Abstract
Ralstonia (Pseudomonas) solanacearum is a soil-borne phytopathogen that causes a wilting disease of many important crops. It makes large amounts of the exopolysaccharide EPS I, which it requires for efficient colonization, wilting, and killing of plants. Transcription of the eps operon, encoding biosynthetic enzymes for EPS I, is controlled by a unique and complex sensory network that responds to multiple environmental signals. This network is comprised of the novel transcriptional activator XpsR, three distinct two-component regulatory systems (VsrAD, VsrBC, and PhcSR), and the LysR-type regulator PhcA, which is under the control of PhcSR. Here we show that the xpsR promoter (PxpsR) is simultaneously controlled by PhcA and VsrD, permitting XpsR to act like a signal integrator, simultaneously coordinating signal input into the eps promoter from both VsrAD and PhcSR. Additionally, we used in vivo expression analysis and in vitro DNA binding assays with substitution and deletion mutants of PxpsR to show the following. (i) PhcA primarily interacts with a typical 14-bp LysR-type consensus sequence around position −77, causing a sixfold activation of PxpsR; a weaker, less-defined binding site between −183 and −239 likely enhances PhcA binding and activation via the −77 site another twofold. (ii) Full 70-fold activation of PxpsR requires the additional interaction of the VsrD response regulator (or its surrogate) with a 14-bp dyadic sequence centered around −315 where it enhances activation (and possibly binding) by PhcA; however, VsrD alone cannot activate PxpsR. (iii) Increasing the distance between the putative VsrD binding site from that of PhcA by up to 232 bp did not dramatically affect PxpsR activation or regulation.
Ralstonia (Pseudomonas) solanacearum (42, 43) is one of the most troublesome prokaryotic phytopathogens in the world (13, 14). It infects plants via wounds or cracks at the emergence point of lateral roots (37). It subsequently spreads into the stem via the vascular system (40, 41), where populations reach >1010 cells per plant, concomitant with wilting and death of the host. R. solanacearum produces a large variety and amount of extracellular products that contribute to disease (34). One of the most important of these is the unusual extracellular polysaccharide EPS I, a large acidic polymer comprised of N-acetylgalactosamine and two derivatives thereof (25, 32). EPS I is required by R. solanacearum for efficient wilting and killing of plants, probably because it restricts water flow in the xylem (5–7). EPS I is also required for efficient and rapid colonization of the plant’s vascular system (28). Production of EPS I requires the products of the 16-kb eps gene cluster (6), which appear to be transcribed from a single promoter into a large polycistronic RNA (19). DNA sequence analysis suggests that the eps operon encodes >12 polypeptides directly involved in biosynthesis and export of EPS I (19).
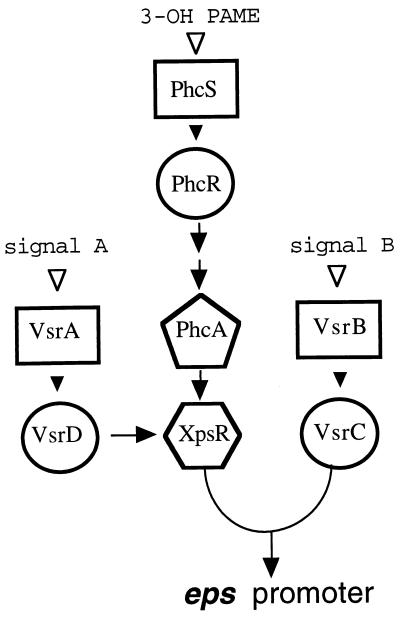
Because it must also survive in the soil outside of a plant host, R. solanacearum has evolved a sophisticated network for controlling expression of genes encoding production of EPS I and its other virulence factors, such as plant cell wall-degrading exoenzymes (4, 18). The network contains at least three distinct signal transduction arrays, each containing a unique two-component system (15, 26), comprised of a membrane-bound kinase sensor and a response regulator (Fig. 1). It is likely that each sensor (VsrA, VsrB, or PhcS) responds to a different environmental signal by phosphorylating its cognate response regulator (VsrD, VsrC, or PhcR), which in turn activates or represses promoters of appropriate virulence genes. While maximal transcription of eps requires all components of the network to be active, previous experiments (18) showed that the control of the eps promoter affected by both the VsrAD and PhcSR/PhcA signal transduction arrays is indirect, because the reduced eps transcription caused by inactivation of either or both of these systems can be overcome by constitutively expressed xpsR. This observation coupled with the fact that the VsrAD and PhcSR/PhcA systems control transcription of xpsR (3, 4, 10, 18) led to the proposals that VsrAD and PhcA control levels of XpsR protein in a signal-dependent manner and that XpsR acts as a signal integrator which in concert with the VsrBC system controls transcription from the eps promoter (35). How XpsR and VsrC activate transcription is unknown. While mutagenesis of the eps promoter has identified a single 20-bp region around position −70 that is essential for activation by both XpsR and VsrC (36), attempts to demonstrate binding of XpsR to the eps promoter have been unsuccessful, suggesting that like some eukaryotic transcription factors, XpsR may bind to another regulatory protein that itself is bound to the eps promoter.
FIG. 1.
Organization of the regulatory network controlling eps and other virulence genes of R. solanacearum. Signal transduction pathways through the three known two-component systems, VsrAD (18, 33), VsrBC (17, 18), and PhcSR (4), that in conjunction with phcA (2) and xpsR control transcription from the eps biosynthetic operon promoter are shown. Rectangles represent membrane-bound kinase sensors; circles represent response regulators. Arrows indicate positive transcriptional control. Closed arrowheads indicate hypothesized, signal-induced phosphoryl transfer between sensory kinase and response regulator; open arrowheads indicate positions of environmental signal input and recognition. 3-Hydroxypalmitic acid methyl ester (3-OH PAME) is proposed to be the signal perceived by the PhcSR system (4, 10); available evidence suggests that PhcR affects phcA expression indirectly. Signals perceived by VsrA and VsrB are unknown.
xpsR is the first of four genes in a 5-kb operon (8, 9, 18) whose transcription appears to be directed by a promoter found directly upstream of xpsR. Although none of the four open reading frames (ORFs) in the operon has any obvious motifs or homologs in sequence databases, their functions have been explored by analysis of site-directed mutants. The first ORF, XpsR, is a 33-kDa positive regulator of eps transcription (18); the second ORF (58 kDa) is Tek, which is processed into a major extracellular protein that is associated with EPS I (8); the third is Ert, a 44-kDa ORF (formerly called Region II or RgnII [6, 32]) which plays an ill-defined and conditional role in EPS I synthesis (9); the fourth ORF is a homolog of Tek. With the exception of XpsR, none of the ORFs appears to be absolutely required for virulence (6, 8, 9). To better understand the function of these genes and how levels of their products are transcriptionally regulated, we used different types of mutagenesis to investigate where and how PhcA and VsrD might interact with the xpsR promoter and found evidence that they use an atypical mechanism to regulate transcription from this promoter.
MATERIALS AND METHODS
Bacteria, plasmids, and media.
Descriptions of the R. solanacearum strains and plasmids used are shown in Table 1; for maps of plasmids, see Fig. 2 and 5. The Escherichia coli host strain used for most recombinant DNA manipulations was DH5α (12). Vectors used were pTZ18U/pTZ19U (22), pRK415 (20), and pRG970 (39). R. solanacearum and E. coli were grown at 30°C in 1% peptone–0.1% Casamino Acids–0.1% yeast extract with 0.5% glucose or 0.5% sucrose (BG medium) and at 37°C in LB medium (23), respectively. Antibiotics were used at 50 μg/ml for kanamycin, 100 μg/ml for ampicillin (20 μg/ml for R. solanacearum), 50 μg/ml for spectinomycin, and 25 μg/ml for tetracycline.
TABLE 1.
R. solanacearum strains and plasmids
| Strain or plasmid | Descriptiona | Reference |
|---|---|---|
| Strain | ||
| AW | Wild-type pathogen; LacZ− | 29 |
| AW201 | epsA1::nptI EPS− LacZ− Kmr | 19 |
| AW-D5 | vsrD5::TnphoA LacZ− Kmr | This study |
| AW1-80 | phcA80::Tn5 LacZ− Kmr | 1 |
| Plasmid | ||
| pCB5 | pLAFR3 cosmid with eps and rgnII; Tcr | 6 |
| pTZRLZ7 | 2.7-kb BamHI fragment of pCB5 on pTZ18U; Apr | This study |
| pTZRLZ3 | 610-bp BamHI fragment of pJH161 on pTZ18U; Apr | This study |
| pTZRLZ1 | BamHI fragment from pRLZ1 on pTZ18U; Apr | This study |
| pRG970 | Transcriptional fusion vector with promoterless lacZ; Spr | 39 |
| pRLZ7 | pRG970 with nt −2400 to +275 of PxpsR fused to lacZ; Spr | This study |
| pRLZ1 | Same as for pRLZ7 except nt −338 to +36 | This study |
| pRLZ8 | Same as for pRLZ7 except nt −310 to +36 | This study |
| pRLZ4 | Same as for pRLZ7 except nt −286 to +36 | This study |
| pRLZ5 | Same as for pRLZ7 except nt −236 to +36 | This study |
| pRLZ2 | Same as for pRLZ7 except nt −117 to +36 | This study |
| pRLZ6 | Same as for pRLZ7 except nt −76 to +36 | This study |
| pRLZ15 | Same as for pRLZ1 except nt −322 CCC→TGG | This study |
| pRLZ16 | Same as for pRLZ1 except nt −319 to −316 (CTAA) deleted | This study |
| pRLZ17 | Same as for pRLZ1 except nt −327 AAT→GTA | This study |
| pRMB-S | Same as for pRLZ2 except nt −82 TAAAAA→AGATCT | This study |
| pRM5-S | Same as for pRLZ2 except nt −73 T→C | This study |
| pRM7-S | Same as for pRLZ2 except nt −83 T→C | This study |
| pRMB | Same as for pRLZ1 except nt −82 TAAAAA→AGATCT | This study |
| pRM7 | Same as for pRLZ1 except nt −83 T→C | This study |
| pRMB-M | Same as for pRMB except nt −286 to +36 | This study |
| pRM7-M | Same as for pRM7 except nt −286 to +36 | This study |
| pRLZ11 | Same as for pRLZ1 except has 4-bp insertion at −183 | This study |
| pRLZ12 | Same as for pRLZ1 except has 118-bp insertion at −183 | This study |
| pRLZ13 | Same as for pRLZ1 except has 232-bp insertion at −183 | This study |
Apr, Kmr, Tcr, and Spr denote resistance to ampicillin, kanamycin, tetracycline, and spectinomycin, respectively. LacZ−, produced no detectable β-galactosidase activity; EPS−, nonmucoid and deficient in production of exopolysaccharide; nt, nucleotide.
FIG. 2.
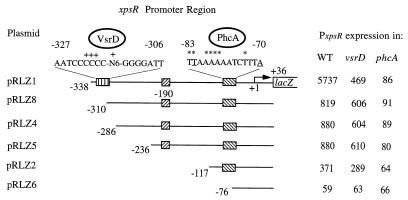
Identification of PxpsR sequences required for transcriptional regulation by VsrD and PhcA. The xpsR promoter and various lengths of upstream sequences were fused to lacZ on pRG970 to generate plasmids pRLZ1 to pRLZ8. PxpsR expression (i.e., transcription directed by each PxpsR fragment) was monitored in the wild type (WT), strain AW-D5 (vsrD mutant), and strain AW1-80 (phcA mutant) of R. solanacearum by measuring LacZ activity (β-galactosidase activity in Miller units) as described in Table 2, footnote a. Nucleotide numbering is relative to the transcription start site of xpsR (+1|→). Striped and hatched boxes and associated sequences illustrate putative binding sites for VsrD and PhcA, respectively. The symbol + above the putative VsrD binding site dyad indicates the positions of mutations that eliminated VsrD activation of PxpsR.; N6 represents the sequence TAAATT. Underlined nucleotides in the PhcA binding site indicate the T-N11-A motif found in the binding sites of nearly all LysR-type activators (31). Asterisks indicate the positions of mutations that affected PhcA binding and activation of PxpsR (Table 3). The complete xpsR promoter region sequence is GenBank under accession no. U18136. pRLZ7 (Table 1) had PxpsR activity similar to that of pRLZ1.
FIG. 5.
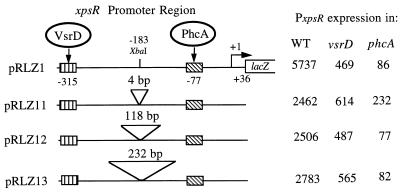
Effect of spacer insertions between the putative VsrD and PhcA binding sites on expression of PxpsR. Various lengths (4, 118, and 232 bp) of spacer DNA were inserted into the XbaI site (at −183) in PxpsR on fusion plasmid pRLZ1 (Fig. 2) to create plasmids pRLZ11 to pRLZ13. Expression directed from each PxpsR derivative was monitored in wild-type (WT), vsrD mutant (AW-D5), and phcA mutant (AW1-80) strains of R. solanacearum by measuring LacZ activity as described in Table 2, footnote a. Striped and hatched boxes represent binding sites for VsrD and PhcA, respectively.
Construction of PxpsR-lacZ fusions.
pRLZ7 and pTZRLZ7 were constructed by inserting the 2.7-kb BamHI fragment of pCB5 (6) into BamHI-digested pRG970 (39) and pTZ18U, respectively. pRLZ3 and pTZRLZ3 were constructed by inserting the 610-bp BamHI fragment of pJH161 (18) into BamHI-digested pRG970 and pTZ18U, respectively. The primer pairs T7/RP2, RP7/RP2, RP3/RP2, RP4/RP2, RP1/RP2, and RP5/RP2 were used in PCR with pTZRLZ3 as template to generate the xpsR promoter fragments with various lengths of upstream regulatory sequences used in construction of pRLZ1-pRLZ8. One-hundred-microliter PCRs contained 1× reaction buffer (Perkin-Elmer), 1.5 mM MgCl2, 200 μM deoxynucleoside triphosphates (dNTPs), 2.5 U of Amplitaq polymerase (Perkin-Elmer), 0.4 μM primer pair, and 100 ng of pTZRLZ3. After denaturation for 4 min at 95°C, 20 thermal cycles (1 min at 94°C, 1 min at 50°C, and 2 min at 72°C) were performed. PCR products were extracted with phenol-chloroform, precipitated with ethanol, digested with BamHI or BamHI-BglII, gel purified, ligated into BamHI-digested pRG970, and transformed into E. coli. Plasmids from transformants that were blue on LB plates with spectinomycin and 5-bromo-3-chloro-indolyl-β-d-galactopyranoside (X-Gal) were isolated, and orientation was confirmed by restriction enzyme analysis. Primers included T7 (5′-TAATACGACTCACTATAGGG-3′), RP1 (5′-CAGGATCCCCGGCTGTGCGACAT-3′), RP2 (5′-TAGGATCCATTGAATCCGTGCAAC-3′), RP3 (5′-CCGGATCCTACGCGTTCAGATATG-3′), RP4 (5′-GAGGATCCCAACACGCTGCTTTAC-3′), RP5 (5′-GAAGATCTATCTTTACTCTCCTTTA-3′), RP7 (5′-GAGGATCCGA TTCGTTTTTTTCTTG-3′), RP8 (5′-GAGGATCCGAATTCCGACAAATCCTGGCTAAATTGGGGAT-3′), RP9 (5′-GAGGATCCGAATTCCGACAGTACCCCCCTAAATTGG-3′), and RP10 (5′-GAGGATCCGAATTCCGACAAATCCCCCΔHATTGGGGATTCGTT-3′); underlining indicates altered nucleotides.
Mutagenesis of putative binding sites.
To alter the PhcA binding site, two PxpsR regions, −338 to −83 and −76 to +36, were amplified by PCR using pTZRLZ3 as template and primer pairs T7/RP6 and RP5/RP2, respectively. Since both RP5 (see above) and RP6 (5′-GAAGATCTACATCACGCCAGCTTTG-3′) contain a 5′ BglII site, ligation of the two BglII-digested PCR fragments together changes nucleotides −82 to −77 from TAAAAA to AGATCT. The ligation products were digested with BamHI and ligated into BamHI-digested pRG970 to give pRMB. To construct pRMB derivatives, PCR fragments generated by primer pairs RP1/RP2 and RP3/RP2 with pRMB as template were digested with BamHI and ligated into BamHI-digested pRG970 to obtain pRMB-S and pRMB-M, respectively. PxpsR fusion plasmids with alterations in the putative VsrD binding site were generated by PCR amplification of sequences between nucleotides −338 and +36 by using a pRLZ1 template and primer pairs RP8/RP2, RP9/RP2, and RP10/RP2. Resultant PCR fragments were digested with BamHI and ligated into BamHI-digested pRG970 to obtain pRLZ15, pRLZ17, and pRLZ16, respectively.
Random PCR mutagenesis of the PxpsR fragment (nucleotides −338 to +36) on pTZRLZ1 was done as described by Muhlard et al. (24). Reaction mixtures (100 μl) contained 10 mM Tris-HCl (pH 8.3), 50 mM KCl, 0.001% gelatin, 1 mM each dGTP, dCTP, and dTTP, 200 μM dATP, 2.5 U of Amplitaq polymerase, 0.4 μM each T7 and M13 forward primers, and either 0.5 mM MnCl2–4 mM MgCl2 or 0.25 mM MnCl2–1.5 mM MgCl2. Thermal cycling consisted of 25 cycles of 1 min at 94°C, 1 min at 50°C, and 2 min at 72°C. PCR products were digested with BamHI, gel purified, ligated into BamHI-digested pRG970, transformed into E. coli, and plated onto LB plates with spectinomycin and X-Gal. Plasmids from pooled blue colonies were isolated and electrotransformed into R. solanacearum AW201. Twenty of the 2,000 transformants obtained on BG plates with spectinomycin and X-Gal showed reduced blue color (LacZ activity). Plasmids individually isolated from these 20 strains were transformed into E. coli, reisolated, and electrotransformed into the wild type and regulatory mutants of R. solanacearum for analysis of PxpsR expression. Derivatives of pRM5 and pRM7 were constructed by using the same strategy as that employed with pRMB to give pRM5-M, pRM5-S, pRM7-M, and pRM7-S. Sequence alterations in plasmids were determined directly by automated sequencing of plasmids prepared with Wizard Minipreps (Promega).
Insertional mutagenesis of PxpsR.
pRLZ11 containing a 4-bp insertion between the VsrD and PhcA binding sites of PxpsR was constructed in three steps: (i) the 374-bp BamHI fragment of pTZRLZ1 was inserted into BamHI-digested pUC9 to give pUCRLZ1; (ii) pUCRLZ1 was linearized with XbaI, filled-in with four dNTPs and Klenow enzyme, and recircularized to give pUCRLZ11; and (iii) the BamHI fragment of pUCRLZ11 was ligated into BamHI-digested pRG970. pRLZ12 and pRLZ13 were constructed by ligating the 114-bp SmaI fragment of pJH161 (18) with pUCRLZ1 which had been digested with XbaI and whose ends had been filled in with four dNTPs and Klenow enzyme followed by recloning of the BamHI fragments of the resultant plasmids into BamHI-digested pRG970. pRLZ12 contains a single 114-bp SmaI fragment; pRLZ13 contains two tandem 114-bp SmaI fragments.
Purification of PhcA and use in mobility shift DNA binding analyses.
Five-hundred-milliliter cultures of E. coli BL21 DE3 containing the pET3d vector (38) or the phcA overexpression plasmid pET3231 (18) were grown to an A600 of 0.3 at 37°C and then transferred to a 25°C shaker. After 10 min, isopropylthiogalactoside was added to 0.5 mM to induce overexpression of phcA and shaking was continued for 4 h. Cells were harvested, washed once with buffer A (10 mM Tris-HCl [pH 7.0], 25 mM KCl, 2 mM mercaptoethanol, 1 mM phenylmethylsulfonylfluoride), resuspended in 5 ml of buffer A, and sonicated for 3 min at 4°C. Broken cells were centrifuged at 15,000 × g for 20 min. Ammonium sulfate (1.4 g) was added to the resultant supernatant (5.5 ml), and after 1 h at 4°C, precipitated proteins were removed by centrifugation (12,000 × g, 10 min). An additional 0.32 g of ammonium sulfate was added to the supernatant, and after 1 h at 4°C, precipitated proteins were recovered by centrifugation, redissolved 1 ml of buffer A, and dialyzed extensively against buffer A. After any precipitate was discarded, the sample was applied to a 1.5-ml phosphocellulose column equilibrated with buffer A. After the column was washed with buffer A with 0.18 M KCl, PhcA was eluted with buffer A with 0.25 M KCl.
Between 0.5 and 24 μg of protein eluted from the phosphocellulose column (estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis to contain >90% PhcA) was incubated with 4,000 cpm of DNA fragment (labeled by filling in with Klenow enzyme and [α-32P]dATP [21]) in 30 μl of a mixture containing 10 mM Tris-HCl (pH 7.5), 50 mM KCl, 1 mM EDTA, 2 mM dithiothreitol, 130 μg of bovine serum albumin per ml, and 100 μg of salmon sperm DNA per ml. Binding was analyzed by electrophoresis and autoradiography as described previously (30). To quantify PhcA binding, dried gels were analyzed with a Molecular Dynamics phosphorimager and ImageQuant software.
Molecular genetic techniques.
The methods used for preparation, analysis, and manipulation of recombinant DNA, fragment purification, CaCl2-mediated transformation, and electrotransformation were standard (17, 18, 21).
RESULTS
Two distinct upstream segments of the xpsR promoter region are required for transcriptional regulation.
Previously (18), we showed that (i) transcription of xpsR is activated >10-fold by PhcA and may involve binding of PhcA to the xpsR promoter; (ii) PhcA-activated transcription of xpsR can be further increased >5-fold by the VsrAD two-component system; and (iii) a phcA mutation eliminates VsrAD activation of xpsR without affecting vsrAD expression, implying that VsrAD alone cannot activate xpsR transcription. However, where and how each protein acts to effect full transcriptional activation was not defined. Therefore, the xpsR promoter region (PxpsR) and various lengths of upstream sequences were joined to a promoterless lacZ gene on pRG970 (41), creating a series of reporters with transcriptional fusions of xpsR to lacZ. Activation of PxpsR by PhcA or VsrD was assessed by comparing β-galactosidase levels in wild-type, phcA mutant, and vsrD mutant strains of R. solanacearum containing each reporter plasmid. Transcription of lacZ directed by a fragment with PxpsR containing 338 bp of sequence upstream of the xpsR transcription start site (pRLZ1) was normally regulated (Fig. 2): inactivation of vsrD reduced expression of the PxpsR::lacZ fusion 12-fold, while inactivation of phcA reduced expression 67-fold. PxpsR expression levels in phcA mutants were not further reduced by additional inactivation of vsrD (data not shown), confirming that phcA is epistatic to vsrD. An analogous fusion plasmid with extended (>2-kb) sequences upstream of PxpsR (pRLZ7) (Table 1) gave the same PxpsR expression levels as pRLZ1 and showed wild-type regulation (data not shown).
Thus, the 338-bp sequence upstream of the PxpsR transcription start site contains all cis-acting elements required for regulation of PxpsR by PhcA and VsrD. However, deletion of sequences between nucleotides −338 and −310 (to give pRLZ8) (Fig. 2) reduced lacZ expression directed by PxpsR in wild-type cells sevenfold and eliminated its ability to be activated by VsrD. In contrast, transcription directed by this −310 to +36 fragment remained phcA dependent since β-galactosidase levels directed by pRLZ8 in a phcA mutant background were ninefold lower than those in the wild type. pRLZ4 and pRLZ5, with shorter PxpsR fragments (−286 to +36 and −236 to +36, respectively), fused to lacZ gave the same phenotype as pRLZ8 (Fig. 2). These results show that the sequences between −338 and −310 are required for activation of xpsR transcription by VsrD. They also show that PhcA can activate PxpsR via the region downstream of −236 and in the absence of VsrD, albeit at a 10-fold-lower level, reaffirming the suggestion that VsrD enhances PhcA-mediated activation. Examination of the PxpsR sequences specifically required for regulation by VsrD revealed a palindromic sequence (AATCCCC-N8-GGGGATT) (Fig. 2) of the type that has been shown to bind transcriptional regulators.
Identification of critical nucleotides required for VsrD activation of PxpsR.
To test the hypothesized involvement of the palindromic sequence between nucleotides −327 and −307 in VsrD activation and more accurately define a putative binding site for VsrD, we used PCR to make three different mutations that destroyed its dyadic structure or spacing and then tested the effect on PxpsR activation. A derivative of pRLZ1 (−338 to +36 of xpsR fused to lacZ) with the CCC between −322 and −320 changed to TGG (pRLZ15) had a PxpsR expression level that was more than sixfold lower than that of the wild type and was not activated by VsrD (Table 2). Similar results were obtained with a derivative that had the distance between two halves of the dyad shortened by 4 bp (i.e., nucleotides −319 to −316 [CTAA] were deleted; pRLZ16) (Table 2). However, when the nucleotides between positions −327 and −325 (AAT) were changed to GTA, there was no effect on PxpsR expression or VsrD activation. In conjunction with the results of the deletion experiments described above, these results strongly suggest that the PxpsR sequences located between −324 and −310, and more specifically the CCCC-N6-GGGG dyadic sequence (−322 to −309) (Fig. 2), are critical for activation of PxpsR by VsrD. This may be the site where VsrD binds.
TABLE 2.
Effect of site-directed mutations in the −315 palindromic region of the xpsR promoter on expression and regulation
| Plasmid | PxpsR nucleotide sequence fused to lacZ | Mutation | Expressiona from PxpsR in:
|
|
|---|---|---|---|---|
| WT | vsrD | |||
| pRLZ1 | −338 to +36 | None | 5,280 | 443 |
| pRLZ15 | −338 to +36 | −322 CCC→TGG | 775 | 577 |
| pRLZ16 | −338 to +36 | −319 CTAA→Δ | 843 | 575 |
| pRLZ17 | −338 to +36 | −327 AAT→GTA | 4,910 | 709 |
Expression (transcription) from PxpsR was monitored by measuring LacZ (β-galactosidase) activity in Miller units as described by Miller (23) in overnight cultures of R. solanacearum wild type (WT) or vsrD mutants (strain AW-D5) carrying pRG970 vectors with PxpsR::lacZ fusions having the indicated mutation. Values are averages from three independent experiments.
Identification of nucleotides involved in PhcA-mediated activation of PxpsR.
A 14-bp site centered around nucleotide −77 of PxpsR (Fig. 2) was previously proposed as a binding site for PhcA (18) because of its conformity with the consensus structure and location of binding sites for LysR-type activators that are similar to PhcA (31). Consistent with this hypothesis, pRLZ6 with nucleotides −76 to +36 of xpsR fused to lacZ, and hence lacking half of this site, showed no activation by PhcA while an xpsR promoter fragment with the putative binding site and only 40 bp of additional upstream sequence (nucleotides −117 to +36; pRLZ2) (Fig. 2) fused to lacZ, showed a sixfold-higher activation of expression by PhcA. When an xpsR promoter fragment with the putative binding site and an additional 160 bp of upstream sequence was fused to lacZ (−236 to +36; pRLZ5), a twofold-higher activation of PxpsR by PhcA was observed. While this suggests that PxpsR sequences between nucleotides −236 and −117 can enhance or assist in PhcA activation mediated via the −77 site, they are clearly not required for PhcA to function at PxpsR.
To further define the sequences between −117 and −76 that are required for PhcA to activate PxpsR and to test the role of the putative PhcA binding site in activation, we constructed pRMB-S, which has −117 to +36 of PxpsR fused to lacZ and also a 5-bp mutation between −82 and −78 that dramatically alters the putative binding site. The 5-bp alteration reduced PhcA-mediated activation of PxpsR by 85% (Table 3), consistent with the predicted requirement for sequence-specific binding of PhcA to the −82 to −70 region.
TABLE 3.
Effect of mutations in the −77 region of the xpsR promoter on its regulated expression and PhcA binding activity
| Plasmid | PxpsR nucleotide sequence fused to lacZ | Mutation | Expressiona from PxpsR in:
|
PhcA bindingb | |
|---|---|---|---|---|---|
| WT | phcA | ||||
| pRLZ2 | −117 to +36 | None | 373 | 64 | 100 |
| pRMB-S | −117 to +36 | −82 TAAAAA→AGATCT | 95 | 62 | 25 |
| pRM7-S | −117 to +36 | −83 T→C | 60 | 62 | <5 |
| pRM5-S | −117 to +36 | −73 T→C | 92 | 65 | 25 |
Expression (transcription) from PxpsR was monitored by measuring LacZ (β-galactosidase) activity in Miller units as described by Miller (23) in cultures of R. solanacearum wild type (WT) or phcA mutants (strain AW1-80) carrying pRG970 with PxpsR::lacZ fusions having the indicated mutation. Values are averages from three independent experiments with <20% variation.
PhcA binding was determined by quantitative phosphorimaging of at least two gel shift DNA binding titrations (see Materials and Methods) similar to those shown in Fig. 3. The amount of purified PhcA protein needed to retard mobility of 50% of a given fragment was determined from a titration curve with at least three points. Each value is equal to [(amount needed for wild-type fragment)/(amount needed for mutant fragment)] × 100.
For a more precise analysis, we used random PCR mutagenesis to screen for nucleotide substitutions that affected activation of PxpsR by PhcA (Materials and Methods). Two important mutant plasmids were obtained: pRM7-S, with a single nucleotide substitution at −83 that completely eliminated activation of PxpsR by PhcA, and pRM5-S, in which a substitution at −73 reduced PhcA-mediated activation by 90% (Table 2). These results are totally consistent with the proposed location and function of the PhcA binding site centered around −77 of PxpsR (Fig. 2).
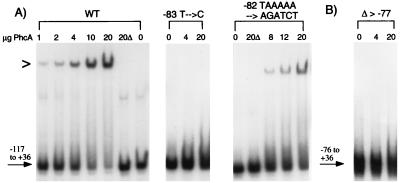
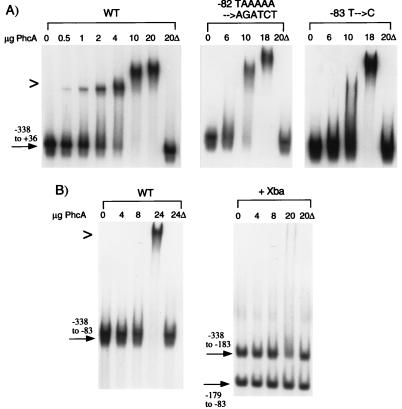
To show that activation of PxpsR by PhcA directly involves and requires binding by PhcA to the −83 to −70 site, we used a gel mobility shift assay to monitor PhcA binding to wild-type and mutant PxpsR fragments (Fig. 3A). Incubation of a wild-type PxpsR fragment (−117 to +36) with increasing amounts of purified PhcA resulted in retardation of the mobility of an increasing proportion of the fragment. With 20 μg, almost all of the PxpsR fragment was bound by PhcA (i.e., had reduced mobility), whereas 20 μg of an identical preparation from E. coli lacking phcA had no effect. Migration of the same PxpsR fragment lacking sequences upstream of −76 (Fig. 3B) was unaffected by incubation with up to 20 μg of purified PhcA. This confirms the presence of a specific binding site for PhcA downstream of −117.
FIG. 3.
Gel mobility shift assays of PhcA binding to −117 to +36 xpsR promoter fragments with mutations in the PhcA binding site. (A) DNA fragments containing the −117 to +36 sequences of wild-type PxpsR (WT) or PxpsR with the indicated sequence alterations were isolated, labeled with [α-32P]dATP, and incubated with 0 to 20 μg of purified PhcA protein or with 20 μg of a similar protein preparation from E. coli lacking phcA (20Δ). Reaction mixtures were electrophoresed, and the dried gels were subjected to autoradiography as described previously (30); quantitative results were obtained by phosphorimaging and are summarized in Table 3. (B) A labeled PxpsR DNA fragment lacking sequences upstream of nucleotide −77 (Δ > −77) was analyzed as described for panel A. The symbol > to the left of the gel indicates the position of retarded (bound) fragment.
The −117 to +36 PxpsR fragment harboring the −83 T→C point mutation that completely eliminated activation by PhcA showed no evidence of mobility retardation (binding) by PhcA, even at the highest levels tested (Fig. 3A). Similarly, binding of PhcA to the −117 to +36 PxpsR fragment with the 5-bp substitution at −82 to −78 was nearly fourfold lower than that to wild-type PxpsR, correlating with its ninefold-lower in vivo activation by PhcA (Fig. 3 and Table 3). Similar results were obtained with the PxpsR fragment with the −73 T→C mutation (Table 3 and data not shown). These results are consistent with the hypothesis that PhcA binds specifically to the −83 to −70 sequence of PxpsR to activate its transcription.
Effect of upstream sequences on expression from PxpsR with PhcA binding site mutations.
Previous genetic analysis (18) suggested that the VsrA/VsrD two-component system by itself cannot activate PxpsR but rather acts in conjunction with PhcA. To confirm this, we assessed the effect of PhcA binding site mutations on in vivo transcription directed by fragments with the complete xpsR promoter region (−338 to +36), which included the putative VsrD binding site. Because phcA is required for PxpsR activation (Fig. 2) and because PhcA binding site mutations reduced PhcA activation of and binding to the −117 to +36 PxpsR fragment (Table 3 and Fig. 3), we expected there would be little or no PhcA-mediated activation of complete PxpsR fragments with the same mutations. However, the PxpsR-lacZ fusion constructs with mutant PhcA binding sites (pRMB and pRM7) showed at least 20-fold PhcA-mediated activation (Table 4). Deletion of sequences upstream of −286 (i.e., those containing the putative VsrD activation or binding site) from the fusion plasmids strongly reduced, but did not eliminate, PxpsR expression (pRMB-M and pRM7-M; Table 4). This shows that even in the absence of VsrD interactions, these larger PxpsR fragments with PhcA binding site mutations can be activated >3.5-fold by PhcA, much higher than the 0.5-fold activation observed when the same mutant xpsR promoters were harbored on the smaller (−117 to +36) fragments. Thus, the sequences between −117 and −286 restored most, but not all, of the PhcA-mediated activation to xpsR promoter fragments with mutations in the PhcA binding site at nucleotide −77. Nonetheless, in all situations examined, full activation still required the presence of VsrD and its putative binding site.
TABLE 4.
Effect of upstream sequences on PhcA binding and transcription activation of PxpsR fragments with PhcA binding site mutations
| Plasmid | PxpsR nucleotide sequence fused to lacZ | Mutation | Expressiona directed from PxpsR in:
|
PhcA bindingb | ||
|---|---|---|---|---|---|---|
| WT | vsrD | phcA | ||||
| pRLZ1 | −338 to +36 | None | 5,737 | 469 | 86 | 100 |
| pRMB | −338 to +36 | −82 TAAAAA→AGATCT | 1,978 | 138 | 90 | 50 |
| pRM7 | −338 to +36 | −83 T→C | 903 | 56 | 43 | 35 |
| pRM7-M | −286 to +36 | Same as that for pRM7 | 225 | 185 | 64 | NT |
| pRMB-M | −286 to +36 | Same as that for pRMB | 290 | 214 | 83 | NT |
Expression (transcription) from PxpsR was monitored by measuring LacZ (β-galactosidase) activity in Miller units in cultures of R. solanacearum wild type (WT), AW1-80 (phcA mutant), and AW-D5 (vsrD mutant) carrying pRG970 vectors with PxpsR::lacZ fusions having the indicated mutation. Values are averages from three independent experiments with <20% variation.
To explore this phenomenon further, we used gel shift assays to compare PhcA binding to complete (−338 to +36) PxpsR fragments harboring mutant PhcA binding sites (Fig. 4A; summarized in Table 4) with its binding to analogous fragments lacking sequences upstream of nucleotide −117 (Fig. 3A; summarized in Table 3). Addition of sequences from nucleotides −118 to −338 greatly (more than sixfold) stimulated binding of PhcA to all PxpsR fragments, especially to the one with the −83 T→C mutation, where PhcA binding was increased from undetectable to >35% that of wild type. However, this binding differed from that observed to analogous wild-type fragments in that it was of lower affinity and highly cooperative (i.e., exhibited a sharp threshold). For example, with 4 μg of PhcA, nearly all of the −338 to +36 wild-type fragment was bound, whereas with 6 μg, there was no binding to either mutant fragment (Fig. 4A). However, increasing the amount of PhcA by only 66% caused complete binding of the fragment with the mutation at nucleotides −82 to −77. Similar behavior was observed for the fragment with the −83 T→C mutation. The same upstream sequences also appear to increase PhcA binding to fragments with wild-type PhcA binding sites, since quantification by phosphorimager that showed fourfold more PhcA is needed to retard the mobility of the −117 to +36 fragment relative to the −338 to +36 fragment (compare Fig. 3A and 4A). In all cases, the sequences between nucleotides −117 and −338 enhance binding and transcriptional activation by PhcA, suggesting that a second PhcA binding site upstream of −117 may be responsible for the restoration of PhcA activation to PxpsR fragments with mutant PhcA binding sites. It is plausible that this site stabilizes or enhances PhcA binding and activation mediated via the primary −77 site.
FIG. 4.
Gel mobility shift assays of PhcA binding to complete xpsR promoter fragments with mutations in the −77 PhcA binding site. (A) Labeled DNA fragments containing PxpsR sequences from −338 to +36 and having the indicated sequence alterations were prepared and incubated with various amounts of purified PhcA or with 20 μg of a similar protein preparation from E. coli lacking phcA (20Δ). (B) DNA fragments containing PxpsR sequences from −338 to −83 were labeled and either used directly as a binding substrate or first digested with XbaI (+Xba) before incubation with purified PhcA or control 20Δ as for panel A. Binding was detected and quantified as described in the legend to Fig. 3; results are summarized in Table 4. WT, wild type. The symbol > to the left of the gels indicates the position of retarded (bound) fragment.
To confirm this second site, we measured PhcA binding to complete PxpsR fragments lacking the primary PhcA binding site around −77. With 24 μg of PhcA, the PxpsR fragment with only nucleotides −338 to −83 showed weak and highly cooperative binding that could be detected only when >18 μg of PhcA was used (Fig. 4B and gels not shown); in comparison, binding to fragments harboring only the −77 site was detected when 1 μg of PhcA was used (Fig. 3A). When the −338 to −83 fragment was cleaved at −183 with XbaI, only the −338 to −183 fragment showed faint evidence of mobility retardation (Fig. 4B). Analysis of this and other gels by phosphorimager consistently showed that incubation with 20 μg of PhcA specifically reduced the amount of the −338 to −183 fragment migrating at the native position by >30%. A fragment with PxpsR sequences between −338 and −239 (from Sau3A digestion) showed no evidence of binding (data not shown). These data suggested that a weak PhcA binding site lies in the −239 to −183 region. Around −190 of PxpsR is a sequence (AATCPyTTA) that exactly matches the −77 to −70 portion of the primary PhcA binding site but that lacks other critical nucleotides (e.g., −83T).
Effect of increased separation between the putative VsrD and PhcA binding sites on PxpsR expression.
The above data suggest that when VsrD protein binds to a site around −315, it affects PhcA function at a site many helical turns downstream. To investigate how critical this spacing is to VsrD/PhcA-mediated activation, we inserted different lengths of spacer DNA (4 to 232 bp) into the XbaI site at −183 of wild-type PxpsR on reporter plasmid pRLZ1 (−338 to +36 of PxpsR fused to lacZ) and assayed the effect on phcA- and vsrD-mediated activation of transcription (Fig. 5). No insertion caused a change in PxpsR expression or regulation of greater than 60%. This effect is insignificant compared to effects of the deletion and substitution mutations described above. Thus, within these limits, the upstream location and distance of the putative VsrD binding site from the PhcA binding site are not critical for normal transcription activation and regulation of PxpsR.
DISCUSSION
xpsR plays a central role in the network that regulates production of the virulence factor EPS I by R. solanacearum (Fig. 1). Previous results (18) implied that the levels of XpsR protein (in conjunction with the response regulator VsrC) determine transcription levels of the eps biosynthetic operon. In turn, levels of XpsR protein are transcriptionally controlled by two independent, signal-responsive regulatory genes: vsrD, encoding a response regulator, and phcA, encoding a global, LysR-type regulator. Increased expression or activity of PhcA, which occurs in response to a cell density signal (3-OH palmitic acid methyl ester) transduced via the PhcSR two-component system (3, 4, 10), enhances transcription of xpsR sixfold. Activation of VsrD (presumably after its phosphorylation by the VsrA sensory kinase in response to an unknown signal) further increases xpsR transcription 12-fold (Fig. 2). Thus, input from two independent sources is summed and transduced into a single output via XpsR.
Here we focused on the mechanism by which the individual signal inputs transmitted through the PhcA and VsrAD systems are summed at PxpsR (Fig. 1). Deletion experiments showed that removal of PxpsR sequences between nucleotides −338 and −310 eliminated VsrAD-dependent activation of PxpsR and reduced, but did not eliminate, PhcA-mediated transcription activation. This suggested that PhcA itself can partially activate PxpsR and that VsrD binding to the −338 to −310 region enhances PhcA-mediated activation. However, it is possible that VsrD does not directly bind here but rather controls production or activity of a regulator that does. Nonetheless, two site-directed mutations that altered the structure of a palindrome centered around −315 (Fig. 2) destroyed activation of PxpsR by VsrD but not the activation mediated by PhcA alone. These results are consistent with the hypothesis that VsrD interacts with the CCCC-N6-GGGG dyad between nucleotides −322 and −307 of PxpsR to enhance its activation by PhcA.
Further deletion of sequences downstream of the putative VsrD binding site reduced PxpsR expression only 2.5-fold. However, when sequences between −117 and −76 were removed, a total loss of PhcA-mediated activation was observed. Moreover, we found that when sequences between −117 and +36 were fused to a reporter, they were sufficient to allow activation of PxpsR by PhcA, implying that these sequences contain an independently functioning PhcA binding and activation site. A likely site centered around −77 was confirmed and further defined by analysis of the effects of mutations in the site on in vivo expression from PxpsR and in vitro binding of PhcA to PxpsR. Mutations in the hypothesized −77 PhcA binding site caused a dramatic reduction in the ability of PhcA to bind to and activate transcription from PxpsR fragments lacking sequences upstream of −117. However, when the mutated PhcA binding sites were placed on complete PxpsR fragments (−338 to +36), high levels of in vivo activation and regulation by PhcA (and VsrD) were observed. Gel shift analyses showed that PhcA binding to these larger fragments was markedly enhanced, although the binding affinity of PhcA for them was still much less than that for wild-type fragments. This implied the existence of another PhcA binding site that can partially suppress the mutations in the primary −77 site. Consistent with this hypothesis, additional DNA binding studies tentatively confirmed the presence of a weak and highly cooperative PhcA binding site between nucleotides −240 and −183. In this region is found an 8-bp sequence (−194 to −187; AATCPyTTA) that exactly matches one-half of the −77 PhcA binding site; the role of this sequence in PhcA activation requires confirmation by site-directed mutagenesis.
These results suggest a model where active PhcA binds to a 14-bp site centered around nucleotide −77 of PxpsR (Fig. 2) and, as a direct result, increases PxpsR transcription at least sixfold. Additional, but less critical, interactions of PhcA with the second site near −190 may stabilize binding and somewhat enhance activation (compare pRLZ2 and pRLZ5) (Fig. 2). Much more important for maximal activation of PxpsR by PhcA is the interaction of signal-activated VsrD with a dyadic site much farther upstream. It is plausible that VsrD (or possibly its surrogate) binds to this dyad centered around position −315 and may increase or stabilize binding of PhcA and/or RNA polymerase. Whatever the mechanism, the observation that PhcA is a prerequisite for any significant expression of PxpsR suggests that PhcA can be thought of as an on-off switch which in the on mode activates a low level of transcription from PxpsR and also allows the possibility of high-level transcription. The extent to which this low-level transcription is turned up is governed by the VsrAD system acting like a rheostat or volume control.
Our experiments imply that VsrD and PhcA (and/or RNA polymerase) interact, even though they are separated by >12 helical turns. Moving the putative VsrD binding site 232 bp upstream did not dramatically affect PxpsR transcription activation or regulation. There are only a few examples of promoters like PxpsR which have distant and separation-insensitive cis-acting sites that bind different regulators (11). By analogy to two of these systems (NtrC and NifA [16, 27]), it is plausible that the DNA between the putative VsrD and PhcA binding sites at PxpsR may become bent, facilitating contacts between the regulatory proteins and/or RNA polymerase. Similar to promoters recognized by NtrC and NifA, additional regulatory proteins such as IHF may assist in DNA bending at PxpsR. Interestingly, there is a consensus IHF binding site (GATCAA-N4-CTG; −239 to −227) (16) between the putative VsrD and PhcA binding sites which requires investigation.
The marked dependence of PhcA on signal-activated VsrD for full transcription activation of PxpsR is unusual, since most other LysR-type regulators activate transcription independent of additional activators (31). PhcA also differs from LysR-type regulators in that it may not require a coinducer and has an unusual hydrophilic tail, a 25-residue, C-terminal extension largely comprised of highly polar residues (2). PhcA also regulates many other target promoters (35), and it will be important to determine if its activation of these is also modulated by additional regulators. Our data suggest that VsrD alone cannot activate PxpsR but rather must work through or with PhcA. In contrast, most other response regulators can directly turn on transcription of their target promoters (15). Moreover, few cases where cooperative regulation of a promoter by a response regulator and a different type of transcriptional activator have been documented (11, 15). Thus, VsrD may be a mechanistically unusual response regulator; whether its behavior at other promoters is as atypical remains to be seen.
ACKNOWLEDGMENTS
We thank K. E. Lee for preliminary work on interaction of VsrD and PhcA with PxpsR and T. Hoover for comments and criticisms on the manuscript.
This research was supported in part by a grant from the National Science Foundation (MCB 94-19582).
REFERENCES
- 1.Brumbley S M, Denny T P. Cloning of wild-type Pseudomonas solanacearum phcA, a gene that when mutated alters expression of multiple traits that contribute to virulence. J Bacteriol. 1990;172:5677–5685. doi: 10.1128/jb.172.10.5677-5685.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brumbley S M, Carney B F, Denny T P. Phenotype conversion in Pseudomonas solanacearum due to spontaneous inactivation of PhcA, a putative LysR transcriptional regulator. J Bacteriol. 1993;175:5477–5487. doi: 10.1128/jb.175.17.5477-5487.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clough S J, Schell M A, Denny T P. Differential expression of virulence genes and motility in Ralstonia (Pseudomonas) solanacearum during exponential growth. Appl Environ Microbiol. 1997;63:844–850. doi: 10.1128/aem.63.3.844-850.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clough S J, Schell M A, Denny T P. A two-component system in Ralstonia (Pseudomonas) solanacearum modulates production of PhcA-regulated virulence factors in response to 3-hydroxypalmitic acid methyl ester. J Bacteriol. 1997;179:3639–3648. doi: 10.1128/jb.179.11.3639-3648.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Denny T P, Carney B F, Schell M A. Inactivation of multiple virulence genes reduces the ability of Pseudomonas solanacearum to cause wilt symptoms. Mol Plant-Microbe Interact. 1990;3:293–300. [Google Scholar]
- 6.Denny T P, Baek S R. Genetic evidence that extracellular polysaccharide is a virulence factor of Pseudomonas solanacearum. Mol Plant-Microbe Interact. 1991;4:198–206. [Google Scholar]
- 7.Denny T P. Involvement of bacterial polysaccharides in plant pathogenesis. Annu Rev Plant Pathol. 1995;33:173–197. doi: 10.1146/annurev.py.33.090195.001133. [DOI] [PubMed] [Google Scholar]
- 8.Denny T P, Ganova-Raeva L, Huang J, Schell M A. Cloning and characterization of tek, the gene encoding the major extracellular protein of Pseudomonas solanacearum. Mol Plant-Microbe Interact. 1996;9:272–281. doi: 10.1094/mpmi-9-0272. [DOI] [PubMed] [Google Scholar]
- 9.Denny, T. P. Unpublished observations.
- 10.Flavier, A. B., S. J. Clough, M. A. Schell, and T. P. Denny. Identification of β-hydroxypalmitic acid methyl ester as a novel autoregulator controlling virulence in Ralstonia solanacearum. Mol. Microbiol., in press. [DOI] [PubMed]
- 11.Gralla J D, Collado-Vides J. Organization and function of transcription regulatory elements. In: Neidhardt F C, Curtis III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: ASM Press; 1996. pp. 1232–1245. [Google Scholar]
- 12.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 13.Hayward A C. Biology and epidemiology of bacterial wilt caused by Pseudomonas solanacearum. Annu Rev Phytopathol. 1991;29:65–87. doi: 10.1146/annurev.py.29.090191.000433. [DOI] [PubMed] [Google Scholar]
- 14.Hayward A C. Hosts of Pseudomonas solanacearum. In: Hayward A C, Hartman G L, editors. Bacterial wilt: the disease and its causative agent Pseudomonas solanacearum. Oxon, United Kingdom: CAB International; 1994. pp. 9–24. [Google Scholar]
- 15.Hoch J A, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. [Google Scholar]
- 16.Hoover T R, Santero E, Porter S C, Kustu S. The integration host factor stimulates the interaction of RNA polymerase with NIFA, the transcriptional activator for nitrogen fixation operons. Cell. 1990;63:11–22. doi: 10.1016/0092-8674(90)90284-l. [DOI] [PubMed] [Google Scholar]
- 17.Huang J, Denny T P, Schell M A. VsrB, a regulator of virulence genes of Pseudomonas solanacearum, is homologous to sensors of the two-component regulatory family. J Bacteriol. 1993;175:6169–6178. doi: 10.1128/jb.175.19.6169-6178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Huang J, Carney B, Denny T P, Weissinger A K, Schell M A. A complex network regulates expression of eps and other virulence genes of Pseudomonas solanacearum. J Bacteriol. 1995;177:1259–1267. doi: 10.1128/jb.177.5.1259-1267.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang J, Schell M A. Characterization of the eps gene cluster of Pseudomonas solanacearum and its transcriptional regulation via a single promoter. Mol Microbiol. 1995;16:977–989. doi: 10.1111/j.1365-2958.1995.tb02323.x. [DOI] [PubMed] [Google Scholar]
- 20.Keen N T, Tamaki S, Kobayashi D, Trollinger D. Improved broad-host-range plasmids for DNA cloning in Gram-negative bacteria. Gene. 1988;70:191–197. doi: 10.1016/0378-1119(88)90117-5. [DOI] [PubMed] [Google Scholar]
- 21.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1982. [Google Scholar]
- 22.Mead D A, Skorupa E S, Kemper B. Single-stranded DNA promoter plasmids for engineering mutant RNAs and protein: synthesis of a ‘stretched’ parathyroid hormone. Nucleic Acids Res. 1985;13:1103–1108. doi: 10.1093/nar/13.4.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Miller J H. Experiments in molecular genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 24.Muhlard D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- 25.Orgambide G, Montrozier H, Servin P, Roussel J, Trigalet-Demery D, Trigalet A. High heterogeneity of the exopolysaccharides of Pseudomonas solanacearum strain GM1000 and the complete structure of the major polysaccharide. J Biol Chem. 1991;266:8312–8321. [PubMed] [Google Scholar]
- 26.Parkinson J S, Kofoid E C. Communication modules in bacterial signaling proteins. Annu Rev Genet. 1992;26:71–112. doi: 10.1146/annurev.ge.26.120192.000443. [DOI] [PubMed] [Google Scholar]
- 27.Porter S C, North A K, Kustu S. Mechanism of transcription activation by NtrC. In: Hoch J, Silhavy T J, editors. Two-component signal transduction. Washington, D.C: ASM Press; 1995. pp. 147–158. [Google Scholar]
- 28.Saile E, McGarvey J, Schell M A, Denny T P. Role of extracellular polysaccharide and endoglucanase in root invasion and colonization of tomato plants by Ralstonia solanacearum. Phytopathology. 1998;87:1264–1271. doi: 10.1094/PHYTO.1997.87.12.1264. [DOI] [PubMed] [Google Scholar]
- 29.Schell M A. Purification and characterization of an excreted endoglucanase from Pseudomonas solanacearum. Appl Environ Microbiol. 1987;53:2237–2241. doi: 10.1128/aem.53.9.2237-2241.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schell M A, Poser E. Demonstration, characterization, and mutational analysis of NahR protein binding to the nah and sal promoters. J Bacteriol. 1989;171:837–846. doi: 10.1128/jb.171.2.837-846.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schell M A. Molecular biology of the LysR family of transcriptional regulators. Annu Rev Microbiol. 1993;47:597–626. doi: 10.1146/annurev.mi.47.100193.003121. [DOI] [PubMed] [Google Scholar]
- 32.Schell M A, Denny T P, Clough S J, Huang J. Further characterization of genes encoding extracellular polysaccharide of Pseudomonas solanacearum and their regulation. In: Nester E W, Verma D P S, editors. Advances in molecular genetics of plant-microbe interactions. Vol. 2. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1993. pp. 231–239. [Google Scholar]
- 33.Schell M A, Denny T P, Huang J. VsrA, a second two-component sensor regulating virulence genes of Pseudomonas solanacearum. Mol Microbiol. 1994;11:489–500. doi: 10.1111/j.1365-2958.1994.tb00330.x. [DOI] [PubMed] [Google Scholar]
- 34.Schell M A, Denny T P, Huang J. Extracellular virulence factors of Pseudomonas solanacearum: role in disease and regulation of expression. In: Kado C I, Crosa J H, editors. Molecular mechanisms of bacterial virulence. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1994. pp. 311–324. [Google Scholar]
- 35.Schell M A. To be or not to be: how Pseudomonas solanacearum decides whether or not to express virulence genes. Eur J Plant Pathol. 1996;102:459–469. [Google Scholar]
- 36.Schell, M. A., and J. Huang. Unpublished observations.
- 37.Schmit J. Proceedings of the Fourth International Conference on Plant Pathogenic Bacteria, Anger, France. 1978. Microscopic study of early stages of infection by Pseudomonas solanacearum EFS on “in vitro” grown tomato seedlings; pp. 841–857. [Google Scholar]
- 38.Studier F W, Rosenberg A, Dunn J, Dubendorf J. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- 39.Van den Eede G, Deblaere R, Goethals K, Van Montegue M, Holsters M. Broad host range and promoter selection vectors for bacteria that interact with plants. Mol Plant-Microbe Interact. 1992;5:228–234. doi: 10.1094/mpmi-5-228. [DOI] [PubMed] [Google Scholar]
- 40.Vasse J, Pascal F, Trigalet A. Microscopic studies of intercellular infection and protoxylem invasion of tomato roots by Pseudomonas solanacearum. Mol Plant-Microbe Interact. 1995;8:241–251. [Google Scholar]
- 41.Wallis F M, Truter S J. Histopathology of tomato plants infected with Pseudomonas solanacearum with emphasis on ultrastructure. Physiol Plant Pathol. 1978;13:307–317. [Google Scholar]
- 42.Yabuuchi E, Kosako Y, Oyaizu H, Yano I, Hotta H, Hashimoto Y, Ezaki T, Arakawa T. Proposal of Burkholderia gen. nov. and transfer of seven species of the genus Pseudomonas group II to the new genus with the type species Burkholderia cepacia Palleroni and Holmes 1981 comb. nov. Microbiol Immunol. 1992;36:1251–1275. doi: 10.1111/j.1348-0421.1992.tb02129.x. [DOI] [PubMed] [Google Scholar]
- 43.Yabuuchi E, Kosako Y, Yano I, Hotta H, Nishiuchi Y. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii Ralston, Palleroni and Doudoroff 1973 comb. nov., Ralstonia solanacearum Smith 1896 comb. nov. and Ralstonia eutropha Davis 1969 comb. nov. Microbiol Immunol. 1995;39:897–904. doi: 10.1111/j.1348-0421.1995.tb03275.x. [DOI] [PubMed] [Google Scholar]