoblotting with anti-VirB2 antibody. The numbers on the left correspond to molecular mass standards. The numbers over each column are the cell incubation temperature and the length of incubation in days (d). VirB2 is indicated by the arrow.
Exocellular VirB2 production requires virB genes.
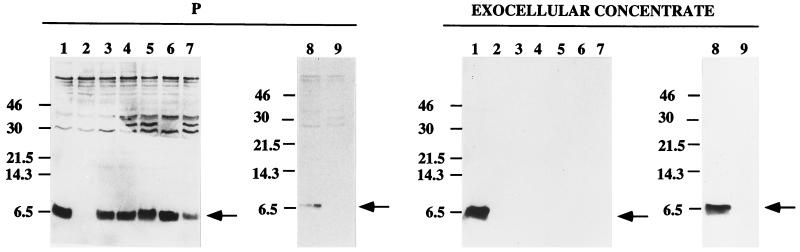
To determine if the production of exocellular VirB2 protein was dependent on certain virB genes, we examined the characteristics of several virB mutants. As shown in Fig. 3, the exocellular presence of VirB2 is strictly dependent on the virB genes, as indicated by the absence of VirB2 in the exocellular preparations from virB mutants and its presence in exocellular preparations from the parental strain, NT1RE(pJK270). Furthermore, when the cellular contents of each mutant strain were examined, VirB2 protein was detected in all these mutants, indicating that VirB2 is still produced within the cellular confines but not exported out of the Agrobacterium cell (Fig. 3). Thus, the processing of VirB2 propilin is not dependent on an intact functional VirB channel. As anticipated, the mutant strain containing a nonpolar mutation in the virB2 gene itself was unable to produce any VirB2 protein.
FIG. 3.
Exocellular VirB2 is not produced in the absence of virB genes. Total cell lysate (P) and exocellular proteins prepared as for Fig. 1 as fractions S1 and S2 were derived from acetosyringone-induced strains grown at 19°C for 3 days. The S1 and S2 fractions were combined and further centrifuged at 100,000 × g for 3 h at 4°C, and the pellet was solubilized in buffer B containing 0.5% sodium deoxycholate (Exocellular Concentrate). The resulting exocellular concentrated proteins and proteins of the total cell lysate were fractionated by Tricine-SDS-PAGE and detected by immunoblotting with anti-VirB2 antibody. Lanes 1, intact virB genes (pJK270); lanes 2, a nonpolar mutation in virB2 (pUCD4606); lanes 3 to 7, Tn5 insertions in virB3 (pJK502), virB4 (pJK190), virB5 (pJK104), virB9 (pJK125), and virB10 (pJK210), respectively; lanes 8, flagellum-free strain NT1REB containing intact virulon (pUCD2614) only; lanes 9, NT1REB without the plasmid. The numbers on the left are molecular mass standards. VirB2 is indicated by the arrows.
To eliminate the possibility of interference caused by appendages such as flagellin and possibly the conjugative pilin presumably encoded by the tra operon of the Ti plasmid, we used the flagellum-free strain NT1REB (6) containing plasmid pUCD2614. This plasmid contains only the vir regulon portion of pTiC58 (26). This strain also produced the exocellular VirB2 protein (Fig. 3). Thus, flagellin encoded by the flagellin genes flaA, flaB, and flaC (6) and proteins encoded by regions outside the vir regulon (“virulon”) of the Ti plasmid do not take part in the exocellular production of VirB2.
Exocellular VirB2 production is correlated with pilus formation.
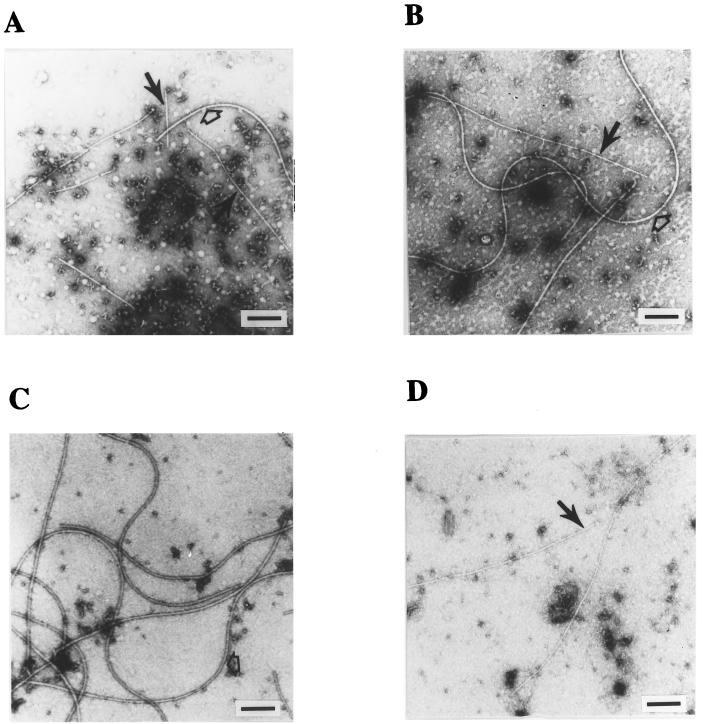
The above genetic and immunological analyses demonstrated that VirB2 is exported, appearing perhaps as structures resembling pilus filaments. Such a finding would support our early hypothesis that the promiscuous pilus is composed of VirB2 as the major pilin subunit (15, 29). This hypothesis is supported by the following electron microscopic evidence. Examination of concentrated exocellular preparations containing VirB2 revealed long thin filaments of variable lengths (Fig. 4, filled arrows). The average width of these filaments was 10 nm, while flagella as an internal control are clearly larger (15 nm in width) (open arrows in Fig. 4A, B, and C). These thin filaments were not observed in exocellular preparations derived from either virB mutant cells or pTiC58-free cells (Fig. 4C). The same pilus was observed in exocellular preparations from NT1REB(pUCD2614) cells (Fig. 4D), confirming the fact that these pili were not encoded by the tra genes of the Ti plasmid.
FIG. 4.
Electron microscopic analysis. The exocellular concentrate derived from acetosyringone-induced cells was negatively stained with 2% uranyl acetate. Thin pilus filaments are indicated by the filled narrow arrow, and the flagella are indicated by the open arrows. (A) Filaments from strain C58; (B) filaments from NT1RE(pJK270); (C) filaments from NT1RE; (D) filaments from NT1REB(pUCD2614). Scale bar = 200 nm.
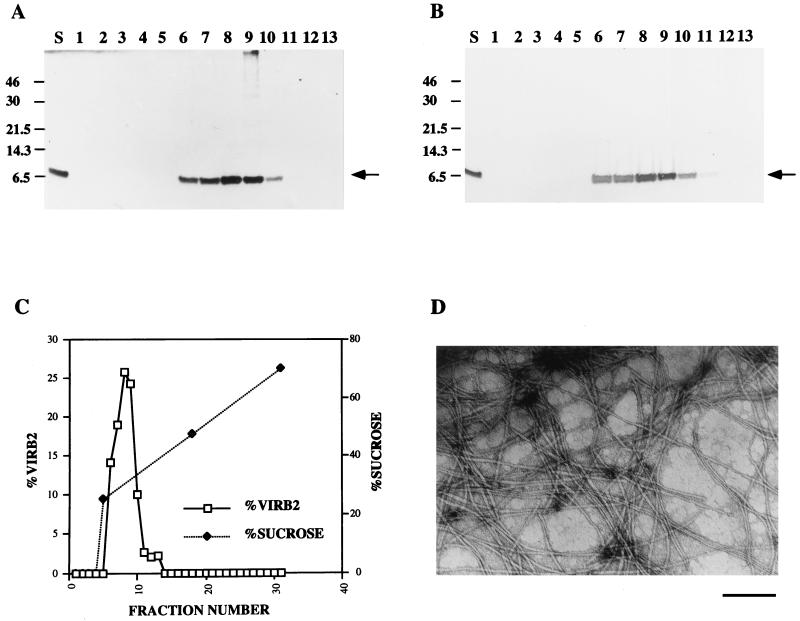
VirB2 is the major component of the pilus.
To further support that our observations were being made on VirB2 as the pilus structural component, the concentrated exocellular protein preparation of A. tumefaciens C58 was fractionated by velocity sedimentation in a 25 to 70% linear gradient of sucrose. Thirty-one fractions were collected and resolved by SDS-PAGE followed by immunoblotting. VirB2 was detected in fractions 6 to 13 (Fig. 5A), with the highest concentration of VirB2 appearing in fractions 8 and 9 (Fig. 5C). As judged by silver staining, these fractions contained a single visible protein as processed VirB2 (Fig. 5B). Transmission electron microscopy confirmed that fractions 8 and 9 contained the majority of aggregated pilus filaments (Fig. 5D).
FIG. 5.
Velocity sedimentation and electron microscopic analyses of VirB2 pili under the conditions described in Materials and Methods. The exocellular concentrate (S) and fractions 1 to 13 were resolved by tricine-SDS-PAGE and analyzed by immunoblotting with anti-VirB2 antibody (A) and by silver staining (B). The numbers on the left are molecular mass standards in kilodaltons. The arrows mark the positions of VirB2 as visualized by both procedures. (C) Velocity sedimentation of VirB2 as a function of sucrose concentration. (D) VirB2 filaments from fraction 9 visualized by electron microscopy. Scale bar = 200 nm.
DISCUSSION
In the present study, we have shown that 7.2-kDa processed VirB2 is present outside the induced Agrobacterium cell as the major exocellular protein, which was previously predicted to constitute the promiscuous pilus (15). As evidenced by velocity gradient sedimentation analysis, this exocellular protein is largely a homocomplex composed of the 7.2-kDa VirB2 “pilin” complexed into thin 10-nm-diameter filaments. This essentially parallels what has been observed in the analysis of purified F pilus, which is composed of a processed 7.2-kDa TraA pilin (9).
Additional support for the presence of the VirB2-specific pilus comes from analyzing the amount of exocellular VirB2 protein as a function of temperature. We found that the increase in exocellular VirB2 protein at 19°C correlates well with the optimal temperature for VirB-specific piliation as observed by Fullner et al. (11). Interestingly, processed VirB2 accumulates in the cell in similar amounts irrespective of the growth temperature (19 versus 28°C). This suggests that the temperature effect is on the exportation of VirB2. Early studies on temperature effects on tumorigenesis demonstrated that crown gall tumor sizes were dramatically decreased when the host plants were inoculated between 28 and 30°C, with no galls appearing at 31°C or above (5, 25). Recent studies on the effect of temperature on the efficiency of transfer of IncQ plasmid RSF1010 by the T-DNA transfer machinery of A. tumefaciens showed that it was severely affected at temperatures above 28°C (10), with optimum transfer occurring at 19°C, correlating well with the amount of piliation observed by Fullner et al. (11). Since piliation is necessary for T-DNA transfer, one would suspect that the efficiency of VirB2-specific piliation might be one of the limiting factors in the transfer process.
The mechanism of bringing the VirB2 subunit to the exterior of the cell is not known, although the virB operon is likely involved. The conservation of plasmid transfer genes based on sequence homologies has been noted between broad-host-range plasmids (20, 21) and extends to the virB operon. The genes of this operon are likely involved in the synthesis and assembly of the promiscuous pilus (15). We find that mutations in the virB genes cause loss of pilus formation. No VirB2 protein and VirB2-specific filaments are detected outside the Agrobacterium cell, and yet, the 7.2-kDa VirB2 protein accumulates within these mutant strains. Thus, there is support for the notion that the proteins encoded by the remaining virB genes make up the transmembrane transport apparatus (or channel) and serve in facilitating the export of the processed VirB2 subunits to the cell exterior. Interestingly, VirB2 is stably expressed and maintained inside the Agrobacterium cell in the absence of other VirB proteins that we have examined so far. Likewise, TraA propilin is processed and localized to the cytoplasmic membrane in the absence of tra genes that are required for F-pilus formation (reviewed in reference 31). The only exception is traQ, which is required for the accumulation of F pilin (reviewed in reference 31). So far no TraQ homolog has been found in A. tumefaciens (13). Jones et al. (13) showed that VirB2 is processed and inserted into the cytoplasmic membrane without the aid of TraQ or vir gene products, suggesting that VirB2 can remain in the cytoplasmic membrane in the absence of other vir genes.
How the processed VirB2 protein is exported is unknown. The structure of the transmembrane transport apparatus is being elucidated by identifying the membrane locations and interplay of VirB proteins. Various interesting hypothetical models of the putative transport complex associated with the Agrobacterium membranes have been proposed (1, 2, 4, 7). Yet, the mechanism of pilin exportation as well as the transfer of the T-DNA complex remains obscure. Whatever structure the transmembrane transporter ends up to be, it is clear that the major component appearing on the surface as thin pili is processed VirB2. This does not rule out other VirB proteins that might be transported out of the cell but to a lesser extent than VirB2. The VirB1 protein, identified as a lytic transglycosylase (3, 24), is processed into a 12-kDa protein termed VirB1* (2). VirB1* has been hypothesized as a potential component of the pilus (2). Since the pilus preparations that we have examined are composed entirely of processed VirB2, we conclude that this protein and not VirB1 is the subunit that is assembled into the promiscuous pilus. However, it remains plausible that VirB1 might be the internal catalytic component that facilitates initial passage of the VirB2 protein through the peptidoglycan barrier of the Agrobacterium cell wall and is cleaved into VirB1* during catalysis, ending up in the VirB-specific pore or channel. The 12-kDa protein is loosened upon lengthy agitation from the pore, making its appearance extracellularly.
Since conjugative pili are entirely composed of conjugative pilin subunits (31), the processed VirB2 subunit may likewise make up the entire promiscuous conjugative pilus of A. tumefaciens. The VirB2-specific pilus is hereafter designated the “T pilus” since accumulating evidence strongly suggests that it is directly involved in T-DNA transfer and since the designation distinguishes the pilus from other pili such as the one presumably encoded by Ti plasmid tra genes. Although we have provided several lines of evidence for the T pilus being composed of primarily processed VirB2, the presence of this structure does not answer the central question of how the T-DNA complex is transported out of the Agrobacterium cell and into the plant cell. In the case of conjugative transfer of F-plasmid DNA, the F-pilus filaments are cylindrical with an outside diameter of 8 nm and a central, hydrophilic lumen of 2 nm (31). Does the pilus mediate close contact between donor and recipient cells? Or once the pilus makes contact with the recipient cell, does it serve as a conduit for the DNA to pass through? The 2-nm lumen is certainly of sufficient size for DNA to traverse it. Harrington and Rogerson (12) have provided evidence that the F pilus is capable of acting as a stable conduit for DNA transfer between donor and recipient, suggesting that the pilus lumen is indeed of sufficient size to accommodate DNA and its piloting proteins. Expanding our knowledge on the physical characteristics of the T pilus would certainly provide interesting insights into and clues to answering the central question of how the T-DNA complex is transferred between Agrobacterium and the plant cell.
ACKNOWLEDGMENTS
We thank John Archdeacon for graciously providing the antibody to purified Ros protein.
This research was supported by NIH grant GM45550 from the National Institute of General Medical Sciences.
REFERENCES
- 1.Baron C, Zambryski P C. Plant transformation: a pilus in Agrobacterium T-DNA transfer. Curr Biol. 1997;6:1567–1569. doi: 10.1016/s0960-9822(02)70773-2. [DOI] [PubMed] [Google Scholar]
- 2.Baron C, Llosa M, Zhou S, Zambryski P C. VirB1, a component of the T-complex transfer machinery of Agrobacterium tumefaciens, is processed to a C-terminal secreted product, VirB1*. J Bacteriol. 1997;179:1203–1210. doi: 10.1128/jb.179.4.1203-1210.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bayer M, Eferl R, Zellnig G, Teferle K, Dijkstra A, Koraimann G, Högenauer G. Gene 19 of plasmid R1 is required for both efficient conjugative DNA transfer and bacteriophage R17 infection. J Bacteriol. 1995;177:4279–4288. doi: 10.1128/jb.177.15.4279-4288.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beijersbergen A, Smith S J, Hooykaas P J J. Localization and topology of VirB proteins of Agrobacterium tumefaciens. Plasmid. 1994;32:212–218. doi: 10.1006/plas.1994.1057. [DOI] [PubMed] [Google Scholar]
- 5.Braun A C. Thermal studies on the factors responsible for tumor initiation in crown gall. Am J Bot. 1947;34:234–240. [PubMed] [Google Scholar]
- 6.Chesnokova O, Coutinho J B, Khan I H, Mikhail M S, Kado C I. Characterization of flagella genes of Agrobacterium tumefaciens, and the effect of a bald strain on virulence. Mol Microbiol. 1997;23:579–590. doi: 10.1046/j.1365-2958.1997.d01-1875.x. [DOI] [PubMed] [Google Scholar]
- 7.Christie P J. Agrobacterium tumefaciens T-complex transport apparatus: a paradigm for a new family of multifunctional transporters in eubacteria. J Bacteriol. 1997;179:3085–3094. doi: 10.1128/jb.179.10.3085-3094.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooley M B, D’Souza M R, Kado C I. The virC and virD operons of the Agrobacterium Ti plasmid are regulated by the ros chromosomal gene: analysis of the cloned ros gene. J Bacteriol. 1991;173:2608–2616. doi: 10.1128/jb.173.8.2608-2616.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frost L S, Paranchych W, Willetts N S. DNA sequence of the F traALE region that includes the gene for F pilin. J Bacteriol. 1984;160:395–401. doi: 10.1128/jb.160.1.395-401.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fullner K J, Nester E W. Temperature affects the T-DNA transfer machinery of Agrobacterium tumefaciens. J Bacteriol. 1996;178:1498–1504. doi: 10.1128/jb.178.6.1498-1504.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fullner K J, Lara L C, Nester E W. Pilus assembly by Agrobacterium T-DNA transfer genes. Science. 1996;273:1107–1109. doi: 10.1126/science.273.5278.1107. [DOI] [PubMed] [Google Scholar]
- 12.Harrington L C, Rogerson A C. The F pilus of Escherichia coli appears to support stable DNA transfer in the absence of wall-to-wall contact between cells. J Bacteriol. 1990;172:7263–7264. doi: 10.1128/jb.172.12.7263-7264.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jones A L, Lai E-M, Shirasu K, Kado C I. VirB2 is a processed pilin-like protein encoded by the Agrobacterium tumefaciens Ti plasmid. J Bacteriol. 1996;178:5706–5711. doi: 10.1128/jb.178.19.5706-5711.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kado C I. Agrobacterium-mediated transfer and stable incorporation of foreign genes in plants. In: Clewell D B, editor. Bacterial conjugation. New York, N.Y: Plenum Press; 1993. pp. 243–254. [Google Scholar]
- 15.Kado C I. Promiscuous DNA transfer system of Agrobacterium tumefaciens: role of the virB operon in sex pilus assembly and synthesis. Mol Microbiol. 1994;12:17–22. doi: 10.1111/j.1365-2958.1994.tb00990.x. [DOI] [PubMed] [Google Scholar]
- 16.Kado C I. T-DNA transfer to plants is mediated by pilus-like apparatus encoded by the Ti plasmid virB operon. Adv Plant Biotechnol. 1994;4:23–36. [Google Scholar]
- 17.Kado C I. Agrobacterium-mediated horizontal gene transfer. In: Setlow J K, editor. Genetic engineering, principles and methods. Vol. 20 1998. , in press. Plenum Press, New York, N.Y. [DOI] [PubMed] [Google Scholar]
- 17a.Kado C I, Jones L, Coutinho J, Shirasu K, Lee L-Y, De La Cruz F, Khan I. Abstracts of the International Conference on Plasmid Biology, Banff, Alberta, Canada. 1994. Transfer of the T-DNA from Agrobacterium to plants appears to be mediated by a conjugative mechanism potentially involving a pilus and promoted by flagella; pp. 28–29. [Google Scholar]
- 18.Kao J, C, Perry K L, Kado C I. Indoleacetic acid complementation and its relation to host range specifying genes on the Ti plasmid of Agrobacterium tumefaciens. Mol Gen Genet. 1982;188:425–432. doi: 10.1007/BF00330044. [DOI] [PubMed] [Google Scholar]
- 19.Langley R A, Kado C I. Studies on Agrobacterium tumefaciens. Conditions for mutagenesis by N-methyl-N′-nitro-N-nitrosoguanidine and relationships of A. tumefaciens mutants to crown-gall tumor induction. Mutat Res. 1971;14:277–286. [Google Scholar]
- 20.Lessl M, Balzer D, Pansegrau W, Lanka E. Sequence similarities between the RP4 Tra2 and the Ti VirB region strongly support the conjugation model for T-DNA transfer. J Biol Chem. 1992;267:20471–20480. [PubMed] [Google Scholar]
- 21.Lessl M, Lanka E. Common mechanisms in bacterial conjugation and Ti-mediated T-DNA transfer to plant cells. Cell. 1994;7:321–324. doi: 10.1016/0092-8674(94)90146-5. [DOI] [PubMed] [Google Scholar]
- 22.Lin B C, Kado C I. Studies on Agrobacterium tumefaciens. VII. Avirulence induced by temperature and ethidium bromide. Can J Microbiol. 1977;23:1554–1561. doi: 10.1139/m77-229. [DOI] [PubMed] [Google Scholar]
- 23.Lundquist R C, Close T C, Kado C I. Genetic complementation of Agrobacterium tumefaciens Ti plasmid mutants in the virulence region. Mol Gen Genet. 1984;193:1–7. doi: 10.1007/BF00327406. [DOI] [PubMed] [Google Scholar]
- 24.Mushegian A R, Fullner K J, Koonin E V, Nester E W. A family of lysozyme-like virulence factors in bacterial pathogens of plants and animals. Proc Natl Acad Sci USA. 1996;93:7321–7326. doi: 10.1073/pnas.93.14.7321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24a.Pohlman R F, Genetti H D, Winans S C. Common ancestry between IncN conjugal transfer genes and macromolecular export systems of plant and animal pathogens. Mol Microbiol. 1994;14:655–668. doi: 10.1111/j.1365-2958.1994.tb01304.x. [DOI] [PubMed] [Google Scholar]
- 25.Riker A J. Studies on the influence of some environmental factors on the development of crown gall. J Agric Res. 1926;32:83–96. [Google Scholar]
- 26.Rogowsky P M, Powell B S, Shirasu K, Lin T-S, Morel P, Zyprian E M, Steck T R, Kado C I. Molecular characterization of the vir regulon of Agrobacterium tumefaciens: complete nucleotide sequence and gene organization of the 28.63-kbp regulon cloned as a single unit. Plasmid. 1990;23:85–106. doi: 10.1016/0147-619x(90)90028-b. [DOI] [PubMed] [Google Scholar]
- 27.Roine E, Wei W, Yuan J, Nurmiaho-Lassila E-L, Kalkkinen N, Romantschuk M, He S Y. Hrp pilus: an hrp-dependent bacterial surface appendage produced by Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA. 1997;94:3459–3464. doi: 10.1073/pnas.94.7.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schägger H, von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range of 1 to 100 kDa. Anal Biochem. 1987;166:368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- 29.Shirasu K, Kado C I. Membrane location of the Ti plasmid VirB proteins involved in the biosynthesis of a pilin-like conjugative structure on Agrobacterium tumefaciens. FEMS Microbiol Lett. 1993;111:287–294. doi: 10.1111/j.1574-6968.1993.tb06400.x. [DOI] [PubMed] [Google Scholar]
- 30.Shirasu K, Koukolíková-Nicola Z, Hohn B, Kado C I. An inner-membrane-associated virulence protein essential for T-DNA transfer from Agrobacterium tumefaciens to plants exhibits ATPase activity and similarities to conjugative transfer genes. Mol Microbiol. 1994;11:581–588. doi: 10.1111/j.1365-2958.1994.tb00338.x. [DOI] [PubMed] [Google Scholar]
- 31.Silverman P M. Towards a structural biology of bacterial conjugation. Mol Microbiol. 1997;23:423–429. doi: 10.1046/j.1365-2958.1997.2411604.x. [DOI] [PubMed] [Google Scholar]
- 32.Stachel S E, Zambryski P C. Agrobacterium tumefaciens and the susceptible plant cell: a novel adaptation of extracellular recognition and DNA conjugation. Cell. 1986;47:155–157. doi: 10.1016/0092-8674(86)90437-x. [DOI] [PubMed] [Google Scholar]