Abstract
Rotation of the bacterial flagellar motor is powered by a transmembrane gradient of protons or, in some species, sodium ions. The molecular mechanism of coupling between ion flow and motor rotation is not understood. The proteins most closely involved in motor rotation are MotA, MotB, and FliG. MotA and MotB are transmembrane proteins that function in transmembrane proton conduction and that are believed to form the stator. FliG is a soluble protein located on the cytoplasmic face of the rotor. Two other proteins, FliM and FliN, are known to bind to FliG and have also been suggested to be involved to some extent in torque generation. Proton (or sodium)-binding sites in the motor are likely to be important to its function and might be formed from the side chains of acidic residues. To investigate the role of acidic residues in the function of the flagellar motor, we mutated each of the conserved acidic residues in the five proteins that have been suggested to be involved in torque generation and measured the effects on motility. None of the conserved acidic residues of MotA, FliG, FliM, or FliN proved essential for torque generation. An acidic residue at position 32 of MotB did prove essential. Of 15 different substitutions studied at this position, only the conservative-replacement D32E mutant retained any function. Previous studies, together with additional data presented here, indicate that the proteins involved in motor rotation do not contain any conserved basic residues that are critical for motor rotation per se. We propose that Asp 32 of MotB functions as a proton-binding site in the bacterial flagellar motor and that no other conserved, protonatable residues function in this capacity.
Many bacteria swim by rotating flagella, each comprising a helical propeller, a flexible hook that functions as universal joint, and a rotary motor in the cell membrane (for recent reviews, see references 1, 2, 32, 33, and 40). The energy for rotation of the flagellar motors comes from the proton gradient across the cytoplasmic membrane (19, 29, 34) or, in some alkaliphilic or marine species, the sodium ion gradient (22). The flagellar motor thus converts electrochemical energy stored in an ion gradient into the mechanical work of rotation. The mechanism of this energy conversion is not understood at the molecular level.
About 50 proteins are needed for the assembly and function of bacterial flagella (32, 33). Most of these function in flagellar assembly or in controlling clockwise (CW)/counterclockwise (CCW) switching. Genetic studies have shown that only five proteins, MotA, MotB, FliG, FliM, and FliN, can be mutated to give a Mot− (immotile) phenotype, in which flagella appear superficially normal but do not rotate (13, 21, 43, 57). These five proteins have been suggested to function, to various extents, in torque generation.
MotA and MotB are integral membrane proteins (9, 11, 45, 54, 55, 60) that together function as a proton channel (4, 16–18, 46). Each motor contains several MotA-MotB complexes, which are arranged circumferentially around the base of each flagellum (25) and function independently to generate torque (3, 7). MotB has a large periplasmic domain (9) that contains a sequence motif believed to bind peptidoglycan (12). MotB has therefore been suggested to function as an anchor to secure the nonrotating parts of the motor, or stator, to the cell body (9, 12). FliG, FliM, and FliN are cytoplasmic proteins (27). They bind to each other in all pairwise combinations (36, 50) and function in a complex, called the switch complex, that is essential for flagellar assembly, rotation, and CW/CCW direction control (53, 57). Ultrastructural studies (14, 15, 24, 26, 58, 59), gene fusion studies (14, 28), protein binding studies (35, 36, 38, 50), and physiological studies (48, 49) suggest that the switch complex proteins are located on the rotor. Among the proteins of the rotor, FliG is the one most extensively involved in torque generation, and mutational studies indicate that a domain near its C terminus is especially important (23, 30, 31). Torque generation is thus likely to occur at an interface between cytoplasmic domains of the MotA-MotB complexes (the stator) and the C-terminal domain of FliG (the rotor).
Charged residues are important for torque generation in the flagellar motor. Recent mutational studies identified functionally important charged residues in both FliG (31) and MotA (61). No single charged residue proved critical for motor function, however. Each could be mutated to a neutral residue, and good motility was preserved. It was suggested that multiple charged residues of each protein function jointly and, to a large extent, redundantly in torque generation (31, 61). Subsequent study showed that the charged residues of FliG interact with those of MotA (62).
Because the flagellar motor is driven by protons (or sodium ions), one or more proton- or sodium-binding sites are probably important to its function. Such sites are most likely formed from acidic groups, because, other factors being equal, acidic groups could transfer protons more rapidly than basic groups. Asp 32 of MotB is a good candidate for such an acidic ion-binding site. This residue is conserved in all MotB sequences presently known. Systematic mutational studies of the membrane-spanning segments of MotA and MotB suggested a structural model for the MotA-MotB complex in which Asp 32 of MotB is located inside the proton channel, near its cytoplasmic end (41, 42). In a random-mutational study of MotB, the conservative mutation D32N was found to abolish motor function (6).
To examine more systematically the role of acidic residues in the function of the flagellar motor, we mutated each of the conserved acidic residues in the five proteins that have been suggested to be involved in torque generation and measured the effects on motility. Additional substitutions of Asp 32 were also characterized, to determine the requirements for function at this position. The results, together with the results from previous mutational studies, suggest that Asp 32 of MotB, and no other acidic residue, has an essential role in proton transfer through the flagellar motor.
MATERIALS AND METHODS
Strains, plasmids, and mutagenesis.
fliG, fliM, and fliN null strains have been described previously (30, 48, 49). The motB strain was RP3087, a gift from J. S. Parkinson (University of Utah). The function of the mutant FliG, FliM, FliN, or MotB proteins was tested by transforming plasmids encoding the mutant proteins into corresponding tester strains and measuring rates of swarming in soft agar. Mutations of acidic residues of MotA and their effects on function were described previously (61).
Plasmid purification and DNA manipulations followed standard methods (39). Site-directed mutagenesis was carried out according to the Altered Sites procedure (Promega, Madison, Wis.). Mutations were confirmed by dideoxy sequencing with reagents from U.S. Biochemical Corp. (Cleveland, Ohio). Mutations in fliG, fliM, and fliN were made in plasmids pSL27 (30), pHT41 (48), and pLS4 (49), respectively, which are all derivatives of pAlter-1 (Promega). pSL27 and pHT41 were also used to express the mutant FliG and FliM proteins for assays of function. For assays of function of the mutant FliN proteins, the fliN mutations were transferred to plasmid pHT39 (30), which allowed controlled expression from the tac promoter. Mutations in motB were made in plasmid pRF4 (a gift from Robert Fazzio, University of Utah), a derivative of pAlter-1 that encodes motA and motB. For tests of motility, motB mutations were transferred to pGM1, which expresses motB from the lac promoter (8). Wild-type pGM1 complemented a motB defect well in the absence of the inducer isopropyl-β-d-thiogalactopyranoside (IPTG), so no IPTG was added for these functional tests. For tests of dominance, motB mutations were transferred to plasmid pJZ18, a derivative of pTM30 (37) that expresses motB from the tac promoter, and the resulting plasmids were transformed into the wild-type strain, RP437 (a gift from J. S. Parkinson). The motility of these strains, relative to that of a control strain expressing wild-type MotB from the same plasmid, was measured at a range of IPTG concentrations. The rates reported are for 80 μM IPTG. For tests of growth impairment, motB mutations were transferred to plasmid pLW3, which overexpresses MotA and a fusion protein containing the first 60 residues of MotB, both from the trp promoter (46, 54). Random mutagenesis of residue 32 of MotB was carried out by using the Altered Sites procedure with an oligonucleotide randomized at the appropriate codon.
Sequence alignment.
Protein sequences were aligned by using the MegAlign program, a part of the package Lasergene (DNAstar, Inc., Madison, Wis.) on a Macintosh computer. Because either aspartic acid or glutamic acid could function in rapid proton transfer, a position was treated as conserved if either Asp or Glu was always present there. Protein sequences were obtained from GenBank. The FliG sequences used were from Escherichia coli, Agrobacterium tumefaciens, Borrelia burgdorferi, Bacillus subtilis, Caulobacter crescentus, and Treponema denticola. The FliG sequence from R. sphaeroides became available after mutations had been made and characterized for four acidic residues that are not conserved in that species but are conserved in all of the others. Data for these residues are included below (Table 1), even though they should provisionally be considered not conserved. FliM sequences were from E. coli, A. tumefaciens, B. subtilis, C. crescentus, and B. burgdorferi. FliN sequences were from E. coli, A. tumefaciens, B. subtilis, C. crescentus, Treponema pallidum, B. burgdorferi, and Pseudomonas aeruginosa. MotA sequences were from E. coli, B. subtilis, Vibrio parahaemolyticus, Bacillus megaterium, Treponema phagedenis (partial sequence only), Rhodobacter sphaeroides, and B. burgdorferi. MotB sequences were from E. coli, B. subtilis, R. sphaeroides, B. megaterium, B. burgdorferi, and Rhizobium meliloti. In some cases, additional protein sequences are available but are not noted here because they did not affect the list of conserved acidic residues.
TABLE 1.
Effects of mutations in conserved acidic residues of FliN, FliM, FliG, and MotA
| Protein and mutation | Swarming rate (concn of IPTG)a |
|---|---|
| FliN | |
| Wild type | 1.00 (10 μM) |
| D55Ab/D59A | 1.21 (50 μM) |
| D90A | 1.23 (25 μM) |
| D95A/D98A | 1.20 (50 μM) |
| E110A | 1.08 (25 μM) |
| D116A | 0.80 (25 μM) |
| D124A | 1.24 (25 μM) |
| FliM | |
| E91A | 1.17 |
| D128A | 1.32 |
| E150A | 0.10 |
| D282A | 1.32 |
| FliG | |
| E33A | 0.89 |
| E58A | 1.00 |
| E108A | 0.85 |
| E125A | 0.0 |
| E201Ac | 0.62 |
| E224Ad | 1.05 |
| E237Ad | 1.16 |
| E272Ac,d | 0.94 |
| D288Ad,e | 0.86 |
| D289Ad | 0.92 |
| E302Ad | 0.94 |
| E320Ac,d,e | 1.04 |
| E327Ad,e | 0.83 |
| D328Ac,d,e | 1.02 |
| MotA | |
| E98Qf | 0.84 |
| E150Qf | 1.12 |
Swarming rates are of strains with chromosomal defects in the genes of interest which express the mutant proteins from plasmids. Rates are relative to control strains which express the wild-type proteins from the plasmids.
Initial alignments indicated that Asp 55 of FliN was conserved, but this proved untrue as more sequences became available.
Not conserved in the reported sequence from R. sphaeroides.
From reference 31. Residue numbers were lower by 2 in that study because of errors in the E. coli fliG sequence initially reported.
Not conserved in A. tumefaciens, in a segment that shows wide divergence from all the other species. Designated “semiconserved” in a previous study (31).
From reference 61.
Assays of motor function, flagellation, and growth impairment.
For assays of swarming motility, plasmids encoding mutant proteins or wild-type control plasmids were transformed into the respective null strains. Overnight cultures of transformants were grown with shaking at 32°C in tryptone broth (1% Bacto-tryptone, 0.5% NaCl) containing ampicillin (100 μg/ml). One microliter of each saturated culture was spotted onto swarm plates containing tryptone broth, 0.28% agar, and ampicillin (100 μg/ml), and the plates were incubated at 32°C. Swarm size was measured at regular time intervals, and plots of diameter versus time were fitted to a line. Swarming rates relative to wild-type controls on the same plates are reported. Swarming of FliN mutants was measured in the same way, except an IPTG-inducible plasmid was used, and swarming rates were measured at several different levels of induction. For both the wild type and the fliN mutants, the swarming rates showed a maximum at a particular IPTG concentration (data not shown). The IPTG concentrations that gave the fastest swarming differed among the mutants and were higher for the mutants than for the wild type. The maximum swarming rate observed for each FliN mutant, relative to the maximum rate observed for the wild type, is reported. For the mutants that failed to swarm at all, swimming motility in liquid medium was also scored, by phase-contrast microscopy. MotB Asp 32 mutants were also studied by tethering cells to coverslips by their flagellar filaments, as described previously (48).
To assay flagellation, cells were stained for flagella by a wet-mount procedure (20). Fields of well-stained cells were located, and the flagella on 50 cells were counted, as described previously (48).
Growth impairments of cells overexpressing MotA together with a fusion protein containing the first 60 residues of MotB were measured essentially as described previously (4). Wild-type cells (strain RP437) were transformed with plasmid pLW3 or with pLW3 variants containing mutations in motB codon 32. Fresh transformants were cultured overnight at 34°C in LB-Ap medium (1% Bacto-tryptone, 0.5% NaCl, 0.5% yeast extract, 100 μg of ampicillin per ml), diluted 100-fold into fresh LB-Ap medium, cultured for 30 min at 34°C, and then induced with 100 μg of indoleacrylic acid (IAA) per ml added as a 10-mg/ml stock in ethanol. Uninduced controls contained just the ethanol. Growth was monitored by measurements of A600 at regular time intervals (typically 30 min). Logarithms of A600 values were plotted versus time, and the linear portion of each growth curve was fitted to a line.
Anti-MotB antibody and immunoblotting.
Antibody was raised against a fusion protein containing residues 1 to 291 of MotB fused to oligohistidine. The fusion protein was expressed, along with the wild-type MotA protein, from plasmid pTB9, which uses the T7 promoter. pTB9 is like the previously described plasmid pTB1 (50) but encodes kanamycin resistance instead of ampicillin resistance. Plasmid pTB1 is a derivative of pET21b (Novagen, Madison, Wis.). E. coli BL21(De3) cells (47) containing plasmid pTB9 were cultured at 37°C in medium (typically a volume of 3 liters) containing 1% Bacto-tryptone, 0.5% yeast extract, 1% NaCl, and 40 μg of kanamycin per ml to an optical density at 600 nm of 1.0 and then were induced with 100 μM IPTG and grown for an additional 4 to 5 h. Cells were pelleted, resuspended in 30 ml of binding buffer (20 mM Tris [pH 7.9], 0.5 M NaCl, 5 mM imidazole) containing 0.6 mg of lysozyme per ml, and incubated on ice for 1 h. Cells were lysed by sonication (Branson model 450, power level of 5, duty cycle of 50%, 2 to 3 min) and by cycles of freezing (−70°C) and thawing (room temperature). Membranes were collected by centrifugation (17,500 × g, 45 min) and resuspended by sonication in binding buffer containing 8 M urea and 2% sodium dodecyl sulfate (SDS). Complete dissociation of MotB from MotA required boiling for 30 min. Insoluble material was removed by brief centrifugation at room temperature. The supernatant was applied to a column containing His-bind resin (Novagen), and the column was washed with ca. 100 volumes of binding buffer containing 6 M urea and 0.5% SDS. The protein was refolded on the column by washing with binding buffer containing 0.25% Triton X-100 and decreasing concentrations of urea and then was washed further with binding buffer containing increased imidazole (30 mM) and 0.25% Triton X-100. The fusion protein was eluted with binding buffer containing 400 mM imidazole and 0.25% Triton X-100. A principal remaining contaminant was removed by diluting the sample 20-fold in water and then precipitating the MotB-His fusion protein with 30% ammonium sulfate.
The purified fusion protein was used to raise antiserum in rabbits (Covance, Denver, Pa.). The serum was purified by preabsorption against proteins from a motAB deletion strain (RP6894; a gift from J. S. Parkinson), as described previously (48). To quantify mutant MotB proteins, derivatives of plasmid pGM1 encoding the mutant proteins were transformed into either the wild-type strain, RP437, or the motAB deletion strain, RP6894, and cells were cultured overnight at 34°C in Luria-Bertani broth (5 ml) under ampicillin selection (100 μg/ml). Cells were collected by centrifugation (2,000 × g, 10 min), resuspended in spheroplast buffer (50 mM Tris [pH 8], 10 mM EDTA, 0.5 M sucrose, 200 μg of lysozyme per ml), and then sonicated. Membranes were collected by centrifugation (16,000 × g, 15 min). Membranes were resuspended in gel loading buffer (4% SDS, 10% 2-mercaptoethanol, 20% glycerol, 0.125 M Tris-Cl [pH 6.8]), and equal amounts (1/50 of the total) were loaded onto 15% polyacrylamide gels. Gels were blotted onto nitrocellulose membranes, and immunoblotting was carried out according to the enhanced chemiluminescence procedure (Amersham, Arlington Heights, Ill.).
RESULTS
Mutations of conserved acidic residues of FliN, FliM, and FliG.
Protein sequence alignments (not shown) showed that FliN has seven conserved acidic residues, FliM has four, FliG has eight (and several more that are conserved in all species except R. sphaeroides or A. tumefaciens, and that are included in this study), and MotA has two. These residues are listed in Table 1. Previous mutational studies examined the effects of replacing the conserved acidic residues in MotA (61) and in the C-terminal domain of FliG that is believed to function most directly in torque generation (31). The present study focused on FliN, FliM, the part of FliG that was not examined in the previous study, and MotB.
The conserved acidic residues were changed to alanine, in most cases one at a time, by site-directed mutagenesis of plasmid-borne genes. The mutant plasmids were used to transform tester strains defective in the corresponding proteins, and the motility of the transformants was assessed by measuring rates of swarming in soft agar. The results are summarized in Table 1.
None of the conserved acidic residues of FliN proved essential for function. Each could be replaced by alanine without seriously affecting swarming motility. FliN was expressed from an IPTG-inducible plasmid for these experiments. For wild-type FliN, swarming was fastest at an induction level of 10 μM IPTG, whereas for the mutants it peaked at higher levels of induction, ranging from 25 to 50 μM IPTG. Thus, although the mutant FliN proteins could all support good motility, this required protein levels somewhat higher than those for the wild-type protein.
FliM contains four conserved acidic residues. Three of these could be replaced by alanine with no ill effects on swarming motility (the swarming rate actually increased significantly in two cases). One mutation, E150A, reduced swarming to about 10% of that of the wild type. Under the microscope, cells of this mutant were observed to swim as fast as wild-type cells, but very smoothly, with no clear episodes of tumbling. The swarming defect of this mutant is therefore due to a defect in CW/CCW motor bias, rather than in torque generation.
The N-terminal part of FliG that was not examined in the previous site-directed mutational study contains five conserved acidic residues. Alanine replacements at four of these positions had mild effects, reducing swarming rates by no more than 40%. One FliG mutation, E125A, abolished swarming. Cells of this mutant appeared immotile when observed under the microscope. The FliG protein is essential for flagellar assembly as well as for motor rotation (56). To determine whether the E125A mutation of FliG affected flagellar assembly, cells were stained for flagella (20). The cells were nonflagellate (no flagella were observed on 50 cells, compared to an average of 5 flagella per cell in a wild-type control). Residue Glu 125 of FliG is thus important for flagellar assembly and is probably not important for torque generation per se.
MotA has two conserved acidic residues. A previous study showed that each could be mutated to a neutral residue with little effect on function (61).
Mutations of residue Asp 32 of MotB.
MotB has a single conserved acidic residue, Asp 32, in the E. coli protein. A conservative mutation of this residue (D32N) abolished motility (6), suggesting that it might have a key role in motor function. To learn more about the requirements for function at residue 32 of MotB, additional substitutions at this position were isolated by localized mutagenesis. Mutagenized plasmids were transformed into a motB-defective strain, and the transformants were plated in soft agar. Both swarming and nonswarming colonies were picked, and plasmid DNA was isolated and sequenced. Mutant plasmids were retransformed into the motB strain, and swarming rates were measured.
Twenty swarming clones were isolated and sequenced; 14 were wild type at codon 32, and 6 were D32E mutants. Approximately 60 nonswarming clones were isolated and sequenced. These contained genes that encoded a total of 15 different replacements at position 32 of MotB (Table 2), including D32E. When the mutants were retested on swarm plates, all of them failed to swarm, except the D32E mutant, which swarmed very slowly (at less than 10% of the wild-type rate [Table 2]). When liquid-grown cells were viewed under the microscope, all of the mutants, including D32E, appeared immotile. If cells of the D32E mutant were allowed to settle on the microscope slide for several minutes, a few slowly tumbling cells could be observed among the population of cells that remained in suspension. The mutants were next tethered to coverslips to examine the performance of individual motors at high load. Cells of the D32E mutant rotated more slowly than wild-type cells, with the motors producing approximately half the normal torque (mean torque ± standard deviation for 30 cells of the mutant, [4.1 ± 1.4] × 10−12 dyne-cm; mean torque ± standard deviation for 33 cells of the wild type, [8.4 ± 3.4] × 10−12 dyne-cm). Cells of the other 14 mutants did not rotate.
TABLE 2.
Effects of replacements in residue Asp 32 of MotB
| Replacement | Swarming ratea | Dominanceb | No. of flagella/cellc |
|---|---|---|---|
| None (wild type) | 1.0 | 1.0 | 3.0 |
| Ala | 0 | 0.06 | 2.3 |
| Arg | 0 | 0.20 | 2.4 |
| Asn | 0 | 0.05 | 2.4 |
| Gln | 0 | 0.04 | 2.3 |
| Glu | 0.08 | 0.07 | 2.1 |
| Gly | 0 | 0.07 | 2.2 |
| His | 0 | 0.06 | 2.0 |
| Ile | 0 | 0.06 | 2.1 |
| Leu | 0 | 0.04 | 2.1 |
| Lys | 0 | 0.26 | 2.1 |
| Pro | 0 | 0.05 | 2.1 |
| Ser | 0 | 0.05 | 2.0 |
| Thr | 0 | 0.06 | 2.0 |
| Trp | 0 | 0.05 | 2.0 |
| Tyr | 0 | 0.05 | 2.1 |
Swarming rates are of the motB strain RP3087 expressing the mutant MotB proteins from plasmid pGM1 and are relative to that of a control strain expressing wild-type MotB from the plasmid.
Swarming rates of wild-type cells expressing the mutant MotB proteins from plasmid pJZ18 relative to that of control cells expressing wild-type MotB from the plasmid. Similar results were observed when the proteins were expressed from plasmid pGM1.
Average value for 50 cells. The strain was RP3087, with the mutant MotB proteins expressed from plasmid pGM1.
Although null mutants of motB are immotile, they assemble flagella that are outwardly normal, so the replacements of Asp 32 in MotB most likely affect motor rotation rather than flagellar assembly. To confirm this, the mutant cells were stained and their flagella were counted. All of the Asp 32 mutants were well flagellated (Table 2), implying that the mutations affect primarily torque generation and not flagellar assembly.
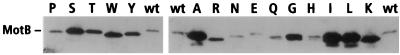
To determine whether the Asp 32 mutations disrupted expression or folding of the proteins, levels of the mutant proteins in saturated cultures were estimated by immunoblotting with a polyclonal anti-MotB antibody. MotB is believed to be stabilized by association with MotA, because when it is overexpressed, it accumulates to high levels only if MotA is also overexpressed (55). MotB levels were therefore tested both in a wild-type strain and in a motAB deletion strain. The results of experiments with the motAB deletion strain are shown in Fig. 1. All of the mutant MotB proteins were observed on immunoblots, in many cases at levels higher than those of the wild-type protein. MotB protein levels were not sensitive to the presence or absence of MotA in the cells, because similar results were obtained when the experiment used a wild-type host strain (not shown). Interestingly, the mutations of Asp 32 altered the mobility of the proteins in SDS-polyacrylamide gels. The wild-type and D32E mutant proteins migrated significantly slower than the others, and the D32R, D32W, and D32K proteins migrated faster.
FIG. 1.
Levels of mutant MotB proteins in cells. Membranes from cells expressing the mutant MotB proteins were collected, treated with loading buffer, and electrophoresed. Proteins were blotted and probed with polyclonal anti-MotB antibody, as described in Materials and Methods. Replacements of Asp 32 are indicated above the lanes according to the standard single-letter code for the amino acids. wt, wild type.
If mutant MotB proteins can accumulate in the cells and can still associate with other flagellar proteins (MotB is known to bind to MotA [50]), then they might have dominant-negative effects when expressed in wild-type cells. To test this, the mutant MotB proteins were expressed in wild-type cells, by using derivatives of plasmid pJZ18 (Materials and Methods). Swarming rates were measured and compared to that of a control expressing wild-type MotB from the plasmid. All of the mutant MotB proteins inhibited the motility of wild-type cells (Table 2). Most reduced swarming to about 5% of control values; the two positively charged replacements D32K and D32R were slightly less dominant than the others, reducing swarming to about 20% of the control values.
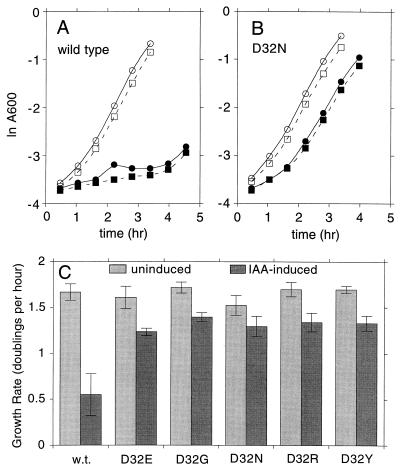
When MotA is overexpressed together with a fusion protein containing the first 60 residues of MotB, the growth rate of cells is reduced significantly (4, 5, 46, 54). This effect is correlated with an increase in the proton permeability of the cell membrane (4). The growth defect can thus be used to assay proton conduction by the MotA-MotB fusion protein complexes, apart from other components of the flagellar motor. A number of mutations in the membrane segments of MotA and MotB have been shown to abolish the growth impairment, presumably because they prevent proton flow through the channel (4–6, 46). Mutations of Asp 32 have not previously been examined in this assay. To determine whether replacements of Asp 32 affect proton conduction as assayed by the growth impairment, selected motB mutations were transferred to plasmid pLW3 (46, 54), which is inducible with IAA, and the growth rates of cells harboring the mutant plasmids were measured. The growth of cells expressing MotA and the wild-type MotB fusion protein was slowed about threefold upon induction with IAA. In contrast, cells overexpressing MotA and MotB fusion proteins with Asp 32 replacements D32E, D32G, D32N, D32R, and D32Y showed much smaller (ca. 20%) growth impairments (Fig. 2).
FIG. 2.
Effect of mutations in Asp 32 of MotB on overexpression-linked growth impairment. MotA and a fusion protein that contains the first 60 residues of MotB (and thus residue 32 and the membrane-spanning segment) were overexpressed from plasmid pLW3 (46, 54), which is inducible with IAA. Growth rates of uninduced cultures and cultures induced with 100 μg of IAA per ml were measured as described in Materials and Methods. (A) Representative duplicate growth curves for the wild type (measured by A600). Open symbols, uninduced; solid symbols, induced with IAA. (B) Representative duplicate growth curves for the D32N mutant. Symbols are as in panel A. (C) Summary of growth rates for the wild type (w.t.) and five Asp 32-replacement mutants. Values are means ± standard deviations (n = 6).
DISCUSSION
To probe the function of acidic residues in the flagellar motor, we mutated to alanine each of the conserved (and also some semiconserved) acidic residues in the five proteins that have been suggested to be involved in torque generation. Of 27 such residues, 26 are not critical for torque generation per se. These include the acidic residues of FliG (Asp 288 and Asp 289) and MotA (Glu 98) shown previously to mediate functionally important electrostatic interactions between the rotor and stator (31, 61, 62). Although those residues jointly are important for function, mutant phenotypes indicate that no single one is critical.
One conserved acidic residue of FliG, Glu 125, proved essential for flagellar assembly. The large effect of the E125A replacement on flagellar assembly is notable, because in an extensive mutational study of FliG, only two point mutations that affected flagellar assembly, R154P and A266D, were found (23). The E125A mutation might disrupt the binding of FliG to another flagellar protein. Known binding partners of FliG include FliF (14, 38), FliM (35, 36, 50), FliN (49, 50), and other copies of itself (50). Mutations of FliG residues I129 and T132, which might be near Glu 125, affected the interaction of FliG with FliM, as measured by the two-hybrid system (35).
One conserved acidic residue of FliM, Glu 150, proved essential for proper CW/CCW motor bias. This residue is in a segment of FliM shown by previous mutational studies to be important in the control of CW/CCW switching (44, 52). Other mutations in conserved acidic residues of FliM actually increased swarming rates. FliM was moderately overexpressed in these experiments, and excess FliM is known to impair motility (10), chiefly by sequestering FliN (49). The enhanced swarming of the D128A and D282A mutants might therefore be due to a weakened interaction with FliN. We have examined the binding of FliM fragments to FliN by using coprecipitation assays and have found that a C-terminal segment of FliM, which includes residue 282, binds to FliN (48a).
In contrast to all other acidic residues examined, Asp 32 of MotB is critical for the function of the flagellar motor. The most important feature of this residue appears to be either a negative side-chain charge or the ability to bind a proton, because among the 15 replacements studied, only the conservative replacement D32E permitted any function. The corresponding mutation in S. typhimurium (D33E) was recently described and also permitted partial function in that species (51). Asp 32 functions specifically in motor rotation, not flagellar assembly, because the mutants were well flagellated. The mutant MotB proteins do not appear to be grossly misfolded, because they accumulated to levels at least as high as those of the wild-type protein and had strong dominant-negative effects when expressed in wild-type cells. The dominant-negative effects suggest that the mutant proteins retain the ability to be installed in the motors and/or the ability to bind to another flagellar component, most likely MotA (50).
Mutations in MotB residue Asp 32 affected not only motor rotation but also proton conduction, as assayed by growth impairments (Fig. 2). This behavior is not unique to mutations of Asp 32. Fourteen different mutations in membrane segments of MotA (4, 5) and two in or near the membrane segment of MotB (6, 46) were previously shown to abolish both motility and the overexpression-linked growth impairment. Certain mutations of MotA residues Pro 173 and Pro 222, which are conserved and are located near the ends of membrane segments, also abolish both motility and the growth impairment (61). These previously characterized mutations are not conservative, however, but involve either the replacement of proline residues that are likely to be important for overall conformation or the introduction of side chains that are larger than those of the wild type and that are, in many cases, branched at the β-carbon or charged (4–6, 46). In contrast, certain replacements of Asp 32 that abolish motility and the growth impairment are conservative (D32N) or are smaller (D32G) than the wild type.
Systematic mutational studies of the membrane segments of MotA and MotB suggested that Asp 32 of MotB is located inside the channel, near its cytoplasmic end (41, 42). Collectively, the available data support the proposal that Asp 32 of MotB has a direct, essential role in proton transfer through the flagellar motor. Moreover, the present results suggest that no other acidic residue of the motor is as important.
The present study focused on acidic residues, because they should allow more rapid proton transfer than basic residues. Basic residues cannot be ruled out on kinetic grounds, however, because basic groups in the motor might be situated near proton acceptors that accelerate proton transfer. The proteins involved in flagellar rotation do not appear to contain any conserved basic residues that are critical for motor rotation, however. MotB, FliM, and FliN have no perfectly conserved basic residues. MotA has one, Arg 90, which participates in functionally important rotor-stator interactions but is not, by itself, critical for function (61, 62). FliG has four conserved basic residues: Lys 9, Arg 160, Lys 264, and Arg 281. Lys 264 and Arg 281 were mutated in an earlier study (they were numbered 262 and 279 in that study, because of errors in the initially reported FliG sequence). Both residues engage in electrostatic interactions between rotor and stator, but neither, by itself, is critical for function (31, 62). Lys 9 is dispensable for function, because it is absent from a FliF-FliG fusion protein that supports motility (14). This leaves only Arg 160. We mutated this residue to alanine and observed a nonflagellate phenotype (data not shown). This suggests that Arg 160 is involved in some aspect of flagellar assembly, but probably not motor rotation per se. We note that mutations of Arg 160 gave an immotile but flagellated phenotype in a mutational study of FliG from Salmonella typhimurium (23). We have previously observed that several mutations in FliG (30), and also in FliM (48), affect flagellar assembly more severely in E. coli than in S. typhimurium.
Because Asp 32 of MotB appears to be the only conserved, protonatable residue critical for motor function, we presently favor the view that protons flow through the MotA-MotB channels and then directly into the cytoplasm, without binding to sites on the rotor. Alternatives can be imagined, however. One possibility is that the proton pathway does include sites on the rotor, but those sites are formed from multiple residues, any one and many pairs of which can be mutated without greatly affecting rates of proton transfer. Such resilience in a proton-transferring site appears unlikely. A second possibility is that N- or C-terminal residues contribute to the proton pathway. In the proteins that have been suggested to function in torque generation, the positions of the N- and C-terminal residues relative to other features of the protein sequences are not well conserved, however. Also, the C-terminal residue of MotA can be deleted without abolishing motility (61), as can the N terminus (14) and C terminus (31a) of FliG, N- and C-terminal segments of FliM (52), and the N terminus of FliN (49). A third possibility is that one or more of the proteins involved in torque generation bind a cofactor that functions in proton transfer. We have no data bearing on this question.
If protons that flow through the stator are not delivered to the rotor, then the stator must communicate with the rotor in some other way. We suggest that the most likely means of communication is via protonation-induced conformational changes in the stator. The aforementioned conserved proline residues of MotA, Pro 173 and Pro 222, were shown recently to be important for motor rotation (61) and may be key determinants of stator conformation. Hydropathy profiles and experimental studies of membrane topology indicate that Pro 173 and Pro 222 of MotA, and also Asp 32 of MotB, are located near the cytoplasmic ends of membrane-spanning segments (9, 11, 45, 60). These three residues may therefore be near each other, possibly forming a site that controls the conformation of the stator and makes it responsive to the protonation state of Asp 32. If the stator assumed different, protonation-regulated conformations that imposed different constraints on the movement of the rotor, this would link rotor movements to proton movements.
ACKNOWLEDGMENTS
We thank Sandy Parkinson and Robert Fazzio for strains and plasmids, Sonya Park for assistance with growth rate measurements, and J. Courtney Empey for assistance with tethering experiments.
This work was supported by grant 2-R01-GM46683 from the National Institute of General Medical Sciences. S.A.L. was supported in part by training grant 5T32-GM08537 from the National Institute of General Medical Sciences. The Protein-DNA Core Facility at the University of Utah receives support from the National Cancer Institute (5P30 CA42014).
REFERENCES
- 1.Berg, H. C. 1995. Torque generation by the flagellar rotary motor. Biophys. J. 68(Suppl. 4):163S–166S. [PMC free article] [PubMed]
- 2.Blair D F. How bacteria sense and swim. Annu Rev Microbiol. 1995;49:489–522. doi: 10.1146/annurev.mi.49.100195.002421. [DOI] [PubMed] [Google Scholar]
- 3.Blair D F, Berg H C. Restoration of torque in defective flagellar motors. Science. 1988;242:1678–1681. doi: 10.1126/science.2849208. [DOI] [PubMed] [Google Scholar]
- 4.Blair D F, Berg H C. The MotA protein of E. coli is a proton-conducting component of the flagellar motor. Cell. 1990;60:439–449. doi: 10.1016/0092-8674(90)90595-6. [DOI] [PubMed] [Google Scholar]
- 5.Blair D F, Berg H C. Mutations in the MotA protein of Escherichia coli reveal domains critical for proton conduction. J Mol Biol. 1991;221:1433–1442. doi: 10.1016/0022-2836(91)90943-z. [DOI] [PubMed] [Google Scholar]
- 6.Blair D F, Kim D Y, Berg H C. Mutant MotB proteins in Escherichia coli. J Bacteriol. 1991;173:4049–4055. doi: 10.1128/jb.173.13.4049-4055.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Block S M, Berg H C. Successive incorporation of force-generating units in the bacterial rotary motor. Nature (London) 1984;309:470–472. doi: 10.1038/309470a0. [DOI] [PubMed] [Google Scholar]
- 8.Boyd A, Mandel G, Simon M I. Integral membrane proteins required for bacterial motility and chemotaxis. Symp Soc Exp Biol. 1982;35:123–137. [PubMed] [Google Scholar]
- 9.Chun S Y, Parkinson J S. Bacterial motility: membrane topology of the Escherichia coli MotB protein. Science. 1988;239:276–278. doi: 10.1126/science.2447650. [DOI] [PubMed] [Google Scholar]
- 10.Clegg D O, Koshland D E., Jr Identification of a bacterial sensing protein and effects of its elevated expression. J Bacteriol. 1985;162:398–405. doi: 10.1128/jb.162.1.398-405.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dean G E, Macnab R M, Stader J, Matsumura P, Burks C. Gene sequence and predicted amino acid sequence of the motA protein, a membrane-associated protein required for flagellar rotation in Escherichia coli. J Bacteriol. 1984;159:991–999. doi: 10.1128/jb.159.3.991-999.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeMot R, Vanderleyden J. The C-terminal sequence conservation between OmpA-related outer membrane proteins and MotB suggests a common function in both gram-positive and gram-negative bacteria, possibly in the interaction of these domains with peptidoglycan. Mol Microbiol. 1994;12:333–334. doi: 10.1111/j.1365-2958.1994.tb01021.x. [DOI] [PubMed] [Google Scholar]
- 13.Enomoto M. Genetic studies of paralyzed mutants in Salmonella. II. Mapping of three mot loci by linkage analysis. Genetics. 1966;54:1069–1076. doi: 10.1093/genetics/54.5.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Francis N R, Irikura V M, Yamaguchi S, DeRosier D J, Macnab R M. Localization of the Salmonella typhimurium flagellar switch protein FliG to the cytoplasmic M-ring face of the basal body. Proc Natl Acad Sci USA. 1992;89:6304–6308. doi: 10.1073/pnas.89.14.6304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francis N R, Sosinsky G E, Thomas D, DeRosier D J. Isolation, characterization and structure of bacterial flagellar motors containing the switch complex. J Mol Biol. 1994;235:1261–1270. doi: 10.1006/jmbi.1994.1079. [DOI] [PubMed] [Google Scholar]
- 16.Garza A G, Harris-Haller L W, Stoebner R A, Manson M D. Motility protein interactions in the bacterial flagellar motor. Proc Natl Acad Sci USA. 1995;92:1970–1974. doi: 10.1073/pnas.92.6.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garza A G, Biran R, Wohlschlegel J, Manson M D. Mutations in motB suppressible by changes in stator or rotor components of the bacterial flagellar motor. J Mol Biol. 1996;258:270–285. doi: 10.1006/jmbi.1996.0249. [DOI] [PubMed] [Google Scholar]
- 18.Garza A G, Bronstein P A, Valdez P A, Harris-Haller L W, Manson M D. Extragenic suppression of motB missense mutations of Escherichia coli. J Bacteriol. 1996;178:6116–6122. doi: 10.1128/jb.178.21.6116-6122.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glagolev A N, Skulachev V P. The proton pump is a molecular engine of motile bacteria. Nature. 1978;272:280–282. doi: 10.1038/272280a0. [DOI] [PubMed] [Google Scholar]
- 20.Heimbrook M E, Wang W L L, Campbell G. Abstracts of the 86th Annual Meeting of the American Society for Microbiology 1986. Washington, D.C: American Society for Microbiology; 1986. Easily made flagella stains, abstr. R-22; p. 240. [Google Scholar]
- 21.Hilmen M, Simon M. Motility and the structure of bacterial flagella. In: Goldman R, Pollard T, Rosenbaum J, editors. Cell motility. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1976. pp. 35–45. [Google Scholar]
- 22.Hirota N, Imae Y. Na+-driven flagellar motors of an alkalophilic Bacillus strain YN-1. J Biol Chem. 1983;258:10577–10581. [PubMed] [Google Scholar]
- 23.Irikura V M, Kihara M, Yamaguchi S, Sockett H, Macnab R M. Salmonella typhimurium fliG and fliN mutations causing defects in assembly, rotation, and switching of the flagellar motor. J Bacteriol. 1993;175:802–810. doi: 10.1128/jb.175.3.802-810.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khan I H, Reese T S, Khan S. The cytoplasmic component of the bacterial flagellar motor. Proc Natl Acad Sci USA. 1992;89:5956–5960. doi: 10.1073/pnas.89.13.5956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Khan S, Dapice M, Reese T S. Effects of mot gene expression on the structure of the flagellar motor. J Mol Biol. 1988;202:575–584. doi: 10.1016/0022-2836(88)90287-2. [DOI] [PubMed] [Google Scholar]
- 26.Khan S, Khan I H, Reese T S. New structural features of the flagellar base in S. typhimurium revealed by rapid-freeze electron microscopy. J Bacteriol. 1991;173:2888–2896. doi: 10.1128/jb.173.9.2888-2896.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kihara M, Homma M, Kutsukake K, Macnab R M. Flagellar switch of Salmonella typhimurium: gene sequences and deduced protein sequences. J Bacteriol. 1989;171:3247–3257. doi: 10.1128/jb.171.6.3247-3257.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kihara M, Francis N R, DeRosier D J, Macnab R M. Analysis of a FliM-FliN flagellar switch fusion mutant of Salmonella typhimurium. J Bacteriol. 1996;178:4582–4589. doi: 10.1128/jb.178.15.4582-4589.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Larsen S H, Adler J, Gargus J J, Hogg R W. Chemomechanical coupling without ATP: the source of energy for motility and chemotaxis in bacteria. Proc Natl Acad Sci USA. 1974;71:1239–1243. doi: 10.1073/pnas.71.4.1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lloyd S A, Tang H, Wang X, Billings S, Blair D F. Torque generation in the flagellar motor of Escherichia coli: evidence of a direct role for FliG but not for FliM or FliN. J Bacteriol. 1996;178:223–231. doi: 10.1128/jb.178.1.223-231.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lloyd S A, Blair D F. Charged residues of the rotor protein FliG essential for torque generation in the flagellar motor of Escherichia coli. J Mol Biol. 1997;266:733–744. doi: 10.1006/jmbi.1996.0836. [DOI] [PubMed] [Google Scholar]
- 31a.Lloyd, S. A., and D. F. Blair. Unpublished results.
- 32.Macnab R. Genetics and biogenesis of bacterial flagella. Annu Rev Genet. 1992;26:129–156. doi: 10.1146/annurev.ge.26.120192.001023. [DOI] [PubMed] [Google Scholar]
- 33.Macnab R M. Flagella and motility. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. 2nd ed. Vol. 1. Washington, D.C: ASM Press; 1996. pp. 123–145. [Google Scholar]
- 34.Manson M D, Tedesco P, Berg H C, Harold F M, van der Drift C. A protonmotive force drives bacterial flagella. Proc Natl Acad Sci USA. 1977;74:3060–3064. doi: 10.1073/pnas.74.7.3060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marykwas D L, Berg H C. A mutational analysis of the interaction between FliG and FliM, two components of the flagellar motor of Escherichia coli. J Bacteriol. 1996;178:1289–1294. doi: 10.1128/jb.178.5.1289-1294.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marykwas D L, Schmidt S A, Berg H C. Interacting components of the flagellar motor of Escherichia coli revealed by the two-hybrid system in yeast. J Mol Biol. 1996;256:564–576. doi: 10.1006/jmbi.1996.0109. [DOI] [PubMed] [Google Scholar]
- 37.Morrison T B, Parkinson J S. Liberation of an interaction domain from the phosphotransfer region of CheA, a signaling kinase of Escherichia coli. Proc Natl Acad Sci USA. 1994;91:5485–5489. doi: 10.1073/pnas.91.12.5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oosawa K, Ueno T, Aizawa S-I. Overproduction of the bacterial flagellar switch proteins and their interactions with the MS ring complex in vitro. J Bacteriol. 1994;176:3683–3691. doi: 10.1128/jb.176.12.3683-3691.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 40.Schuster S C, Khan S. The bacterial flagellar motor. Annu Rev Biophys Biomol Struct. 1994;23:509–539. doi: 10.1146/annurev.bb.23.060194.002453. [DOI] [PubMed] [Google Scholar]
- 41.Sharp L L, Zhou J, Blair D F. Features of MotA proton channel structure revealed by tryptophan-scanning mutagenesis. Proc Natl Acad Sci USA. 1995;92:7946–7950. doi: 10.1073/pnas.92.17.7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharp L L, Zhou J, Blair D F. Tryptophan-scanning mutagenesis of MotB, an integral membrane protein essential for flagellar rotation in Escherichia coli. Biochemistry. 1995;34:9166–9171. doi: 10.1021/bi00028a028. [DOI] [PubMed] [Google Scholar]
- 43.Silverman M, Simon M. Operon controlling motility and chemotaxis in E. coli. Nature. 1976;264:577–580. doi: 10.1038/264577a0. [DOI] [PubMed] [Google Scholar]
- 44.Sockett H, Yamaguchi S, Kihara M, Irikura V M, Macnab R M. Molecular analysis of the flagellar switch protein FliM of Salmonella typhimurium. J Bacteriol. 1992;174:793–806. doi: 10.1128/jb.174.3.793-806.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Stader J, Matsumura P, Vacante D, Dean G E, Macnab R M. Nucleotide sequence of the Escherichia coli motB gene and site-limited incorporation of its product into the cytoplasmic membrane. J Bacteriol. 1986;166:244–252. doi: 10.1128/jb.166.1.244-252.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stolz B, Berg H C. Evidence for interactions between MotA and MotB, torque-generating elements of the flagellar motor of Escherichia coli. J Bacteriol. 1991;173:7033–7037. doi: 10.1128/jb.173.21.7033-7037.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Studier F W, Moffatt B A. Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J Mol Biol. 1986;189:113–130. doi: 10.1016/0022-2836(86)90385-2. [DOI] [PubMed] [Google Scholar]
- 48.Tang H, Blair D F. Regulated underexpression of the FliM protein of Escherichia coli and evidence for a location in the flagellar motor distinct from the MotA/MotB torque generators. J Bacteriol. 1995;177:3485–3495. doi: 10.1128/jb.177.12.3485-3495.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48a.Tang, H., and D. F. Blair. Unpublished results.
- 49.Tang H, Billings S, Wang X, Sharp L, Blair D F. Regulated underexpression and overexpression of the FliN protein of Escherichia coli and evidence for an interaction between FliN and FliM in the flagellar motor. J Bacteriol. 1995;177:3496–3503. doi: 10.1128/jb.177.12.3496-3503.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang H, Braun T F, Blair D F. Motility protein complexes in the bacterial flagellar motor. J Mol Biol. 1996;261:209–221. doi: 10.1006/jmbi.1996.0453. [DOI] [PubMed] [Google Scholar]
- 51.Togashi F, Yamaguchi S, Kihara M, Aizawa S-I, Macnab R M. An extreme clockwise switch bias mutation in fliG of Salmonella typhimurium and its suppression by slow-motile mutations in motA and motB. J Bacteriol. 1997;179:2994–3003. doi: 10.1128/jb.179.9.2994-3003.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Toker A S, Kihara M, Macnab R M. Deletion analysis of the FliM flagellar switch protein of Salmonella typhimurium. J Bacteriol. 1996;178:7069–7079. doi: 10.1128/jb.178.24.7069-7079.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Welch M, Oosawa K, Aizawa S-I, Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci USA. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wilson M L, Macnab R M. Overproduction of the MotA protein of Escherichia coli and estimation of its wild-type level. J Bacteriol. 1988;170:588–597. doi: 10.1128/jb.170.2.588-597.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wilson M L, Macnab R M. Co-overproduction and localization of the Escherichia coli motility proteins MotA and MotB. J Bacteriol. 1990;172:3932–3939. doi: 10.1128/jb.172.7.3932-3939.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamaguchi S, Fujita H, Ishihara A, Aizawa S-I, Macnab R M. Subdivision of flagellar genes of Salmonella typhimurium into regions responsible for assembly, rotation, and switching. J Bacteriol. 1986;166:187–193. doi: 10.1128/jb.166.1.187-193.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yamaguchi S, Aizawa S-I, Kihara M, Isomura M, Jones C J, Macnab R M. Genetic evidence for a switching and energy-transducing complex in the flagellar motor of Salmonella typhimurium. J Bacteriol. 1986;168:1172–1179. doi: 10.1128/jb.168.3.1172-1179.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhao R, Schuster S C, Khan S. Structural effects of mutations in S. typhimurium flagellar switch complex. J Mol Biol. 1995;251:400–412. doi: 10.1006/jmbi.1995.0443. [DOI] [PubMed] [Google Scholar]
- 59.Zhao R, Pathak N, Jaffe H, Reese T S, Khan S. FliN is a major structural protein of the C-ring in the Salmonella typhimurium flagellar basal body. J Mol Biol. 1996;261:195–208. doi: 10.1006/jmbi.1996.0452. [DOI] [PubMed] [Google Scholar]
- 60.Zhou J, Fazzio R T, Blair D F. Membrane topology of the MotA protein of Escherichia coli. J Mol Biol. 1997;251:237–242. doi: 10.1006/jmbi.1995.0431. [DOI] [PubMed] [Google Scholar]
- 61.Zhou J, Blair D F. Residues of the cytoplasmic domain of MotA essential for torque generation in the bacterial flagellar motor. J Mol Biol. 1997;273:428–439. doi: 10.1006/jmbi.1997.1316. [DOI] [PubMed] [Google Scholar]
- 62.Zhou, J., S. A. Lloyd, and D. F. Blair. Electrostatic interactions between rotor and stator in the bacterial flagellar motor. Proc. Natl. Acad. Sci. USA, in press. [DOI] [PMC free article] [PubMed]