Abstract
Pulsed-field gel electrophoresis was used to analyze the genomic organization of 16 bacteria belonging or related to the family Rhizobiaceae of the alpha subgroup of the class Proteobacteria. The number and sizes of replicons were determined by separating nondigested DNA. Hybridization of an rrn gene probe was used to distinguish between chromosomes and plasmids. Members of the genus Agrobacterium all possess two chromosomes, and each biovar has a specific genome size. As previously demonstrated for Agrobacterium tumefaciens C58, the smaller chromosomes of Agrobacterium biovar 1 and Agrobacterium rubi strains appear to be linear. The genomes of Rhizobium strains were all of similar sizes but were seen to contain either one, two, or three megareplicons. Only one chromosome was present in the member of the related genus Phyllobacterium. We found one or two chromosomes in Rhodobacter and Brucella species, two chromosomes in Ochrobactrum anthropi, and one chromosome in Mycoplana dimorpha and Bartonella quintana; all of these genera are related to the Rhizobiaceae. The presence of multiple chromosomes is discussed from a phylogenetic and taxonomic point of view.
Bacterial genomes were long considered to consist of a single circular chromosome. With the discovery of the existence of multiple circular chromosomes or a linear chromosome in some bacteria, this paradigm is no longer valid. Two different circular chromosomes were reported for Rhodobacter sphae-roides (39), Brucella melitensis 16M (27), and Leptospira interrogans (45), while three chromosomes are present in the genomes of Rhizobium meliloti (38), Burkholderia cepacia (7), and related species (33). A linear chromosome was reported first for the spirochete Borrelia burgdorferi (3, 11) and then for the gram-positive organisms Streptomyces lividans (25) and Rhodococcus fascians (8). We subsequently demonstrated that the genome of the gram-negative bacterium Agrobacterium tumefaciens C58 consisted of two chromosomes, one circular and the other linear (1). Most of the organisms presenting a multipartite genomic organization are confined to certain species within the purple bacteria (or Proteobacteriaceae), a phylum of the Bacteria, and perhaps this feature is correlated with the phylogeny of these bacteria. In the present study, we have investigated the genomic organization of organisms belonging to the alpha subgroup of the class Proteobacteria, particularly members of the genera Mycoplana, Ochrobactrum, Rhodobacter, Phyllobacterium, Rhizobium, and Agrobacterium. Although the first three genera do not belong to the family Rhizobiaceae, 16S rRNA sequence comparisons suggest that they belong to a tight phylogenetic group which also includes the genera Brucella and Bartonella (Rochalimaea) (9, 43).
MATERIALS AND METHODS
Strains and growth conditions.
The strains used in this study are listed in Tables 1 and 2. Three well-studied laboratory strains, Agrobacterium tumefaciens C58, Agrobacterium rhizogenes K84, and Rhizobium meliloti 2011, were gifts from X. Nesmes and M. Fernandez (Laboratoire d’Ecologie Microbienne du Sol, Université Claude Bernard Lyon I, Villeurbanne, France). Brucella melitensis 16M is from our laboratory collection. These strains were grown as previously described (1, 27, 38). Strains originating from the American Type Culture Collection (ATCC) or the Collection Française des Bactéries Phytopathogènes (CFBP) were grown as recommended by the suppliers.
TABLE 1.
Strain designations, number and sizes of replicons, and estimated genome sizes for organisms belonging to the family Rhizobiaceae
| Bacterium | Strain and origin | No. of repliconsa | Sizes (kb) of repliconsb | Estimated genome size (kb) | Reference(s) |
|---|---|---|---|---|---|
| A. radiobacter bv. 1 | CFBP 2414T | 3 (2) | 3,000*, 2,100* (L), 680 | 5,780 | This study |
| A. tumefaciens bv. 1 | ATCC 23308T | 4 (2) | 3,000*, 2,100* (L), 550, 250 | 5,900 | This study |
| C58c | 4 (2) | 3000* 2100* (L), 550, 250 | 5,900 | 1 | |
| A. tumefaciens bv. 3 (vitis) | CFBP 2721 | 4 (2) | 3,500*, 1,100*, 500, 250, 200 | 5,550 | This study |
| CFPB 2607 | 4 (2) | 3,500*, 1,050*, 250, 180 | 4,980 | This study | |
| A. rhizogenes bv. 2 | ATCC 11325T | 4 (1) | 4,000*, 2,700, 285 (NS), 250 (NS) | 7,235 | This study |
| K84c | 4 (1) | 4,000*, 2,700, 365, 200 | 7,265 | This study | |
| A. rubi | ATCC 13335T | 4 (2) | 3,100*, 1,800* (L), 550, 285 | 5,735 | This study |
| Rhizobium meliloti | 1021 and 2011c | 3 (1) | 3,500*, 1,700, 1,400 | 6,600 | 38 and this study |
| Rhizobium fredii | ATCC 35423T | 3 (1) | 4,000*, 2,200, 450 | 6,650 | This study |
| Rhizobium leguminosarum bv. trifolii | ATCC 14480T | 4 (1) | 4,600*, 1,100, 650, 450 | 6,800 | This study |
| Rhizobium leguminosarum bv. phaseoli | ATCC 14482T | 4 (NE) | 4,600, 1,100, 450, 285 | 6,435 | This study |
| Bradyrhizobium japonicum | 110 | 1 (1) | 8,700* | 8,700 | 22 |
| P. myrsinacearum | ATCC 43590T | 5 (1) | 3,500*, 680, 500, 365, 285 | 5,330 | This study |
The numbers in parentheses are the numbers of replicons labeled by the 16S rRNA probe; NE, not examined.
Asterisks indicate replicons labeled by 16S rRNA probe; L, linear replicon; NS, not shown.
Strain from Laboratoire d’Ecologie Microbienne du Sol, Lyon, France.
TABLE 2.
Strain designation, number and sizes of replicons, and estimated genome sizes for organisms related to the family Rhizobiaceae
| Bacterium | Strain | No. of repliconsa | Sizes (kb) of repliconsb | Genome size estimate (kb) | Reference |
|---|---|---|---|---|---|
| M. dimorpha | ATCC 4279T | 3 (NE) | 3,200, 300, 150 | 3,150 | This study |
| Rhodobacter capsulatus | ATCC 11166 | 2 (1) | 3,800*, 150 | 3,950 | This study |
| SB1003 | 2 (1) | 3,800*, 134 | 3,934 | 12 | |
| Rhodobacter spheroides | 2.4.1 | 7 (2) | 3,000*, 900*; 5 plasmids | 4,350 | 39 |
| Brucella melitensis | 16M | 2 (2) | 2,050*, 1,150* | 3,200 | 27 |
| O. anthropi | ATCC 49188T | 4 (2) | 2,700*, 1,900*, 150, 100 | 4,850 | This study |
| LMG 3301 | 4 (NE) | 2,700, 1,900, 50, <50 | 4,700 | This study | |
| Bartonella quintana | VR960 | 1 | 1,700 | 1,700 | 32 |
The numbers in parentheses are the numbers of replicons labeled with the 16S rRNA probe; NE, not examined.
Asterisks indicate replicons labeled by the 16S rRNA probe.
Preparation of the rRNA probe.
This probe was prepared by PCR amplification as previously described (1).
Preparation of high-molecular-weight genomic DNAs.
Intact genomic DNAs were prepared in agarose plugs as usually described for gram-negative bacteria (1) except for those of some strains, which were better lysed by proteinase K.
PFGE of intact DNAs.
Pulsed-field gel electrophoresis (PFGE) was performed in a contour-clamped homogeneous electric field apparatus in 0.5× TBE (36), using the Gene Navigator system from Pharmacia (Saint-Quentin-Yvelines, France). Saccharomyces cerevisiae, Shizosaccharomyces pombe, Henselae wingei (Bio-Rad), A. tumefaciens C58, and/or R. meliloti DNA (38) and multimers of phage λ DNA were used as molecular size markers. Different pulsing conditions were used to separate either the larger molecules (above 1 Mb) or the smaller ones (below 1 Mb) (1). Gels were stained with ethidium bromide and photographed under short-wavelength UV light. The sizes of replicons were determined by averaging the measurements from several gels.
RESULTS AND DISCUSSION
To examine the diversity in replicon number and size and to distinguish between the linear and circular forms, we employed PFGE. Except for some randomly linearized forms (originating from the preparation of DNA), which generate faint bands in PFGE, circular molecules do not enter the gel (24, 38). In contrast, linear molecules give rise to a marked increase in the thickness and intensity of ethidium bromide-stained bands.
The sizes of entire replicons can be estimated by comparing their migration to that of different high-molecular-weight markers, although for very large fragments the degree of accuracy is rather low. The migration distance in the gel depends on not only the pulse time but also the G+C content of the molecule (23, 30). Nondigested DNAs were submitted to PFGE to investigate the genomic organization of 16 organisms belonging to six genera (Tables 1 and 2).
Bacteria belonging to the Rhizobiaceae.
In the family Rhizobiaceae are found bacteria which live in association with plant cells. The classification of Agrobacterium and Rhizobium species is based on both phenotypic traits and plasmid-encoded characteristics inducing symbiosis or tumorigenesis (44), rather than chromosomal genes. Moreover, in these genera, extrachromosomal elements represent a major part of the genome (26).
(i) The genus Agrobacterium.
The genus Agrobacterium consists of several genetically and phenotypically different groups or clusters (21). Differences in 16S rRNA sequences clearly separated strains of biovar 1, biovar 2, biovar 3 (Agrobacterium vitis), and Agrobacterium rubi (37).
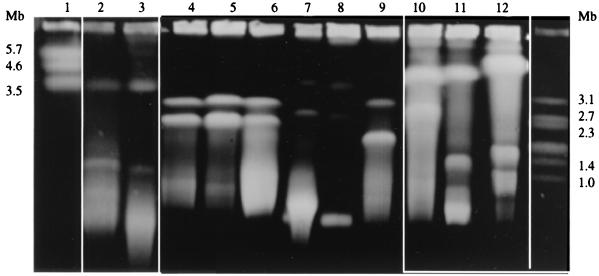
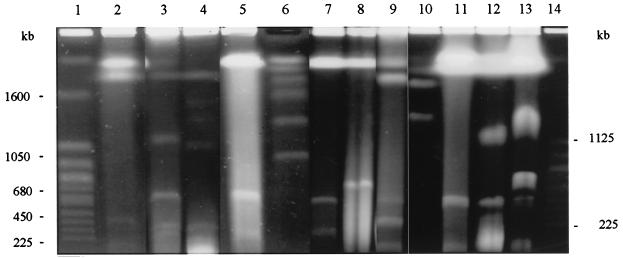
We have previously shown that the biovar 1 strain A. tumefaciens C58 contains four replicons: two megabase-sized chromosomes (one of which is linear) and two plasmids (1). Two other strains belonging to this biovar, Agrobacterium radiobacter CFBP 2414T and A. tumefaciens ATCC 23308T, were tested. Two megabase-sized replicons, both hybridizing with the rRNA probe, were seen in these strains (Fig. 1 and 2 and Table 1). The smaller band was more intense and more diffuse than the larger one, suggesting that, as in A. tumefaciens C58, this replicon is linear. Electrophoresis under different pulsing conditions revealed the presence of small replicons corresponding to plasmids; there were two in A. tumefaciens ATCC 23308T but only one in A. radiobacter CFBP 2414T, which is not a pathogen (Fig. 3 and Table 1).
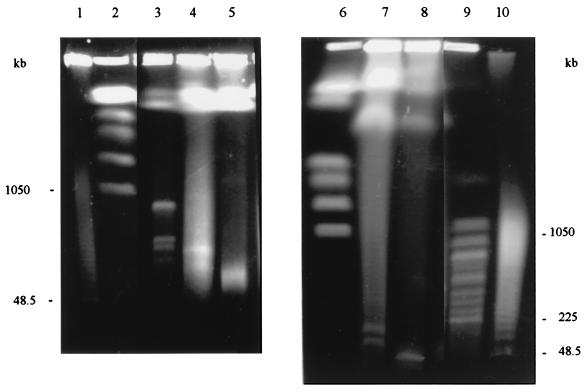
FIG. 1.
PFGE of intact DNAs of bacterial species belonging to the family Rhizobiaceae: separation of large replicons. Lanes: 1, Saccharomyces pombe; 2, A. vitis CFBP 2721; 3, A. vitis CFBP 2607; 4, A. tumefaciens C58; 5, A. tumefaciens ATCC 23308T; 6, A. radiobacter CFBP 2414T; 7, A. rhizogenes ATCC 11325T; 8, A. rhizogenes K84; 9, A. rubi ATCC 13335T; 10, Rhizobium fredii ATCC 35423T; 11, Rhizobium leguminosarum bv. phaseoli ATCC 14482T; 12, Rhizobium leguminosarum bv. trifolii ATCC 14480T; far right, H. wingei. The positions of molecular size markers are indicated on both sides of the gel.
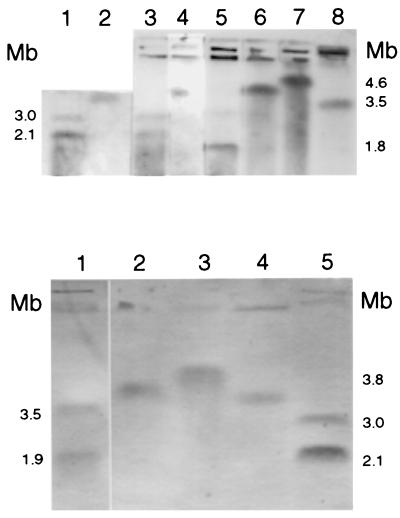
FIG. 2.
Hybridization of large replicons with the 16S rRNA probe. (Upper panel) 1, A. radiobacter CFBP 2414T; 2, A. rhizogenes K84; 3, A. tumefaciens C58; 4, A. rhizogenes ATCC 11325T; 5, A. rubi ATCC 13335T; 6, Rhizobium fredii ATCC 35423T; 7, Rhizobium leguminosarum bv. trifolii ATCC 14480T; 8, Rhizobium meliloti 2011. (Lower panel) Lanes: 1, O. anthropi ATCC 49188T; 2, P. myrsinacearum ATCC 43590T; 3, Rhodobacter capsulatus ATCC 11166; 4, Rhizobium meliloti 2011; 5, A. tumefaciens C58. The positions of molecular size markers are shown on both sides of the two panels.
FIG. 3.
PFGE of intact DNAs of bacterial species belonging to the family Rhizobiaceae: separation of small replicons. Lanes: 1, Saccharomyces cerevisiae; 2, A. rhizogenes K84; 3, A. vitis CFBP 2721; 4, A. vitis CFBP 2607; 5, A. tumefaciens C58; 6, H. wingei; 7, A. tumefaciens ATCC 23308T; 8, A. radiobacter CFBP 2414T; 9, A. rubi ATCC 13335T; 10, Rhizobium meliloti 2011; 11, Rhizobium fredii ATCC 35423T; 12, Rhizobium leguminosarum bv. phaseoli ATCC 14482T; 13, R. leguminosarum bv. trifolii ATCC 14480T; 14, Saccharomyces cerevisiae. The positions of molecular size markers are shown on both sides of the gel.
The results for A. rubi ATCC 13335T, which is one of the three strains forming this cluster (21), were similar to those for biovar 1 strains: it contained one circular and one apparently linear chromosome plus two plasmids (Fig. 1 to 3 and Table 1).
Two strains belonging to biovar 2, A. rhizogenes ATCC 11325T and A. rhizogenes K84, were studied. In both strains, two megareplicons, both apparently circular, were separated, but only the larger one hybridized with the rRNA probe (Fig. 1 and 2 and Table 1). Also present in the ATCC 11325T and the K84 strains were two smaller replicons which correspond to the previously reported agrocine and tumor-inducing plasmids of the latter strain (21) (Fig. 3 and Table 1).
Strains of biovar 3 are tumorigenic for grape vines (21). Two of them, A. tumefaciens CFBP 2721 and CFBP 2607, were analyzed. Two megareplicons, both hybridizing with the rRNA probe and both apparently circular, could be seen, together with three plasmids for the former strain and two plasmids for the latter (Fig. 1 and 3 and Table 1).
The unusual genomic organization of A. tumefaciens C58 was first suspected because of the greater intensity and the diffuse aspect of one of the two megabase-sized bands, the one corresponding to all of the linear molecules when, under the same conditions, only some randomly linearized forms of the circular replicons enter the gel (38). Further evidence was established by insertion of a unique restriction site into each of the chromosomes that led, after enzymatic digestion, to the linearization of the circular molecules and the generation of two fragments from the linear molecules (18). In the other biovar 1 strains and A. rubi, two megabase-sized replicons were present, with the smaller molecule probably being linear. Thus, this multipartite genome with different topologies appears to be a common feature of strains of biovar 1 and A. rubi.
Biovar 2 and 3 strains also possess two megareplicons; however, their intensity in the PFGE gel suggests that they are both circular. Interestingly, while both megareplicons of biovar 3 hybridized with the rRNA probe, only the larger one of biovar 2 hybridized with it. Nevertheless, the presence of other essential housekeeping genes on the second molecule is possible, as was shown for Rhizobium meliloti (35). The sizes of the biovar 2 strain genomes (>7,200 kb) are larger than those of the biovar 1 and A. rubi strains (5,900 to 5,735 kb) as well as those of biovar 3 strains (5,500 or 4,980 kb).
(ii) The genus Rhizobium.
Sobral et al. have described the presence of three megabase-sized replicons in Rhizobium meliloti 1021, one chromosome of 3.5 Mb and two megaplasmids of 1.7 and 1.3 Mb, so called because they did not hybridized with an rRNA probe (38). Nevertheless, essential housekeeping genes such as the GroEL chaperonin-encoding genes are present on these molecules, thus raising questions about the chromosomal status of these replicons (35). We tested a second strain, R. meliloti 2011, and found three replicons of sizes similar to those of strain 1021, again with only the larger one hybridizing with the rRNA probe (Fig. 2 and 3, and Table 1).
For Rhizobium fredii ATCC 35423T, two megabase-sized replicons and a small replicon were separated. Only the largest one was shown to hybridize with the rRNA probe (Fig. 1 to 3 and Table 1).
In Rhizobium leguminosarum ATCC 14480T and ATCC 14482T (corresponding to the biovars trifolii and phaseoli), we found two circular megareplicons plus the two large plasmids previously described for this species (29). We hybridized an rRNA probe with separated replicons of R. leguminosarum bv. trifolii. Again, only one (the largest) contains rRNA genes (Fig. 1 to 3 and Table 1).
The organization of the genome of Bradyrhizobium japonicum is different; this genome has a single chromosome that is larger (8,700 kb) than that of the other Rhizobium species (6,400 to 6,700 kb) (6, 22).
(iii)The genus Phyllobacterium.
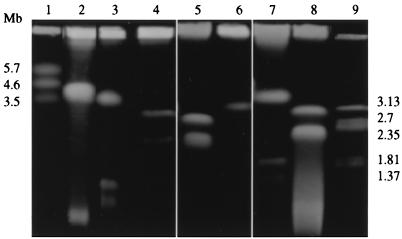
For Phyllobacterium myrsinacearum ATCC 43590T, five replicons were separated, with the larger (megabase-sized) one hybridizing with the rRNA probe (Fig. 2, 4, and 5 and Table 1). The genomic organization for the genus Phyllobacterium also seems different from that of the other genera of the Rhizobiaceae, with there being only one megareplicon (but several large plasmids) and a smaller total genome size (5,330 kb).
FIG. 4.
PFGE of intact DNAs of bacterial species related to the Rhizobiaceae: separation of large replicons. Lanes: 1, Schizosaccharomyces pombe; 2, Rhodobacter capsulatus ATCC 11166; 3, P. myrsinacearum ATCC 43590T; 4, O. anthropi ATCC 49188T; 5, O. anthropi LMG 3301; 6, M. dimorpha ATCC 4279T; 7, Rhizobium meliloti 2011; 8, A. tumefaciens C58; 9, H. wingei. The positions of molecular size markers are shown on both sides of the gel.
FIG. 5.
PFGE of intact DNAs of bacterial species related to the Rhizobiaceae: separation of small replicons. Lanes: 1, lambda DNA ladder; 2, H. wingei; 3, P. myrsinacearum ATCC 43590T; 4, M. dimorpha ATCC 4279T; 5, Rhodobacter capsulatus ATCC 11166; 6, H. wingei; 7, O. anthropi ATCC 49188T; 8, O. anthropi LMG 3301; 9, Saccharomyces cerevisiae; 10, lambda DNA ladder. The positions of molecular size markers are indicated to the left and right.
Related bacteria belonging to others genera and families.
Members of the Rhizobiaceae are also related to taxonomically different organisms (9, 43). Rhodobacter sphaeroides is found on a distant branch of rRNA superfamily IV, which comprises Agrobacterium species, Rhizobium species, and Brucella abortus (9). Other bacteria that are closely related to rRNA superfamily IV are Mycoplana dimorpha, Ochrobactrum anthropi, and Bartonella quintana (37, 40, 43). These related bacteria represent a heterogeneous group whose members have few common features; Rhodobacter sphaeroides is a facultative photosynthetic bacterium (39), M. dimorpha is a soil-living organism (43), Brucella and Bartonella species are animal pathogens (42, 43), and O. anthropi is a rare opportunistic pathogen of immunocompromised patients (2).
(i)The genus Rhodobacter.
Suwanto and Kaplan have shown that Rhodobacter spheroides 2.4.1 possesses two chromosomes, one of 3,000 kb and the other of 900 kb (39). However, the chromosomal structure of Rhodobacter capsulatus SB1003 is quite different, consisting of a unique 3,800-kb chromosome and a 134-kb plasmid (12). We investigated another strain of Rhodobacter capsulatus (ATCC 11166) and found only one megabase-sized replicon, which hybridized to the rRNA probe, and a small replicon (Fig. 2, 4, and 5 and Table 2).
(ii) The genera Ochrobactrum and Brucella.
Two strains of O. anthropi were studied. Two megareplicons of similar sizes were found in both strains ATCC 49188T and LMB 3301, and two small replicons of different sizes were found in each of these strains (Fig. 2, 4, and 5 and Table 2). Only the two larger bands hybridized with the rRNA probe. The physical map of Brucella melitensis has been constructed, and it demonstrated the presence of two circular chromosomes (27). These two chromosomes are also present in the other species of this genus (28), with the exception of one biovar (see below).
(iii) The genera Mycoplana and Bartonella.
The type strain M. dimorpha ATCC 4279T had only one (megabase-sized) replicon and two plasmids (Fig. 4 and 5 and Table 2). Bartonella quintana was shown to possess only one chromosome (reference 32 and unpublished data).
Does genomic organization have phylogenetic significance?
Using highly conserved sequences such as the rRNA genes or housekeeping proteins such as the GroEL chaperonin and RecA, phylogenetic trees have been constructed which have allowed the definition of the alpha subgroup of the Proteobacteria (9, 10, 40–42). The genomic organization of bacteria belonging to this group has been studied to see if a correlation with the phylogeny could be demonstrated.
Genome size differences, increasing with the evolutionary genetic distance between lineages, were shown to exist for the major subgroups of Escherichia coli, which suggests that there is a phylogenic component to this variation (4). The genome of A. rhizogenes K84 (7,265 kb) is 1.45 times larger than that of A. vitis CFBP 2607 (4,980 kb). This degree of variation is comparable to that seen for different strains of Burkholderia cepacia (13). Strains of Agrobacterium biovars 1 and 2 exhibit only 15% DNA homology (21). Our results show that their genome sizes and organizations are also very different, thus providing further evidence that they are genetically distinct. Sawada et al. place Agrobacterium biovar 2 closer to Rhizobium fredii, and this is again supported by the genomic organization (37).
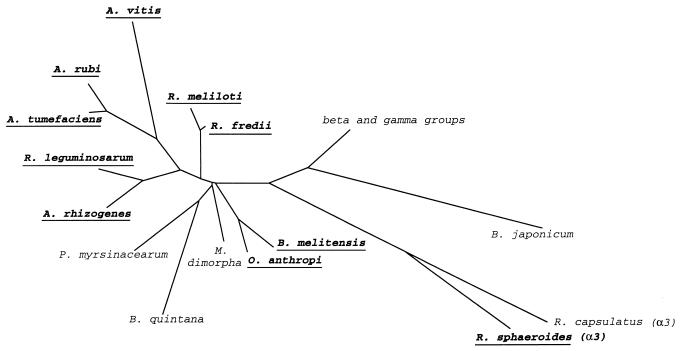
Most of the organisms possessing several megabase-sized replicons belong to the alpha subgroup of the Proteobacteria (Fig. 6); the exceptions are Burkholderia (Pseudomonas) cepacia, which is classified in the β2 subgroup (42), and L. interrogans, which is a spirochete (45). The members of subgroup α2 form a tight cluster, while the β subgroup constitutes a quite phylogenetically diverse class (20, 42). The existence of a more complex genomic architecture (with two or three chromosomes) may have phylogenetic significance if this trait is also found in other organisms of the same lineage. Among the alpha-subgroup genera that we have investigated, this feature is present in all of the species of Agrobacterium, Rhizobium, Brucella (except one [see below]), and Ochrobactrum. On the contrary, Bradyrhizobium, Phyllobacterium, Mycoplana, and Bartonella species have only one chromosome. Finally, in the genus Rhodobacter, R. sphaeroides has two chromosomes while R. capsulatus has only one.
FIG. 6.
Phylogenetic tree showing the genomic organization of organisms belonging to the α2 subgroup of the Proteobacteria (plus Rhodobacter species, which belong to the α3 subgroup) (redrawn from reference 43). Organisms with complex genomes are indicated in boldface and underlined.
The Rhizobiaceae can be divided into two groups. The fast-growing strains (Rhizobium meliloti, Rhizobium fredii, and Rhizobium leguminosarum) all have complex genomes, while the slow-growing species Bradyrhizobium japonicum has a single, very large chromosome (22). The genus Bradyrhizobium, however, is only remotely related to the other genera of the Rhizobiaceae (41). The deeper branching found for Bradyrhizobium japonicum with both the 16S rRNA and the GroEL sequences (9, 40, 43) could mean that the origin of this lineage is close to the single-chromosome ancestor. This taxonomically different genus (17) represents a separate line of descent in the α2 subgroup of the Proteobacteria (41), one which is also remote from the Agrobacterium rRNA branch in rRNA superfamily IV (16). In contrast, Rhodobacter capsulatus and Rhodobacter sphaeroides, with one and two chromosomes, respectively, branch together on the phylogenetic tree (10). In this case of two species belonging to the same lineage, it is difficult to explain how two organisms with such different genomic organizations could have a common ancestor unless this feature is not linked with the phylogeny. Moreover, within the same species—Brucella suis—the genome of the biovar 3 reference strain is composed of a single chromosome of 3.2 Mb while biovar 1 members each possess two chromosomes, of 2.1 and 1.15 Mb, and biovar 2 and 4 members each have two chromosomes, of 1.85 and 1.35 Mb. The four biovars are phenotypically very similar, and the restriction maps of their genomes are also very similar except for the distribution of the same sequences on different linkage groups (19). Other evidence is from outside of the α-proteobacteria, for the gram-positive bacterium Bacillus cereus, whose different strains vary with respect to their chromosome sizes and genome organizations. Within this species, the genome may exist either as one large chromosome with small plasmids or as a small chromosome with stably maintained large extrachromosomal elements which may be considered as fragments of a secondary chromosome (5).
Thus, the presence of multiple chromosomes in α-proteobacterial genomes is not related to a common phylogeny, since it is not always shared either by all of the members of a same clade (e.g., Rhodobacter genus) or even by all of the strains of the same species (e.g., Brucella suis). This trait, found inconstantly among different bacterial lineages, rather seems to have been acquired independently. Where does this complex organization originate?
The classical model of genome evolution involves gene duplication followed by divergence. This can occur via a tandem duplication in the genome, achieved by recombination between repeated sequences (34). Such repeats could be rRNA operons. Following this, a second intrachromosomal recombination event, occurring anywhere within the duplicated region, will result in the formation of two stable replicons if both molecules have an origin of replication; alternatively, the second origin of replication could be acquired by lateral transfer from a different organism. A comparison of the sequences of these molecules will distinguish between these two possibilities. Nevertheless, there is no known environment shared by these different organisms which could explain their “infection” by a new origin. In the case of the genus Brucella, we have shown that the different species exhibit differences in genomic organization. The differences in chromosome size and number can be explained by the occurrence of rearrangements at chromosomal regions containing the three rrn genes. The location and orientation of these genes confirmed that these rearrangements are due to homologous recombination at the rrn loci (19). This phenomenon occurred naturally in the genus Brucella; however, recently the 4,188-kb circular genome of Bacillus subtilis was artificially dissected into two stable circular chromosomes in vivo by such a mechanism (15).
The coexistence of linear and circular chromosomes in the same bacterial cell raises another question. The chromosome of Streptomyces lividans probably oscillates between linear and circular forms, and this may also occur in other bacteria (31). It has been suggested by Hinnebusch and Tilly (14) that one of the origins of linear DNA in bacteria could be genetic exchange between procaryotes and eucaryotes. These authors also add that the most evident example of this gene exchange is the transfer of DNA from the phytopathogen A. tumefaciens into a plant cell to induce the formation of a crown gall tumor. It is perhaps not a coincidence that in this species the chromosomes exhibit both types of structures.
The reason for the presence of a complex genomic organization in many members of the α-proteobacteria remains to be determined. While we have shown that the possession of a complex genome does not have a clear phylogenetic significance, we can speculate that there are structures in or functions of the genome of the alpha subgroup which favor their appearance.
REFERENCES
- 1.Allardet-Servent A, Michaux-Charachon S, Jumas-Bilak E, Karayan L, Ramuz M. Presence of one linear and one circular chromosome in the Agrobacterium tumefaciens C58 genome. J Bacteriol. 1993;175:7869–7874. doi: 10.1128/jb.175.24.7869-7874.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alnor D, Frimodt-Moller N, Espersen F, Frederiksen W. Infections with the unusual human pathogens Agrobacterium species and Ochrobactrum anthropi. Clin Infect Dis. 1994;18:914–920. doi: 10.1093/clinids/18.6.914. [DOI] [PubMed] [Google Scholar]
- 3.Baril C, Richaud C, Baranton G, Saint Girons I. Linear chromosome of Borrelia burgdorferi. Res Microbiol. 1989;140:507–516. doi: 10.1016/0923-2508(89)90083-1. [DOI] [PubMed] [Google Scholar]
- 4.Bergthorsson U, Ochman H. Heterogeneity of genome sizes among natural isolates of Escherichia coli. J Bacteriol. 1995;177:5784–5789. doi: 10.1128/jb.177.20.5784-5789.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carlson C R, Kolsto A-B. A small (2.4 Mb) Bacillus cereus chromosome corresponds to a conserved region of a larger (5.3 Mb) Bacillus cereus chromosome. Mol Microbiol. 1994;13:161–169. doi: 10.1111/j.1365-2958.1994.tb00411.x. [DOI] [PubMed] [Google Scholar]
- 6.Chakrabarti S K, Mishra A K, Chakrabartty P K. Genome size variation of rhizobia. Experientia. 1983;40:1290–1291. [Google Scholar]
- 7.Cheng H-P, Lessie T G. Multiple replicons constituting the genome of Pseudomonas cepacia 17616. J Bacteriol. 1994;176:4034–4042. doi: 10.1128/jb.176.13.4034-4042.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Crespi M, Messens E, Caplan A B, Van Montagu M, Desomer J. Fasciation induction by the phytopathogen Rhodococcus fascians depends upon a linear plasmid encoding a cytokinin synthase gene. EMBO J. 1992;11:795–804. doi: 10.1002/j.1460-2075.1992.tb05116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.De Ley J, Mannheim W, Segers P, Lievens A, Denijn M, Vanhoucke M, Gillis M. Ribosomal ribonucleic acid cistron similarities and taxonomic neighborhood of Brucella and CDC group Vd. Int J Syst Bacteriol. 1987;37:35–42. [Google Scholar]
- 10.Eisen J A. The RecA protein as a model molecule for molecular systematic studies of bacteria: comparison of trees of RecAs and 16S rRNAs from the same species. J Mol Evol. 1995;41:1105–1123. doi: 10.1007/BF00173192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferdows M S, Barbour A G. Megabase-sized linear DNA in the bacterium Borrelia burgdorferi, the Lyme disease agent. Proc Natl Acad Sci USA. 1989;86:5969–5973. doi: 10.1073/pnas.86.15.5969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fonstein M, Zheng S, Haselkorn R. Physical map of the genome of Rhodobacter capsulatus SB 1003. J Bacteriol. 1992;174:4070–4077. doi: 10.1128/jb.174.12.4070-4077.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hendrickson W, Hubner A, Kavanaugh-Black A. Proceedings of the 35th Hanford Symposium on Health and the Environment. Microbial genome research and its application. 1996. Multiple chromosomes of Burkholderia cepacia; pp. 88–89. [Google Scholar]
- 14.Hinnebusch J, Tilly K. Linear plasmids and chromosomes in bacteria. Mol Microbiol. 1993;10:917–922. doi: 10.1111/j.1365-2958.1993.tb00963.x. [DOI] [PubMed] [Google Scholar]
- 15.Itaya M, Tanaka T. Experimental surgery to create subgenomes of Bacillus subtilis 168. Proc Natl Acad Sci USA. 1997;94:5378–5382. doi: 10.1073/pnas.94.10.5378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jarvis B D W, Gillis M, De Ley J. Intra- and intergeneric similarities between the ribosomal ribonucleic acid cistrons of Rhizobium and Bradyrhizobium species and some related bacteria. Int J Syst Bacteriol. 1986;36:129–138. [Google Scholar]
- 17.Jordan D C. Family III. Rhizobiaceae Conn 1938, 321AL. In: Holt J G, editor. Bergey’s manual of determinative bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. pp. 234–256. [Google Scholar]
- 18.Jumas-Bilak E, Maugard C, Michaux-Charachon S, Allardet-Servent A, Perrin A, O’Callaghan D, Ramuz M. Study of the organization of the genomes of Escherichia coli, Brucella melitensis and Agrobacterium tumefaciens by insertion of a unique restriction site. Microbiology. 1995;141:2425–2432. doi: 10.1099/13500872-141-10-2425. [DOI] [PubMed] [Google Scholar]
- 19.Jumas-Bilak E, Michaux-Charachon S, Bourg G, O’Callaghan D, Ramuz M. Differences in chromosome number and genome rearrangements in the genus Brucella. Mol Microbiol. 1998;27:99–106. doi: 10.1046/j.1365-2958.1998.00661.x. [DOI] [PubMed] [Google Scholar]
- 20.Karlin S, Weinstock G M, Brendel V. Bacterial classifications derived from RecA protein sequence comparisons. J Bacteriol. 1995;177:6881–6893. doi: 10.1128/jb.177.23.6881-6893.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kerters K, De Ley J. Genus III. Agrobacterium. In: Holt J G, editor. Bergey’s manual of systematic bacteriology. Vol. 1. Baltimore, Md: The Williams & Wilkins Co.; 1984. p. 244. [Google Scholar]
- 22.Kündig C, Hennecke H, Göttfert M. Correlated physical and genetic map of the Bradyrhizobium japonicum 110 genome. J Bacteriol. 1993;175:613–622. doi: 10.1128/jb.175.3.613-622.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee J J, Smith H O, Redfield R J. Organization of the Haemophilus influenzae Rd genome. J Bacteriol. 1989;171:3016–3024. doi: 10.1128/jb.171.6.3016-3024.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levene S D, Zimm B H. Separation of open circular DNA using pulsed-field electrophoresis. Proc Natl Acad Sci USA. 1987;84:4054–4057. doi: 10.1073/pnas.84.12.4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin Y-S, Kieser H M, Hopwood D A, Chen C W. The chromosomal DNA of Streptomyces lividans 66 is linear. Mol Microbiol. 1993;10:923–933. doi: 10.1111/j.1365-2958.1993.tb00964.x. [DOI] [PubMed] [Google Scholar]
- 26.Martinez-Romero E. Recent developments in Rhizobium taxonomy. Plant Soil. 1994;161:11–20. [Google Scholar]
- 27.Michaux S, Paillisson J, Carles-Nurit M-J, Bourg G, Allardet-Servent A, Ramuz M. Presence of two independent chromosomes in the Brucella melitensis 16M genome. J Bacteriol. 1993;175:701–705. doi: 10.1128/jb.175.3.701-705.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Michaux-Charachon S, Bourg G, Jumas-Bilak E, Guigue-Talet P, O’Callaghan D, Allardet-Servent A, Ramuz M. Genome structure and phylogeny in the genus Brucella. J Bacteriol. 1997;179:3244–3249. doi: 10.1128/jb.179.10.3244-3249.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Prakash R K, Schilperoort R A, Nuti M P. Large plasmids of fast-growing rhizobia: homology studies and location of structural nitrogen fixation (nif) genes. J Bacteriol. 1981;145:1129–1136. doi: 10.1128/jb.145.3.1129-1136.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pyle L E, Corcoran L N, Bergeman B J, Withley J C, Finch L R. Pulsed-field electrophoresis indicates larger than expected sizes for Mycoplasma genomes. Nucleic Acids Res. 1988;16:6015–6025. doi: 10.1093/nar/16.13.6015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Redenbach M, Flett F, Piendl W, Glocker I, Rauland U, Wafig O, Kliem R, Leblond P, Cullum J. The Streptomyces lividans 66 chromosome contains a 1 Mb deletogenic region flanked by two amplifiable regions. Mol Gen Genet. 1993;241:255–262. doi: 10.1007/BF00284676. [DOI] [PubMed] [Google Scholar]
- 32.Reschke D K, Frazier M E, Mallavia L P. Transformation and genomic restriction mapping of Rochalimaea spp. Acta Virol. 1991;35:519–525. [PubMed] [Google Scholar]
- 33.Rodley P D, Römling U, Tümmler B. A physical genome map of the Burkholderia cepacia type strain. Mol Microbiol. 1995;17:57–67. doi: 10.1111/j.1365-2958.1995.mmi_17010057.x. [DOI] [PubMed] [Google Scholar]
- 34.Roth J R, Benson N, Galitski T, Haack K, Lawrence J G, Miesel L. Rearrangements of the bacterial chromosome: formation and applications. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechtera M, Umbarger H E, editors. Escherichia coli and salmonella: cellular and molecular biology. Vol. 2. Washington, D.C: ASM Press; 1996. pp. 2256–2276. [Google Scholar]
- 35.Rusanganwa E, Gupta R S. Cloning and characterization of multiple groEL chaperonin-encoding genes in Rhizobium meliloti. Gene. 1993;126:67–75. doi: 10.1016/0378-1119(93)90591-p. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook J, Fritsh E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 37.Sawada H, Ieki H, Oyaizu H, Matsumoto S. Proposal for rejection of Agrobacterium tumefaciens and revised descriptions for the genus Agrobacterium and for Agrobacterium radiobacter and Agrobacterium rhizogenes. Int J Syst Bacteriol. 1993;43:694–702. doi: 10.1099/00207713-43-4-694. [DOI] [PubMed] [Google Scholar]
- 38.Sobral B W S, Honeycutt R J, Atherly A G, McClelland M. Electrophoretic separation of the three Rhizobium meliloti replicons. J Bacteriol. 1991;173:5173–5180. doi: 10.1128/jb.173.16.5173-5180.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suwanto A, Kaplan S. Physical and genetic mapping of the Rhodobacter sphaeroides 2.4.1. genome: presence of two unique circular chromosomes. J Bacteriol. 1989;171:5850–5859. doi: 10.1128/jb.171.11.5850-5859.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Viale A M, Arakaki A K, Soncini F C, Ferreyra R G. Evolutionary relationships among eubacterial groups as inferred from GroEL (chaperonin) sequence comparisons. Int J Syst Bacteriol. 1994;44:527–533. doi: 10.1099/00207713-44-3-527. [DOI] [PubMed] [Google Scholar]
- 41.Willems A, Collins M D. Phylogenetic analysis of rhizobia and agrobacteria based on 16S rRNA gene sequences. Int J Syst Bacteriol. 1993;43:305–313. doi: 10.1099/00207713-43-2-305. [DOI] [PubMed] [Google Scholar]
- 42.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yanagi M, Yamasato K. Phylogenetic analysis of the family Rhizobiaceae and related bacteria by sequencing of 16S rRNA gene using PCR and a DNA sequencer. FEMS Microbiol Lett. 1993;107:115–120. doi: 10.1111/j.1574-6968.1993.tb06014.x. [DOI] [PubMed] [Google Scholar]
- 44.Young J P W. Molecular phylogeny of rhizobia and their relatives. In: Palacio R, Mora J, Newton W E, editors. New horizons in nitrogen fixation. Dordrecht, The Netherlands: Kluwer Academic Publishers; 1993. pp. 587–592. [Google Scholar]
- 45.Zuerner R L, Herrmann J L, Saint Girons I. Comparison of genetic maps for two Leptospira interrogans serovars provides evidence for two chromosomes and intraspecies heterogeneity. J Bacteriol. 1993;175:5445–5451. doi: 10.1128/jb.175.17.5445-5451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]