Abstract
rRNA plays a central role in protein synthesis and is intimately involved in the initiation, elongation, and termination stages of translation. However, the mode of its participation in these reactions, particularly as to the decoding of genetic information, remains elusive. In this paper, we describe a new approach that allowed us to identify an rRNA segment whose function is likely to be related to translation termination. By screening an expression library of random rRNA fragments, we identified a fragment of the Escherichia coli 23S rRNA (nucleotides 74 to 136) whose expression caused readthrough of UGA nonsense mutations in certain codon contexts in vivo. The antisense RNA fragment produced a similar effect, but in neither case was readthrough of UAA or UAG observed. Since termination at UGA in E. coli specifically requires release factor 2 (RF2), our data suggest that the fragments interfere with RF2-dependent termination.
The rRNAs have been implicated in all three stages of translation (1, 4, 8, 16, 22, 30, 33), and some experiments suggest a direct catalytic participation in peptide bond formation (29). The involvement of rRNA in the last stage of translation, peptide chain termination, was indicated by several studies (1, 4, 8, 22, 33). In a ribosome that has just completed translation of an mRNA into a protein, termination occurs when one of the three termination codons encounters the decoding site (A site) and a release factor (RF) binds to the ribosome and triggers the hydrolysis of the ester bond between the last tRNA and the completed polypeptide (35). In Escherichia coli, there are two termination codon-dependent RFs: RF1 works at UAA and UAG, and RF2 functions at UAA and UGA (35). Large rRNAs have been implicated in RF binding (4, 8) and catalysis of peptidyl-tRNA hydrolysis during termination (1).
The complexity of the large rRNAs makes it difficult to study their structures and functions in the ribosome, especially considering that rRNA folding and performance can be influenced by the numerous ribosomal proteins. However, some rRNA fragments seem to have properties of the whole ribosome, such as the ability to interact with translation factors (26), antibiotics (14, 21, 34), or mRNA and tRNA analogs (34). To simplify the analysis of rRNA involvement in termination, we screened an rRNA random fragment library (37) to identify rRNA fragments that allowed readthrough of a termination codon in vivo, presumably due to inhibition of translation termination. Such rRNA fragments may interfere with the binding of RF to the ribosome or with the correct positioning and folding of rRNA segments that are important for normal termination.
In this paper, we report that the expression of a small fragment of E. coli 23S rRNA or its antisense causes ribosomes to read through UGA, but not UAA or UAG, in vivo. Since termination at UGA in E. coli is driven by RF2, our data suggest that the fragments interfere with RF2-dependent termination.
MATERIALS AND METHODS
Library screening.
The construction of the rRNA random fragment library that expresses rRNA fragments from the tac promoter in the plasmid pPOT1 has been described elsewhere (37). E. coli AL1 [glyV55 recA/F′ trpA(UGA115)] has a UGA nonsense mutation at position 115 of trpA (28), the gene for the α subunit of tryptophan synthetase, and therefore requires readthrough of that UGA to grow on medium without tryptophan (Trp). The mutant trpA(UGA115) was derived in one step from the ocher mutant trpA7 (40), kindly provided by V. Horn and C. Yanofsky. DNA sequence analysis of the ocher mutant trpA gene revealed the ocher codon TAA at codon position 115 (32). The TAA115 was then converted to TGA115 in vivo, in the presence of a glycine tRNA mutant that suppresses UGA mutations (32). AL1 cells transformed with the plasmid library were grown on glucose minimal medium (GM) supplemented with 10 μg of indole (Ind) per ml and 100 μg of ampicillin per ml (27). Since the trpB polypeptide, namely, the tryptophan synthetase β subunit, can convert Ind to Trp, the AL1 transformants were able to grow on GM with Ind regardless of whether the UGA in trpA was read through or not. The transformants were then screened by replica plating to GM containing ampicillin and 800 μg of isopropyl-β-d-thiogalactopyranoside (IPTG), an inducer of transcription from the tac promoter, applied to the surface of 30 ml of the solid medium contained in one plate. The screening was performed at three different temperatures, 25, 31, and 37°C, to accommodate the possibility that some fragments might acquire an active conformation at temperatures other than 37°C. However, the only IPTG-dependent Trp+ clone that we found was obtained at 37°C (see Results).
In vivo tests.
The UGA-suppressing RNA fragment and its antisense were tested for readthrough of all three nonsense codons at each of four codon positions in trpA, 15, 115, 211, and 234, as well as of UAG at position 243 (the UAA and UGA codons were not available). The E. coli strains used for the tests contained the mutant trpA genes on the Fredericq episome (15) and have been described elsewhere (27). The mutations used for the frameshift tests were the +1 mutations trpA8 (38) and trpE9777 (3), on the chromosome, and the −1 mutation trpE91, on an F′ (2). The method for detection of readthrough and frameshift on plates has been described elsewhere (6, 38) and was used here with some modifications as follows. Each mutant strain was transformed with the fragment-containing pPOT1 or pPOT19 (pPOT19 is identical to pPOT1 except for the two mutations in the replication origin that increase the plasmid copy number about 10-fold [data not shown]). As controls, pPOT1 and pPOT19 (without inserts) were introduced into each mutant strain. The transformants were selected on L agar with ampicillin at 37°C and then single colony purified at 37°C on GM with Ind and with 19 amino acids other than Trp plus ampicillin, and patches from each strain were placed on the growth plate (GM supplemented with Ind and ampicillin). For the experiments with pPOT1 constructs, the patches were grown at 37°C. For the experiments where strains containing the pPOT19 constructs were compared with those containing pPOT1 constructs, the patches were grown at 31°C. The plates with grown patches were replicated to a series of plates in the following order: (i) GM supplemented with a low level of Trp, 40 ng/ml, plus ampicillin; (ii) the same as plate i plus 1 mM IPTG; (iii) GM with Ind, ampicillin, and 1 mM IPTG; and (iv) GM with Ind and ampicillin. The indication of readthrough or frameshifting was the observation of IPTG-induced growth on GM with low Trp and ampicillin (see Results). To grow efficiently on GM with the low level of Trp, strains with the nonsense mutations in trpA required readthrough of the nonsense codons and the frameshift mutants required frameshifting. This medium proved to be more sensitive in displaying the Trp+ phenotype than just GM because, to grow efficiently on GM with this little an amount of exogenous Trp, the bacteria needed to synthesize less Trp than they did on GM without Trp. Growth on the Ind plates corresponded to general growth since neither readthrough of nonsense codons in trpA nor frameshifting in the frameshift mutants was required for growth on these plates. For the experiments with the pPOT1 constructs, the replica plates were incubated at 37°C for up to 6 days. The replica plates with strains containing the pPOT19 constructs together with the pPOT1 constructs were incubated at 31°C for up to 8 days. As expected, neither pPOT1 nor pPOT19 by itself caused either readthrough or frameshifting in the presence of IPTG (see Results and data not shown).
RESULTS
Identification of an rRNA fragment that causes UGA readthrough.
To identify fragments of rRNA that inhibit termination, we screened a random rRNA fragment library (37). The library expresses rRNA fragments from the tac promoter in the plasmid pPOT1 in the presence of IPTG, an inducer of transcription from the tac promoter. The rRNA fragments transcribed from the tac promoter are flanked by short vector sequences that form hairpin structures, one an 8-bp stem-loop at the beginning of the transcript, the other a 7-bp stem-loop downstream, corresponding to the trp terminator (see Fig. 1B in reference 37). The small hairpins should not interfere with folding of the inserted rRNA fragments and may even increase the stability of the transcript (13, 25, 37). The plasmid library was introduced into E. coli AL1, which, to grow on medium without Trp, required readthrough of UGA at codon position 115 (UGA115) of the reporter trpA mRNA (Fig. 1). Of approximately 40,000 transformants screened, only one required IPTG to grow on medium without Trp, i.e., showed an IPTG-dependent Trp+ phenotype. Isolation of the plasmid from this clone and reintroduction of it into AL1 conferred on that strain the ability to grow on medium without Trp. That ability was IPTG dependent, indicating that the Trp+ phenotype was caused by the transcription of a plasmid-borne ribosomal gene segment from the tac promoter. Indeed, sequence analysis revealed that a 63-nucleotide fragment from domain I of 23S rRNA (nucleotides 74 to 136) in direct orientation (Fig. 2) was inserted immediately downstream of the tac promoter. To ensure that no mutations that might have spontaneously occurred in the vector portion of the fragment-containing plasmid (isolated from the library) were responsible for the readthrough of UGA, the fragment was reintroduced into the pPOT1 vector. The IPTG-induced expression of the recloned fragment caused readthrough of UGA115. Consistent with its functional importance, the nucleotide 74–136 segment contained several conserved nucleotides (Fig. 2). The segment has been proposed to be involved in formation of a pseudoknot (18, 19, 23) via RNA-RNA tertiary interaction as indicated in Fig. 2 (top).
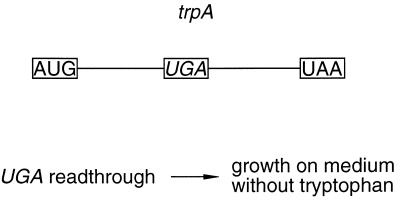
FIG. 1.
Diagram of the reporter system used for screening a random rRNA fragment library. In the reporter gene, trpA, the codon UAU (codes for Tyr) at codon position 115 (28) is replaced by UGA (see Materials and Methods). Readthrough of the UGA nonsense codon is required for growth on medium lacking tryptophan.
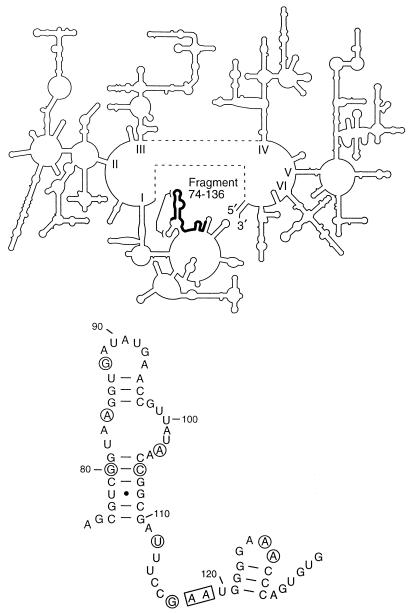
FIG. 2.
Location of the 23S rRNA fragment isolated from the library. (Top) The secondary structure of 23S rRNA with the 74–136 segment indicated in boldface. A proposed RNA-RNA tertiary interaction (18, 19, 23) that involves the segment is indicated. (Bottom) The 74–136 fragment in detail. The nucleotides absolutely conserved among bacteria (9) are circled. The universally conserved nucleotides (24) are boxed.
Readthrough and frameshifting tests.
We tested the ability of the UGA-suppressing fragment to cause readthrough of all three termination codons, represented by nonsense mutations at several positions in trpA (see Materials and Methods). The ability of the fragment to cause ribosomal frameshifts was tested with three strains that require their ribosomes to shift translational frame in mutant trpA or trpE genes to be Trp+ (see Materials and Methods). Expression of the fragment in pPOT1 caused readthrough of UGA at two codon positions, 15 and 115, of trpA but not of UAA or UAG at the same positions. No readthrough was observed with nonsense codons at three other positions in trpA, and the fragment did not cause frameshifts.
Since the level of fragment expression from pPOT1 (present at about 20 copies per cell) might not be sufficient to register either readthrough of UAA and UAG or frameshift, we cloned the fragment into the higher-copy-number plasmid pPOT19, which is identical to pPOT1 (37) except for the two mutations in the replication origin that increase the plasmid copy number about 10-fold. The IPTG-induced expression of the fragment in pPOT19 was lethal to bacteria at 37°C but not at 31°C. Therefore, the tests of the ability of the fragment cloned in pPOT19 to cause readthrough of nonsense codons and frameshifts were performed at 31°C, in parallel with the same tests for the fragment in pPOT1 at 31°C. As was true of fragment expression from pPOT1 at 37°C, readthrough of UGA but not UAA or UAG at the two trpA positions (15 and 115) previously examined was observed with the fragment expressed from both pPOT1 and pPOT19 at 31°C. Readthrough of UGA15 was much more efficient when the fragment was expressed from pPOT19 than when it was expressed from pPOT1 (data not shown), indicating that the amount of fragment produced from pPOT19 was indeed higher than that from pPOT1. As was observed with the fragment in pPOT1 at 37°C, expression of the fragment at 31°C from either pPOT1 or pPOT19 resulted neither in readthrough of any of the nonsense codons available at three other positions (211, 234, and 243) in trpA nor in detectable ribosome frameshifting. Failure of the fragment to cause readthrough of UGA at positions 211 and 234 (UGA was not available at position 243) may be another example of the well-documented phenomenon referred to as codon context effects (reviewed in references 5, 6, and 36), in which nucleotides adjacent to stop codons, i.e., the mRNA context, in some way affect the efficiency of stop codon readthrough. The sequences immediately preceding and following codon positions 211 and 234 contain differences from those associated with positions 15 and 115 (28) that are potentially relevant to termination or readthrough efficiency.
The results described here suggest that the expression of the rRNA fragment caused a defect in termination of translation at UGA rather than a decrease in the general accuracy of translation. If the fragment had decreased general ribosomal accuracy, one would have seen readthrough of all stop codons, as well as frameshifting, in the presence of the fragment. The validity of this reasoning was verified recently for two rRNA mutants that cause UGA-specific readthrough in vivo and were observed, in a new in vitro termination assay, to be preferentially defective in RF2-dependent peptidyl-tRNA hydrolysis (1).
UGA readthrough caused by the antisense fragment.
If the 23S rRNA segment corresponding to the fragment (nucleotides 74 to 136) is normally involved in UGA termination in the ribosome, one may expect that an antisense RNA, complementary to that segment, can cause UGA readthrough. Consequently, the DNA fragment that corresponds to that rRNA segment was cloned in the reverse orientation under the tac promoter in pPOT1 and pPOT19. Readthrough of the same nonsense codons tested with the sense fragment was tested with the antisense fragment. The ability of the antisense fragment to cause frameshift in all the strains used for the frameshift test with the sense fragment was determined also. The antisense fragment cloned in pPOT1, when expressed from the tac promoter, caused readthrough of UGA115 but not of UAA or UAG at the same position (Fig. 3). Figure 3 also shows UGA115 readthrough caused by the sense fragment, which was detectable after 1 day of growth. UGA readthrough caused by the antisense fragment was weaker than that caused by the sense fragment and was detectable after 2 days of growth. The antisense fragment caused neither readthrough of the nonsense codons at the other positions nor frameshifting. The antisense fragment cloned into pPOT19 caused readthrough of UGA115 but not of the other nonsense codons, and it did not cause frameshifting (data not shown).
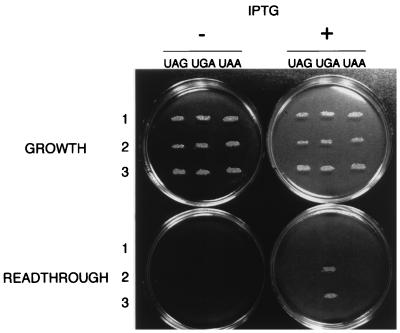
FIG. 3.
UGA readthrough caused by the antisense and sense fragments. “GROWTH” plates are the GM plates supplemented with Ind (see Materials and Methods). On these plates, general growth is monitored. “READTHROUGH” plates are the GM plates supplemented with a low level of Trp, to detect readthrough (see Materials and Methods). Bacterial patches growing on an Ind plate were replicated to the low-Trp plates (with and without IPTG) and to Ind plates (with and without IPTG) (see Materials and Methods). The “−” column corresponds to the plates without IPTG, and the “+” column corresponds to the plates with 1 mM IPTG. The “UAG,” “UGA,” and “UAA” columns correspond to the isogenic strains with the UAG, UGA, and UAA nonsense codons, respectively, at position 115 of trpA. Rows 1, 2, and 3 correspond to the strains transformed with pPOT1, pPOT1 with the sense fragment, and pPOT1 with the antisense fragment, respectively. The “GROWTH” plates were photographed after 24 h of incubation at 37°C. The “READTHROUGH” plates were photographed after 42 h of incubation at 37°C. (A negative of the photograph was scanned [SprintScan 35; Polaroid], and the figure was generated with the help of Adobe Photoshop 3.0 [Macintosh].)
DISCUSSION
We used a library expressing random rRNA fragments to identify those that may affect ribosome functions. Conceptually, this approach is similar to a selection of DNA fragments encoding either peptides that act as dominant inhibitors of protein function or antisense RNA fragments that inhibit gene expression (17, 20). In experiments described in this report, we looked for rRNA fragments that can cause readthrough of termination codons, that is, suppression of nonsense mutations in trpA.
The data presented in this study show that the expression of a small fragment of E. coli 23S rRNA and its antisense causes ribosomes to read through one of the termination codons, UGA, but not the other two, UAA and UAG, in vivo. Since RF2 is the only RF required for termination at UGA, the data are consistent with the possibility that both fragments interfere with RF2-dependent termination, as has been demonstrated for rRNA mutations that cause UGA-specific readthrough (1).
The ability of both the sense and the antisense fragments to cause readthrough of UGA effectively argues against the possibility that some peptides encoded in both the sense and the antisense fragments are produced and cause the readthrough. Although from the sequence of the sense construct one can deduce several open reading frames, the only peptide that might be produced from the antisense construct is dissimilar to all of the several hypothetical peptides from the sense construct (data not shown). Furthermore, each of the potential peptide-encoding sequences, in either construct, has an apparently weak ribosome-binding (Shine-Dalgarno) site and so should not be translatable efficiently. Therefore, it is most likely that both the sense and antisense RNA fragments themselves interfered with termination.
One can suggest a few different mechanisms by which the fragments may cause UGA readthrough. First, the sense fragment may mimic a binding site for RF2 and compete with the ribosome for binding to the RF. The antisense fragment then may prevent RF2 binding to the ribosome by base pairing with this putative binding site for RF2. Another possibility is that the sense fragment may titrate a ribosomal protein that may be needed for termination at UGA and the antisense fragment may prevent binding of that ribosomal protein to the ribosome by base pairing with the natural binding site for this protein. Indeed, ribosomal proteins L24 and L29 have been cross-linked to the 74–136 segment (31); however, such experiments indicate proximity of the proteins to the segment, not necessarily contact. Furthermore, footprint experiments on the accessibility of domain I of 23S rRNA to chemical reagents and RNases in the presence of L24 strongly indicate that the 74–136 segment is not a binding site for L24 (10). There is, in fact, no direct evidence that the 74–136 segment is a binding site for any ribosomal protein. Finally, the fragments may impair termination by interfering with the correct positioning and folding of the 74–136 segment of rRNA during ribosome assembly.
The 74–136 segment of E. coli 23S rRNA corresponds to a part of 5.8S rRNA in eukaryotes. The 5.8S rRNA has been implicated in translation (11, 12, 39). In particular, it was shown that mutations at or near the region of Schizosaccharomyces pombe 5.8S rRNA that corresponds to the 74–136 segment from E. coli 23S rRNA inhibited cell growth and in vitro protein synthesis (11). The possible functional importance of the E. coli 74–136 segment, as suggested by the studies of 5.8S rRNA (11, 12, 39), is supported by the recently identified proximity of the segment to ribosome-bound tRNA (7) and by the results of our study, which demonstrate that the 74–136 fragment and its antisense enhance UGA readthrough in vivo.
ACKNOWLEDGMENTS
We thank Måns Ehrenberg and Lev Kisselev for critical reading of the manuscript and for discussions. We also thank Klas Hedenstierna for help with generating Fig. 2 and for discussions. We are grateful to Vildan Dinçbas, David Freistroffer, Diarmaid Hughes, Reza Karimi, Frances Pagel, Johan Paulsson, Michael Pavlov, Tanel Tenson, Wenbing Xu, and Song Zhao for discussions and Walter J. Pagel for expert editorial consultation.
This work was supported by a grant to E.J.M. from the National Institute of General Medical Sciences (GM21499) and a grant to A.M. from the National Science Foundation (MCB9420768).
REFERENCES
- 1.Arkov A L, Freistroffer D V, Ehrenberg M, Murgola E J. Mutations in RNAs of both ribosomal subunits cause defects in translation termination. EMBO J. 1998;17:1507–1514. doi: 10.1093/emboj/17.5.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atkins J F, Nichols B P, Thompson S. The nucleotide sequence of the first externally suppressible −1 frameshift mutant, and of some nearby leaky frameshift mutants. EMBO J. 1983;2:1345–1350. doi: 10.1002/j.1460-2075.1983.tb01590.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bronson M J, Yanofsky C. Characterization of mutations in the tryptophan operon of Escherichia coli by RNA nucleotide sequencing. J Mol Biol. 1974;88:913–916. doi: 10.1016/0022-2836(74)90407-0. [DOI] [PubMed] [Google Scholar]
- 4.Brown C M, McCaughan K K, Tate W P. Two regions of the Escherichia coli 16S ribosomal RNA are important for decoding stop signals in polypeptide chain termination. Nucleic Acids Res. 1993;21:2109–2115. doi: 10.1093/nar/21.9.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buckingham R H. Codon context. Experientia. 1990;46:1126–1133. doi: 10.1007/BF01936922. [DOI] [PubMed] [Google Scholar]
- 6.Buckingham R H, Sörensen P, Pagel F T, Hijazi K A, Mims B H, Brechemier-Baey D, Murgola E J. Third position base changes in codons 5′ and 3′ adjacent UGA codons affect UGA suppression in vivo. Biochim Biophys Acta. 1990;1050:259–262. doi: 10.1016/0167-4781(90)90177-4. [DOI] [PubMed] [Google Scholar]
- 7.Bullard J M, van Waes M A, Bucklin D J, Hill W E. Regions of 23 S ribosomal RNA proximal to transfer RNA bound at the P and E sites. J Mol Biol. 1995;252:572–582. doi: 10.1006/jmbi.1995.0521. [DOI] [PubMed] [Google Scholar]
- 8.Caskey C T, Bosch L, Konecki D S. Release factor binding to ribosome requires an intact 16S rRNA 3′ terminus. J Biol Chem. 1977;252:4435–4437. [PubMed] [Google Scholar]
- 9.de Peer Y V, Chapelle S, De Wachter R. A quantitative map of nucleotide substitution rates in bacterial rRNA. Nucleic Acids Res. 1996;24:3381–3391. doi: 10.1093/nar/24.17.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Egebjerg J, Leffers H, Christensen A, Andersen H, Garrett R A. Structure and accessibility of domain I of Escherichia coli 23S RNA in free RNA, in the L24-RNA complex and in 50S subunits. J Mol Biol. 1987;196:125–136. doi: 10.1016/0022-2836(87)90515-8. [DOI] [PubMed] [Google Scholar]
- 11.Elela S A, Good L, Melekhovets Y F, Nazar R N. Inhibition of protein synthesis by an efficiently expressed mutation in the yeast 5.8S ribosomal RNA. Nucleic Acids Res. 1994;22:686–693. doi: 10.1093/nar/22.4.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elela S A, Nazar R N. Role of the 5.8S rRNA in ribosome translocation. Nucleic Acids Res. 1997;25:1788–1794. doi: 10.1093/nar/25.9.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emory S A, Bouvet P, Belasco J G. A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992;6:135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- 14.Fourmy D, Recht M I, Blanchard S C, Puglisi J D. Structure of the A site of Escherichia coli 16S ribosomal RNA complexed with an aminoglycoside antibiotic. Science. 1996;274:1367–1371. doi: 10.1126/science.274.5291.1367. [DOI] [PubMed] [Google Scholar]
- 15.Fredericq P. The recombination of colicinogenic factors with other episomes and plasmids. In: Wolstenholme G E, O’Connor M, editors. Bacterial plasmids and episomes. Boston, Mass: Little, Brown & Co.; 1969. pp. 163–174. [Google Scholar]
- 16.Gualerzi C O, Pon C L. mRNA-ribosome interaction during initiation of protein synthesis. In: Zimmermann R A, Dahlberg A E, editors. Ribosomal RNA: structure, evolution, processing, and function in protein biosynthesis. Boca Raton, Fla: CRC Press, Inc.; 1996. pp. 259–276. [Google Scholar]
- 17.Gudkov A V, Kazarov A R, Thimmapaya R, Axenovich S A, Mazo I A, Roninson I B. Cloning mammalian genes by expression selection of genetic suppressor elements: association of kinesin with drug resistance and cell immortalization. Proc Natl Acad Sci USA. 1994;91:3744–3748. doi: 10.1073/pnas.91.9.3744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gutell R R, Larsen N, Woese C R. Lessons from an evolving rRNA: 16S and 23S rRNA structures from a comparative perspective. Microbiol Rev. 1994;58:10–26. doi: 10.1128/mr.58.1.10-26.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gutell R R, Woese C R. Higher order structural elements in ribosomal RNAs: pseudo-knots and the use of noncanonical pairs. Proc Natl Acad Sci USA. 1990;87:663–667. doi: 10.1073/pnas.87.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holzmayer T A, Pestov D G, Roninson I B. Isolation of dominant negative mutants and inhibitory antisense RNA sequences by expression selection of random DNA fragments. Nucleic Acids Res. 1992;20:711–717. doi: 10.1093/nar/20.4.711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Howard B, Thom G, Jeffrey I, Colthurst D, Knowles D, Prescott C. Fragmentation of the ribosome to investigate RNA-ligand interactions. Biochem Cell Biol. 1995;73:1161–1166. doi: 10.1139/o95-125. [DOI] [PubMed] [Google Scholar]
- 22.Jemiolo D K, Pagel F T, Murgola E J. UGA suppression by a mutant RNA of the large ribosomal subunit. Proc Natl Acad Sci USA. 1995;92:12309–12313. doi: 10.1073/pnas.92.26.12309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leffers H, Kjems J, Østergaard L, Larsen N, Garrett R A. Evolutionary relationships amongst archaebacteria. A comparative study of 23S ribosomal RNAs of a sulphur-dependent extreme thermophile, an extreme halophile and a thermophilic methanogen. J Mol Biol. 1987;195:43–61. doi: 10.1016/0022-2836(87)90326-3. [DOI] [PubMed] [Google Scholar]
- 24.Maidak B L, Olsen G J, Larsen N, Overbeek R, McCaughey M J, Woese C R. The RDP (Ribosomal Database Project) Nucleic Acids Res. 1997;25:109–110. doi: 10.1093/nar/25.1.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLaren R S, Newbury S F, Dance G S C, Causton H C, Higgins C F. mRNA degradation by processive 3′-5′ exoribonucleases in vitro and implications for prokaryotic mRNA decay in vivo. J Mol Biol. 1991;221:81–95. [PubMed] [Google Scholar]
- 26.Munishkin A, Wool I G. The ribosome-in-pieces: binding of elongation factor EF-G to oligoribonucleotides that mimic the sarcin/ricin and thiostrepton domains of 23S ribosomal RNA. Proc Natl Acad Sci USA. 1997;94:12280–12284. doi: 10.1073/pnas.94.23.12280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Murgola E J, Pagel F T, Hijazi K A, Arkov A L, Xu W, Zhao S Q. Variety of nonsense suppressor phenotypes associated with mutational changes at conserved sites in Escherichia coli ribosomal RNA. Biochem Cell Biol. 1995;73:925–931. doi: 10.1139/o95-100. [DOI] [PubMed] [Google Scholar]
- 28.Nichols B P, Yanofsky C. Nucleotide sequences of trpA of Salmonella typhimurium and Escherichia coli: an evolutionary comparison. Proc Natl Acad Sci USA. 1979;76:5244–5248. doi: 10.1073/pnas.76.10.5244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Noller H F, Hoffarth V, Zimniak L. Unusual resistance of peptidyl transferase to protein extraction procedures. Science. 1992;256:1416–1419. doi: 10.1126/science.1604315. [DOI] [PubMed] [Google Scholar]
- 30.Noller H F, Powers T, Allen P N, Moazed D, Stern S. rRNA and translation: tRNA selection and movement in the ribosome. In: Zimmermann R A, Dahlberg A E, editors. Ribosomal RNA: structure, evolution, processing, and function in protein biosynthesis. Boca Raton, Fla: CRC Press, Inc.; 1996. pp. 259–276. [Google Scholar]
- 31.Osswald M, Greuer B, Brimacombe R. Localization of a series of RNA-protein cross-link sites in the 23S and 5S ribosomal RNA from Escherichia coli, induced by treatment of 50S subunits with three different bifunctional reagents. Nucleic Acids Res. 1990;18:6755–6760. doi: 10.1093/nar/18.23.6755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pagel, F. T., and E. J. Murgola. Unpublished data.
- 33.Pagel F T, Zhao S Q, Hijazi K A, Murgola E J. Phenotypic heterogeneity of mutational changes at a conserved nucleotide in 16S ribosomal RNA. J Mol Biol. 1997;267:1113–1123. doi: 10.1006/jmbi.1997.0943. [DOI] [PubMed] [Google Scholar]
- 34.Purohit P, Stern S. Interaction of a small RNA with antibiotic and RNA ligands of the 30S subunit. Nature. 1994;370:659–662. doi: 10.1038/370659a0. [DOI] [PubMed] [Google Scholar]
- 35.Tate W P, Poole E S, Mannering S A. Hidden infidelities of the translational stop signal. Prog Nucleic Acid Res Mol Biol. 1996;52:293–335. doi: 10.1016/s0079-6603(08)60970-8. [DOI] [PubMed] [Google Scholar]
- 36.Tate W P, Mannering S A. Three, four or more: the translational stop signal at length. Mol Microbiol. 1996;21:213–219. doi: 10.1046/j.1365-2958.1996.6391352.x. [DOI] [PubMed] [Google Scholar]
- 37.Tenson T, DeBlasio A, Mankin A. A functional peptide encoded in the Escherichia coli 23S rRNA. Proc Natl Acad Sci USA. 1996;93:5641–5646. doi: 10.1073/pnas.93.11.5641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tucker S D, Murgola E J, Pagel F T. Missense and nonsense suppressors can correct frameshift mutations. Biochimie. 1989;71:729–739. doi: 10.1016/0300-9084(89)90089-8. [DOI] [PubMed] [Google Scholar]
- 39.Walker K, Elela S A, Nazar R N. Inhibition of protein synthesis by anti-5.8S rRNA oligodeoxyribonucleotides. J Biol Chem. 1990;265:2428–2430. [PubMed] [Google Scholar]
- 40.Yanofsky C, Helinski D R, Maling B D. The effects of mutation on the composition and properties of the A protein of Escherichia coli tryptophan synthetase. Cold Spring Harbor Symp Quant Biol. 1961;26:11–23. doi: 10.1101/sqb.1961.026.01.006. [DOI] [PubMed] [Google Scholar]