Abstract
A periplasmic protein has been found to prevent aggregation of the acid-unfolded dimethyl sulfoxide reductase (DMSOR), the periplasmic terminal reductase of dimethyl sulfoxide respiration in the phototroph Rhodobacter sphaeroides f. sp. denitrificans, in a manner similar to that of the Escherichia coli chaperonin GroEL (Matsuzaki et al., Plant Cell Physiol. 37:333–339, 1996). The protein was isolated from the periplasm of the phototroph. It had a molecular mass of 58 kDa and had no subunits. The sequence of 14 amino-terminal residues of the protein was completely identical to that of the periplasmic dipeptide transport protein (DppA) of E. coli. The 58-kDa protein prevented aggregation to a degree comparable to that of GroEL on the basis of monomer protein. The 58-kDa protein also decreased aggregation of guanidine hydrochloride-denatured rhodanese, a mitochondrial matrix protein, during its refolding upon dilution. The 58-kDa protein is a kind of molecular chaperone and could be involved in maintaining unfolded DMSOR, after secretion of the latter into the periplasm, in a competent form for its correct folding.
The periplasmic space is the region between the inner and outer membranes of gram-negative bacteria. The periplasmic space contains many proteins, such as solute transport proteins, and their functional expression in the space is essential for bacterial growth. Recently, the periplasm of Escherichia coli has been shown to play an important role in the expression of a wide variety of recombinant proteins from different sources. However, the mechanism of protein folding in the periplasm is less well understood (reviewed in references 15 and 27). None of the classical molecular chaperones such as GroEL and DnaK, which are located in the cytoplasm, have been identified in the periplasm, and no general molecular chaperone-like proteins, which play roles in the correct folding of proteins by preventing side reactions, have been found. Most recently, however, some periplasmic substrate-binding proteins, the oligopeptide-binding protein OppA of E. coli, the maltose-binding protein MalE of E. coli, and the galactose-binding protein MglB of Salmonella typhimurium, have been demonstrated to have chaperone activities (19). On the other hand, some enzymes that facilitate protein folding in the periplasm of E. coli have been found. Dsb proteins (required for the correct formation of disulfide bonds of periplasmic proteins), PPIase (peptidyl prolyl cis-trans isomerase), and RotA (catalyzes cis-trans isomerization of proline) are enzymes which modify the structures of amino acids within polypeptides (15, 27). Specialized periplasmic chaperones such as PapD are also known to serve in pilin assembly into pili (7).
The denitrifying phototrophic bacterium Rhodobacter sphaeroides f. sp. denitrificans (20) is capable of dimethyl sulfoxide respiration as one of its anaerobic respiration systems. The terminal reductase of the respiration, dimethyl sulfoxide reductase (DMSOR), is a periplasmic soluble protein with a molecular weight of 84,903. It consists of a single polypeptide and contains one molecule of molybdenum (Mo) cofactor and nine cysteine residues per molecule (21, 28). Recently, its three-dimensional structure has been determined by X-ray crystallography (22). We reported previously that spheroplasts prepared from a Mo cofactor-deficient mutant secrete an unfolded form of DMSOR into the medium (11) and that there are no disulfide bonds in the native conformation of DMSOR (12). In an in vitro refolding experiment, we have shown that the acid-unfolded DMSOR prepared by releasing the Mo cofactor from DMSOR by acidification is protected from aggregation by GroEL or a protein(s) in the periplasm (13). Therefore we thought that a molecular chaperone-like protein(s) is present in the periplasm.
In this study we isolated a periplasmic chaperone-like protein from the periplasm of R. sphaeroides by monitoring its ability to prevent aggregation of the acid-unfolded DMSOR. This protein was homologous to DppA, the periplasmic dipeptide transport protein of E. coli. This fact agrees with the findings that some periplasmic substrate-binding proteins have molecular chaperone-like activities (19) but suggests that most other periplasmic proteins do not have chaperone activity since only DppA was isolated by the refolding activity assay.
MATERIALS AND METHODS
Bacteria and growth conditions.
A green mutant strain of Rhodobacter sphaeroides f. sp. denitrificans IL106 (20) was used. The medium and conditions for growth of the photodenitrifier were described previously (29).
Preparation of DMSOR and the periplasmic fraction.
DMSOR was purified according to published protocols (21). The periplasmic fraction was obtained as described previously (13).
Acid denaturation.
DMSOR (100 μg/ml) was denatured by elimination of Mo cofactor by acidification as described elsewhere (13).
Measurement of prevention of aggregation of unfolded DMSOR.
Acid-denatured DMSOR was diluted 2.5-fold with refolding buffer (125 mM Tris-HCl [pH 8.0], 12.5 mM KCl). An aliquot (15 μl) of the acid-unfolded DMSOR in refolding buffer was mixed with an equal volume of each fraction to be assayed. The mixture was incubated at 30°C for 3 h, and aliquots (20 μl) were subjected to electrophoresis on 7.5% nondenaturing polyacrylamide gels at 4°C. DMSOR on the gels was transferred to nitrocellulose membranes as described elsewhere (11) and analyzed immunologically as described elsewhere (29). The acid-unfolded DMSOR in refolding buffer aggregated during incubation for 3 h and disappeared on the nondenaturing gel because the protein did not enter the gel. When a protein with the molecular chaperone-like activity was present during incubation, the unfolded form became visible on the gel. However, it was difficult to quantitatively determine the activity.
Protein determination.
Protein concentration was determined by using a protein assay kit (Bio-Rad Laboratories) with bovine serum albumin (BSA) as a standard.
Purification of a protein capable of preventing aggregation.
The periplasmic fraction obtained from 10-liter bacterial cultures was precipitated with ammonium sulfate at 34 to 85% saturation. The precipitates were dissolved in a minimum quantity of 50 mM Tris-HCl buffer (pH 7.5) and applied to a column (3.0 by 100 cm) of Sephacryl S-200 equilibrated with 50 mM Tris-HCl buffer (pH 7.5) containing 0.1 M NaCl. The proteins were eluted with the same buffer, and the eluate was collected in 4.7-ml fractions. The activity was recovered between fractions 57 and 72. These fractions were pooled and applied to a column (1.5 by 10 cm) of DEAE-Sephacel equilibrated with 50 mM Tris-HCl buffer (pH 7.5) containing 0.1 M NaCl. The proteins were eluted with a linear NaCl gradient of 0.1 to 0.3 M in 80 ml of the same buffer. The activity was recovered in fractions eluted at NaCl concentrations of 0.17 to 0.21 M. These fractions were pooled, concentrated by ultrafiltration, resuspended in 50 mM potassium phosphate buffer (pH 7.0) containing 2 M ammonium sulfate, and applied to a column (1.5 by 6 cm) of phenyl Sepharose CL-4B equilibrated with 50 mM potassium phosphate buffer (pH 7.0) containing 2 M ammonium sulfate. The proteins were eluted with a linear ammonium sulfate gradient from 2 to 0 M in 80 ml of buffer. The protein with the activity was not eluted at ammonium sulfate concentrations from 2 to 0 M. Then, the protein was eluted with 50 mM potassium phosphate buffer (pH 7.0) containing 40% ethyleneglycol.
Two-dimensional PAGE.
Two-dimensional polyacrylamide gel electrophoresis (PAGE) was carried out as described previously (11) except that the nondenaturing gel in the first dimension was immersed in denaturing buffer containing 2% sodium dodecyl sulfate (SDS) and 10% 2-mercaptoethanol followed by heating in a microwave oven for 20 s.
Determination of amino acid sequence.
The amino-terminal amino acid sequence of the purified protein was determined by using a gas phase protein sequencer (Applied Biosystems 473A). The internal amino acid sequence was determined as follows. The purified protein (4.5 μg) was digested with 2.0 μg of V8 protease in 40 μl of 50 mM Tris-HCl (pH 7.5) at 37°C for 3 h and electrophoresed on an SDS-polyacrylamide gel. Then, the amino acid sequences of the two major polypeptides were determined.
In vitro assay of aggregation of rhodanese and refolding of MDH.
Rhodanese (0.393 mg/ml) was denatured in 80 mM potassium phosphate buffer, pH 7.6, containing 10 mM dithiothreitol–5.6 M guanidine hydrochloride (Gdn-HCl) for 30 min at room temperature. Denatured rhodanese was diluted 100-fold into 50 mM Tris-HCl buffer, pH 7.2. Aggregation of denatured rhodanese was monitored at 320 nm as the increase in turbidity at room temperature as previously described (25). Malate dehydrogenase (MDH) (36 μg/ml) was denatured by a procedure similar to that used for rhodanese. Refolding of MDH was measured by monitoring MDH activity after dilution of 5 μl of the denatured MDH solution with 195 μl of dilution buffer (100 mM potassium phosphate, pH 7.6) (8).
Materials.
Sephacryl S-200, DEAE-Sephacel, and phenyl Sepharose CL-4B were obtained from Pharmacia Biotech. GroEL was purchased from Takara Biochemicals. Bovine liver mitochondrial rhodanese and BSA (fraction V) were from Sigma. MDH was from Oriental Yeast.
RESULTS
Purification of the protein capable of preventing aggregation of acid-unfolded DMSOR.
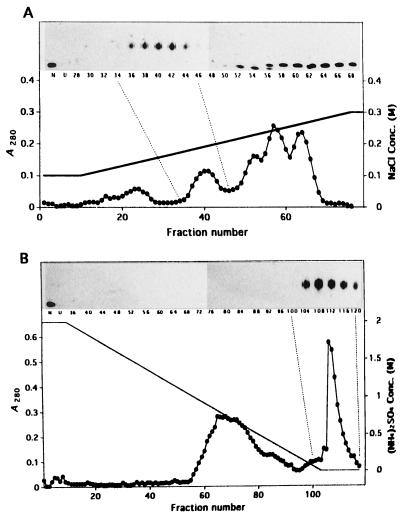
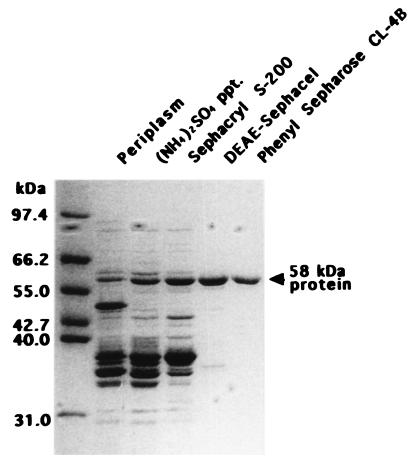
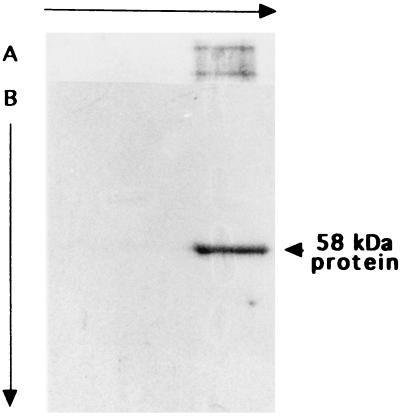
Previously (13), we showed that acid-unfolded DMSOR aggregated during incubation in refolding buffer and that the aggregation was suppressed by incubation with GroEL, one of the best-characterized cytoplasmic molecular chaperonins from E. coli, or a protein(s) with a molecular mass of about 40 kDa from the periplasm of R. sphaeroides f. sp. denitrificans. The periplasmic protein has been further purified. The proteins were precipitated with ammonium sulfate at 34 to 85% saturation and chromatographed on a column of Sephacryl S-200, DEAE-Sephacel, and phenyl Sepharose CL-4B. The results of the DEAE-Sephacel and phenyl Sepharose CL-4B chromatographies are presented in Fig. 1. Although the data in the insets are not quantitatively exact, the peaks of activity appeared around fractions 34 to 46 in DEAE-Sephacel chromatography (Fig. 1A) and 104 to 116 in phenyl-Sepharose CL-4B chromatography (Fig. 1B), differing from the peaks of absorbance at 280 nm of 52, 57, and 64 (Fig. 1A) and 65 and 72 (Fig. 1B), respectively. These results indicate that a specific protein took part in the activity. The protein was purified to almost one polypeptide by phenyl Sepharose CL-4B chromatography (Fig. 2). The protein consisted of a single polypeptide with a molecular mass of 58 kDa, since the molecular mass was about 40 kDa by gel filtration. The 58-kDa protein was clearly visible in the protein profile of crude periplasmic extracts (Fig. 2), showing that it was an abundant periplasmic protein under the conditions used.
FIG. 1.
DEAE-Sephacel and phenyl Sepharose CL-4B chromatography profiles. The periplasmic protein was precipitated with 34 to 85% ammonium sulfate and applied to a column of Sephacryl S-200. Then, the fractions with the activity were applied to columns of DEAE-Sephacel and phenyl Sepharose CL-4B as described in Materials and Methods. Absorbance at 280 nm (•) and the ability to prevent aggregation of acid-unfolded DMSOR (insets) were determined. (A) DEAE-Sephacel chromatography. (B) Phenyl Sepharose CL-4B chromatography. N, native DMSOR; U, acid-unfolded DMSOR with no sample.
FIG. 2.
Purification of the protein capable of preventing aggregation. Fractions from the purification steps were separated by SDS-PAGE and stained with Coomassie brilliant blue. The amounts of protein applied to the lanes of the gel from left (periplasm) to right (phenyl Sepharose) were 20, 15, 15, 5, and 5 μg, respectively. The migration positions of molecular mass marker proteins are shown to the left of the gel.
Amino-terminal and internal amino acid sequences of the 58-kDa protein.
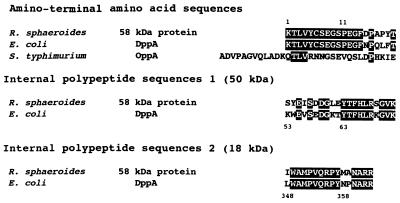
The amino-terminal and internal amino acid sequences of the 58-kDa protein were determined (Fig. 3). A comparison of the sequence with proteins in the DNA Data Base of Japan revealed that the sequence of the 14 amino-terminal amino acid residues was completely identical to that of DppA, a periplasmic dipeptide transport protein of E. coli, and that 16 of 20 amino-terminal amino acid residues were identical between these proteins. The internal amino acid sequences of 50- and 18-kDa polypeptides were also highly homologous to DppA, i.e., 13 of 20 residues and 13 of 16 residues were identical, respectively. E. coli DppA was reported to be more similar to Salmonella typhimurium OppA, the oligopeptide-binding protein, than any other protein or region encoded by an open reading frame in the data base, but it has weak similarity to OppA (24% identity) (17). Between the amino-terminal sequences of the 58-kDa protein and OppA, only 5 of 20 residues were identical. The 58-kDa protein was thus referred to as a DppA-like protein.
FIG. 3.
Amino-terminal and internal amino acid sequences of R. sphaeroides 58-kDa protein compared with those of E. coli DppA and S. typhimurium OppA. Reverse-contrast letters show identical residues.
Specific activity.
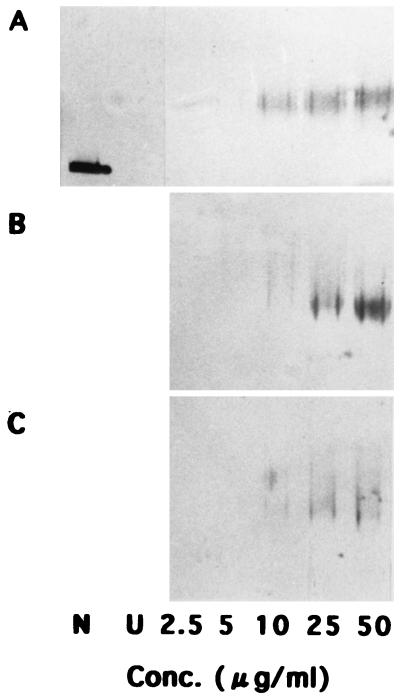
We previously showed that GroEL strongly prevented aggregation of acid-unfolded DMSOR. The specific ability to prevent aggregation of the 58-kDa protein was determined in comparison with those of GroEL and BSA (Fig. 4). At concentrations of 50 and 25 μg/ml in the reaction mixture, GroEL appeared to have the most intense activity among the three molecules examined, with the 58-kDa protein showing a little weaker activity than that of GroEL. BSA had low activity. At a concentration of 10 μg/ml in the reaction mixture, the 58-kDa protein appeared to have stronger activity than that of GroEL. Since GroEL is an oligomeric protein composed of 14 subunits, each with a mass of 57 kDa (reviewed in reference 4), these results indicate that the 58-kDa protein had activity comparable to that of GroEL on a monomeric basis. However, the activity of GroEL on a molar basis was about 14-fold stronger than that of the 58-kDa protein.
FIG. 4.
Specific ability to prevent aggregation of the 58-kDa protein. An aliquot (15 μl) of the acid-unfolded DMSOR in refolding buffer was mixed with an equal volume of the purified protein (A), GroEL (B), or BSA (C) at final concentrations of 2.5, 5, 10, 25, and 50 μg per ml in 30-μl reaction mixtures. N, native DMSOR; U, acid-unfolded DMSOR with no sample.
Aggregation of the 58-kDa protein.
We found that the 58-kDa protein migrated to make a broad band on nondenaturing gels. We examined whether the purified 58-kDa protein preparation was contaminated with heterologous proteins or if the protein formed aggregates by two-dimensional electrophoresis (Fig. 5). On the first nondenaturing gel, the DppA-like protein migrated as a broadened band (Fig. 5A). The protein migrated to one sharp band in the second round of SDS-PAGE after the first gel was heated in a microwave oven in denaturing buffer containing SDS and 2-mercaptoethanol. These results indicate that the final preparation was composed of only 58-kDa protein and that the protein had a tendency to aggregate.
FIG. 5.
Two-dimensional electrophoresis of the 58-kDa protein. The purified 58-kDa protein (10 μg) was first subjected to nondenaturing PAGE (A) and then to SDS-PAGE (B). The gels were stained with Coomassie brilliant blue.
Suppression of aggregation by the 58-kDa protein of rhodanese.
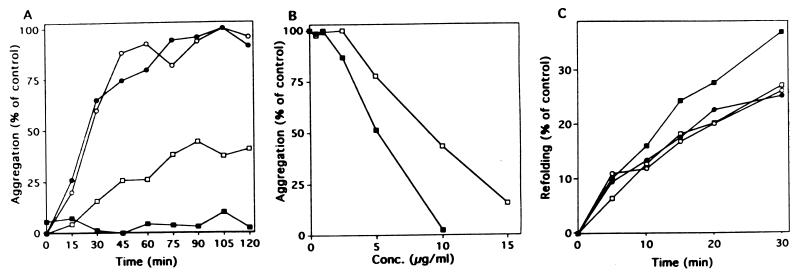
The major role of molecular chaperones in protein folding is considered to be protection of the unfolded proteins from aggregation (1, 9). To investigate whether the 58-kDa protein has the chaperone-like effect on other proteins, MDH from yeast (8) and bovine rhodanese (26) were selected as unfolded protein substrates, since both enzymes have been used as useful models for studying protein folding and the functional roles of protein dynamics (14). Gdn-HCl-denatured rhodanese and MDH were diluted into a large excess of dilution buffer containing the 58-kDa protein, GroEL, or BSA, and aggregation of rhodanese and reactivation of MDH were examined (Fig. 6). Dilution of Gdn-HCl-denatured rhodanese in this buffer resulted in its aggregation. The aggregation was efficiently suppressed when GroEL and the 58-kDa protein were present, although the 58-kDa protein (10 μg/ml) was around 60% as effective as GroEL (Fig. 6A). BSA in the buffer had no effect. The ability to prevent aggregation of rhodanese was plotted as a function of 58-kDa protein and GroEL concentrations (Fig. 6B). The 58-kDa protein had a lower activity than that of GroEL by about 40% on the basis of protein quantity. This result is comparable to that in Fig. 4 which shows that the 58-kDa protein had a little weaker activity than that of GroEL on the denatured DMSOR. Reactivation of Gdn-HCl-denatured MDH was rapid even in the absence of molecular chaperones, and the 58-kDa protein and BSA appeared to have little effect on reactivation (Fig. 6C). GroEL promoted reactivation by about 130% of that of the control.
FIG. 6.
Effects of R. sphaeroides 58-kDa protein on aggregation of rhodanese and reactivation of MDH. (A) Time course of aggregation upon dilution of Gdn-HCl-denatured rhodanese in the absence of added proteins (•) and in the presence of BSA (○), GroEL (▪), and R. sphaeroides 58-kDa protein (□) as monitored by light scattering. BSA, GroEL, and the 58-kDa protein were added at a concentration of 10 μg/ml. The concentration of rhodanese was 3.93 μg/ml. (B) Plot of the prevention of aggregation of rhodanese as a function of GroEL (▪) or R. sphaeroides 58-kDa protein (□) concentration. (C) Time course of reactivation of denatured MDH in the absence of added proteins (•) and in the presence of BSA (○), GroEL (▪), and R. sphaeroides 58-kDa protein (□). The concentration of BSA, GroEL, and the 58-kDa protein used was 10 μg/ml. The concentration of MDH was 0.9 μg/ml. At the indicated times aliquots were withdrawn and MDH activities were measured.
DISCUSSION
We have found that acid-unfolded DMSOR aggregates during incubation in the refolding buffer and that GroEL and a periplasmic protein prevent aggregation (13). This provides a good experimental system in which to identify proteins which function like molecular chaperones in the periplasm, and we have attempted to isolate such proteins. In all chromatographies, the 58-kDa protein was recovered at specific fractions and identified. It is an abundant periplasmic protein and appears to be hydrophobic with a tendency to aggregate.
The most noteworthy characteristic of the 58-kDa protein is its amino acid sequence homology with E. coli DppA. E. coli DppA has been suggested to be involved in the ability of cells to utilize and undergo chemotaxis toward dipeptides (10) and is a periplasmic dipeptide transport protein (17). The purified 58-kDa protein has almost the same molecular size as E. coli DppA. The first 14 amino-terminal amino acid residues are completely identical to those of E. coli DppA, and the internal sequences also show high degrees of similarity to those of E. coli DppA. Recently, the structure of the unbound form of E. coli DppA was determined (16). Arg355 and Tyr357 in E. coli DppA are located in the middle of a loop which has been shown to be involved in the binding site for peptides (16). These amino acid residues are also present in the internal polypeptide 2 (18 kDa) obtained from the R. sphaeroides 58-kDa protein. Thus, the purified 58-kDa protein can be called R. sphaeroides DppA.
Richarme and Caldas (19) have shown that the periplasmic-binding proteins E. coli OppA, E. coli MalE, and S. typhimurium MglB are able to behave as general molecular chaperones even in the absence of ATP. In our study, however, only DppA was isolated as the chaperone for the unfolded DMSOR from the cell. We found that DctP (23), the periplasmic dicarboxylate-binding protein purified from this phototroph, also had the activity to prevent aggregation of the unfolded DMSOR, but this activity was comparable to only that of BSA (data not shown). Therefore, all of the periplasmic substrate-binding proteins cannot have chaperone-like functions. It is likely that R. sphaeroides DppA is the only protein playing a role of molecular chaperone for at least the unfolded DMSOR in the periplasm. However, it is not clear whether R. sphaeroides DppA serves as molecular chaperone for other periplasmic proteins or if there is a specific chaperone for each of the other proteins in the periplasm. We are now isolating mutants of the dppA gene in order to clarify the role of DppA for other periplasmic proteins.
Protein disulfide isomerase (PDI), which acts as a catalyst of disulfide bond formation (5), has also recently been shown to exhibit a chaperone-like activity in the in vitro refolding of proteins (2, 18, 24). Furthermore, PDI is suggested to be involved in quality control (3, 30) and also accelerates protein folding within cells (17). Its chaperone-like activity, however, appears to be dependent on the proteins used as substrates. No significant chaperone-like activity of PDI for dihydrofolate reductase is evident, but PDI suppressed aggregation of denatured rhodanese (6). These differences indicate that proteins such as rhodanese, which are liable to aggregate and are unable to refold spontaneously, are preferable substrates for PDI, whereas proteins such as dihydrofolate reductase and RNase A, which spontaneously refold, are not (18). This also seems to be the case for R. sphaeroides DppA; R. sphaeroides DppA had little effect on the folding of MDH but effectively suppressed the aggregation of rhodanese and DMSOR. The unfolded DMSOR is liable to aggregate, and the efficiency of folding is very poor (13).
Zahn et al. (30) reported that a monomeric polypeptide of the chaperonin GroEL corresponding to residues 191 to 345 also has the activity of the tetradecamer in facilitating the refolding of rhodanese in the absence of ATP. The weaker binding of denatured rhodanese to the monomeric polypeptide is adequate for chaperonin activity and weak enough to allow the subsequent refolding of rhodanese (3). R. sphaeroides DppA may show activity similar to that of this fragment of GroEL.
ACKNOWLEDGMENTS
We are grateful to T. Hiyama (Saitama University, Urawa, Japan) for his valuable advice.
This work was partly supported by a grant-in-aid from the Ministry of Education, Science and Culture, Japan (no. 8640830). M. Matsuzaki was the recipient of a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists.
REFERENCES
- 1.Buchner J, Schmidt M, Fuchs M, Jaenicke R, Rudolph R, Schmid F X, Kiefhaber T. GroE facilitates refolding of citrate synthase by suppressing aggregation. Biochemistry. 1991;30:1586–1591. doi: 10.1021/bi00220a020. [DOI] [PubMed] [Google Scholar]
- 2.Cai H, Wang C C, Tsou C L. Chaperone-like activity of protein disulfide isomerase in the refolding of a protein with no disulfide bonds. J Biol Chem. 1994;269:24550–24552. [PubMed] [Google Scholar]
- 3.Corrales F J, Fersht A R. Toward a mechanism for GroEL/GroES chaperone activity: an ATPase-gated and -pulsed folding and annealing cage. Proc Natl Acad Sci USA. 1996;93:4509–4512. doi: 10.1073/pnas.93.9.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fenton W A, Horwich A L. GroEL-mediated protein folding. Protein Sci. 1997;6:743–760. doi: 10.1002/pro.5560060401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goldberger R F, Epstein C J, Anfinsen C B. Acceleration of reactivation of reduced bovine pancreatic ribonuclease by a microsomal system from rat liver. J Biol Chem. 1963;238:628–635. [PubMed] [Google Scholar]
- 6.Hayano T, Hirose M, Kikuchi M. Protein disulfide isomerase mutant lacking its isomerase activity accelerates protein folding in the cell. FEBS Lett. 1995;377:505–511. doi: 10.1016/0014-5793(95)01410-1. [DOI] [PubMed] [Google Scholar]
- 7.Hultgren S J, Norwack S, Abgaham S N. Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu Rev Microbiol. 1991;45:363–415. doi: 10.1146/annurev.mi.45.100191.002123. [DOI] [PubMed] [Google Scholar]
- 8.Kawata Y, Nosaka M, Hongo K, Mizobata T, Nagai J. Chaperonin GroE and ADP facilitate the folding of various proteins and protect against heat inactivation. FEBS Lett. 1994;345:229–232. doi: 10.1016/0014-5793(94)00456-0. [DOI] [PubMed] [Google Scholar]
- 9.Kiefhaber T, Rudolph R, Kohler H-H, Buchner J. Protein aggregation in vitro and in vivo: a quantitative model of the kinetic competition between folding and aggregation. Bio/Technology. 1991;9:825–829. doi: 10.1038/nbt0991-825. [DOI] [PubMed] [Google Scholar]
- 10.Manson M D, Blank V, Brade G. Peptide chemotaxis in E. coli involves the Tap signal transducer and the dipeptide permease. Nature. 1986;321:253–256. doi: 10.1038/321253a0. [DOI] [PubMed] [Google Scholar]
- 11.Masui H, Satoh M, Satoh T. Secretion of both partially unfolded and folded apoproteins of dimethyl sulfoxide reductase by spheroplasts from a molybdenum cofactor-deficient mutant of Rhodobacter sphaeroides f. sp. denitrificans. J Bacteriol. 1994;176:1624–1629. doi: 10.1128/jb.176.6.1624-1629.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matsuzaki M, Satoh T. Detection of a compact folding intermediate of dimethyl sulfoxide reductase secreted from a molybdenum cofactor-deficient mutant of Rhodobacter sphaeroides f. sp. denitrificans. Plant Cell Physiol. 1997;38:1286–1290. [Google Scholar]
- 13.Matsuzaki M, Yamaguchi Y, Masui H, Satoh T. Stabilization by GroEL, a molecular chaperone, and a periplasmic fraction, as well as refolding in the presence of dithiothreitol, of acid-unfolded dimethyl sulfoxide reductase, a periplasmic protein of Rhodobacter sphaeroides f. sp. denitrificans. Plant Cell Physiol. 1996;37:333–339. [Google Scholar]
- 14.Mendoza J A, Lorimer G H, Horowitz P M. Chaperonin cpn60 from Escherichia coli protects the mitochondrial enzyme rhodanese against heat inactivation and supports folding at elevated temperatures. J Biol Chem. 1992;267:17631–17634. [PubMed] [Google Scholar]
- 15.Missiakas D, Raina S. Protein folding in the bacterial periplasm. J Bacteriol. 1997;179:2465–2471. doi: 10.1128/jb.179.8.2465-2471.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nickitenko A V, Trakhonov S, Quiocho F A. 2 Å resolution structure of DppA, a periplasmic dipeptide transport/chemosensory receptor. Biochemistry. 1995;34:16585–16595. doi: 10.1021/bi00051a006. [DOI] [PubMed] [Google Scholar]
- 17.Olson E R, Dunyak D S, Jurss L M, Poorman R A. Identification and characterization of dppA, an Escherichia coli gene encoding a periplasmic dipeptide transport protein. J Bacteriol. 1991;173:234–244. doi: 10.1128/jb.173.1.234-244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Puig A, Gilbert H F. Protein disulfide isomerase exhibits chaperone and anti-chaperone activity in the oxidative refolding of lysozyme. J Biol Chem. 1994;269:7764–7771. [PubMed] [Google Scholar]
- 19.Richarme G, Caldas T D. Chaperone properties of the bacterial periplasmic substrate-binding proteins. J Biol Chem. 1997;272:15607–15612. doi: 10.1074/jbc.272.25.15607. [DOI] [PubMed] [Google Scholar]
- 20.Satoh T, Hoshino Y, Kitamura H. Rhodopseudomonas sphaeroides f. sp. denitrificans, a denitrifying strain as a subspecies of Rhodopseudomonas sphaeroides. Arch Microbiol. 1976;108:265–269. doi: 10.1007/BF00454851. [DOI] [PubMed] [Google Scholar]
- 21.Satoh T, Kurihara F N. Purification and properties of dimethylsulfoxide reductase containing a molybdenum cofactor from a photodenitrifier, Rhodopseudomonas sphaeroides f. sp. denitrificans. J Biochem. 1987;102:191–197. doi: 10.1093/oxfordjournals.jbchem.a122032. [DOI] [PubMed] [Google Scholar]
- 22.Schindelin H, Kisker C, Hilton J, Rajagopalan K V, Rees D C. Crystal structure of DMSO reductase: redox-linked changes in molybdopterin coordination. Science. 1996;272:1615–1621. doi: 10.1126/science.272.5268.1615. [DOI] [PubMed] [Google Scholar]
- 23.Shaw J G, Hamblin M J, Kekky D J. Purification, characterization and nucleotide sequence of the periplasmic C4-dicarboxylate-binding protein (DctP) from Rhodobacter capsulatus. Mol Microbiol. 1991;5:3055–3062. doi: 10.1111/j.1365-2958.1991.tb01865.x. [DOI] [PubMed] [Google Scholar]
- 24.Song J, Wang C. Chaperone-like activity of protein disulfide-isomerase in the refolding of rhodanese. Eur J Biochem. 1995;231:312–316. doi: 10.1111/j.1432-1033.1995.tb20702.x. [DOI] [PubMed] [Google Scholar]
- 25.Taguchi H, Konishi J, Ishii N, Yoshida M. A chaperonin from a thermophilic bacterium, Thermus thermophilus, that controls refoldings of several thermophilic enzymes. J Biol Chem. 1991;266:22411–22418. [PubMed] [Google Scholar]
- 26.Taguchi H, Makino Y, Yoshida M. Monomeric chaperonin-60 and its 50-kDa fragment possess the ability to interact with non-native proteins, to suppress aggregation, and to promote protein folding. J Biol Chem. 1994;269:8529–8534. [PubMed] [Google Scholar]
- 27.Wülfing C, Plückthun A. Protein folding in the periplasm of Escherichia coli. Mol Microbiol. 1994;12:685–692. doi: 10.1111/j.1365-2958.1994.tb01056.x. [DOI] [PubMed] [Google Scholar]
- 28.Yamamoto I, Wada N, Ujiiye T, Tachibana M, Matsuzaki M, Kajiwara H, Watanabe Y, Hirano H, Okubo A, Satoh T, Yamazaki S. Cloning and nucleotide sequence of the gene encoding dimethyl sulfoxide reductase from Rhodobacter sphaeroides f. sp. denitrificans. Biosci Biotech Biochem. 1995;59:1850–1855. doi: 10.1271/bbb.59.1850. [DOI] [PubMed] [Google Scholar]
- 29.Yoshida Y, Takai M, Satoh T, Takami S. Molybdenum requirement for translocation of dimethyl sulfoxide reductase to the periplasmic space in a photodenitrifier, Rhodobacter sphaeroides f. sp. denitrificans. J Bacteriol. 1991;173:3277–3281. doi: 10.1128/jb.173.11.3277-3281.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zahn R, Buckle A M, Perrett S, Johnson C M, Corrales F J, Golbik R, Fersht A R. Chaperone activity and structure of monomeric polypeptide binding domains of GroEL. Proc Natl Acad Sci USA. 1996;93:15024–15029. doi: 10.1073/pnas.93.26.15024. [DOI] [PMC free article] [PubMed] [Google Scholar]