Significance
SARS-CoV-2 uses human ACE2 as the cell surface receptor to infect host cells, ultimately leading to the development of COVID-19. The complex progression of the disease and its impact on multiple organs suggest that additional cellular factors may be involved in the interaction with SARS-CoV-2. Integrin α5β1, a cell adhesion molecule that is widely expressed in various tissues, has emerged as one such factor. Our study has revealed that SARS-CoV-2 spike protein exploits α5β1 signaling to facilitate cell–cell fusion and trigger inflammatory responses through the interaction with α5β1. Both processes may contribute to the infection and pathogenesis of SARS-CoV-2.
Keywords: SARS-CoV-2, integrin, cell fusion, inflammation
Abstract
The Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) virus infects host cells by engaging its spike (S) protein with human ACE2 receptor. Recent studies suggest the involvement of integrins in SARS-CoV-2 infection through interaction with the S protein, but the underlying mechanism is not well understood. This study investigated the role of integrin α5β1, which recognizes the Arg-Gly-Asp (RGD) motif in its physiological ligands, in S-mediated virus entry and cell–cell fusion. Our results showed that α5β1 does not directly contribute to S-mediated cell entry, but it enhances S-mediated cell–cell fusion in collaboration with ACE2. This effect cannot be inhibited by the putative α5β1 inhibitor ATN-161 or the high-affinity RGD-mimetic inhibitor MK-0429 but requires the participation of α5 cytoplasmic tail (CT). We detected a direct interaction between α5β1 and the S protein, but this interaction does not rely on the RGD-containing receptor binding domain of the S1 subunit of the S protein. Instead, it involves the S2 subunit of the S protein and α5β1 homo-oligomerization. Furthermore, we found that the S protein induces inflammatory responses in human endothelial cells, characterized by NF-κB activation, gasdermin D cleavage, and increased secretion of proinflammatory cytokines IL-6 and IL-1β. These effects can be attenuated by the loss of α5 expression or inhibition of the α5 CT binding protein phosphodiesterase-4D (PDE4D), suggesting the involvement of α5 CT and PDE4D pathway. These findings provide molecular insights into the pathogenesis of SARS-CoV-2 mediated by a nonclassical RGD-independent ligand-binding and signaling function of integrin α5β1 and suggest potential targets for antiviral treatment.
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the virus that causes COVID-19, uses the same cell entry receptor, angiotensin-converting enzyme 2 (ACE2), as SARS-CoV. However, it has a broader cell tropism, higher transmission rates, and a more complex pathogenesis, suggesting that additional host factors and mechanisms specific to SARS-CoV-2 are involved (1, 2). The severity of COVID-19 is associated with dysregulated inflammatory responses, characterized by elevated levels of cytokines such as IL-6 and IL-1β, as well as injury to vascular endothelial cells (3, 4). Beyond respiratory epithelial cells primarily targeted by SARS-CoV-2, a wide range of cell types in vascular, immune, and even central nervous systems experience dysfunction, contributing to the pathogenesis of COVID-19 (5–7). While innate immune receptors such as toll-like receptors (TLRs) may play a role in COVID-19 inflammation (8), they function as intrinsic receptors against most bacterial and viral infections. The specific host receptors responsible for the intricated pathogenesis of COVID-19 have yet to be fully delineated.
SARS-CoV-2 uses its spike (S) protein to bind to ACE2 for cell entry (9). Recent studies have indicated potential roles of integrins in SARS-CoV-2 infection (10–16) and COVID-19 pathogenesis (17–23). In humans, there are 24 α/β integrin heterodimers formed by the combination of 18 α and 8 β subunits (24). Integrins play critical roles in various biological processes, including leukocyte recirculation and migration, wound healing, blood clotting, and immune response (24). Some viruses use integrins as receptors or coreceptors to facilitate infection (25–27). The consideration of integrins as putative receptors for SARS-CoV-2 was primarily based on the presence of an integrin-binding RGD (Arg-Gly-Asp) motif on the SARS-CoV-2 S (SARS2-S) protein (28, 29). Among the eight integrins that recognize the RGD motif, α5β1, αVβ3, and αIIbβ3 have been reported to directly interact with the SARS2-S protein in studies using purified proteins (12, 14, 17–19). However, the results of these studies have shown inconsistencies and controversies. Some indirect evidence for the interaction between integrins and SARS2-S was obtained from cell-based assays such as cell adhesion (19, 30). Of note, the potential ligand-binding inhibitor ATN-161 for α5β1 has been used in SARS-CoV-2 infection and cellular function assays (10, 20, 31). However, ATN-161 is not a direct RGD-blocking inhibitor, and its mechanism of inhibition remains undefined.
The objective of this study was to investigate the potential role of α5β1 in SARS-CoV-2 spike-mediated infection, its interaction with SARS2-S, and the resulting cellular response. Our data showed that α5β1 does not play a direct role in virus cell entry, but instead facilitates cell–cell fusion mediated by the SARS2-S protein. We also detected a direct interaction between α5β1 and SARS2-S, which does not depend on the RGD motif. This interaction involves the non-receptor-binding S2 subunit of SARS2-S and the homo-oligomerization of α5β1. Furthermore, our study demonstrated that SARS2-S induces proinflammatory responses in human endothelial cells, evidenced by NF-κB activation, cytokine release, and cleavage of gasdermin D (GSDMD). These processes were found to be mediated by α5β1 signaling through the PDE4D pathway. α5β1 is widely expressed in immune cells, lung, heart, and endothelial cells. The SARS2-S-induced and α5β1-mediated cellular responses may contribute to the complex pathogenesis of COVID-19.
Results
Integrin α5β1 Does Not Play a Role in S-Mediated Virus Entry.
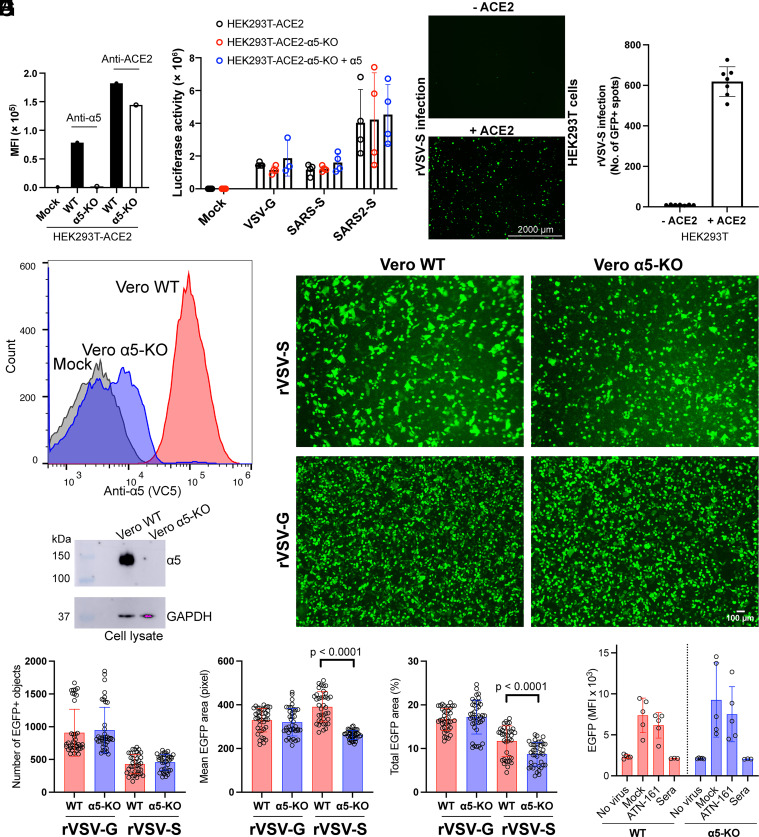
To investigate the involvement of α5β1 in SARS2-S mediated cell entry, we used S-pseudotyped lentivirus and a replication-competent recombinant vesicular stomatitis virus (rVSV) encoding enhanced green fluorescent protein (EGFP) and SARS2-S protein (rVSV-S) or VSV G protein (rVSV-G) for virus infection assays. HEK293T-ACE2 stable cells naturally express high levels of α5β1, which was knocked out (KO) using CRISPR/cas9 (Fig. 1A). Lentivirus and rVSV-S infection were assessed by measuring luciferase activity and EGFP expression, respectively. Notably, the absence of α5 expression in HEK293T-ACE2 cells had no impact on SARS2-S-pseudotyped lentivirus infection, similar to the control lentivirus pseudotyped with VSV G or SARS-CoV S (SARS-S) protein (Fig. 1B). Additionally, overexpression of α5β1 in HEK293T-ACE2-α5-KO cells also did not affect lentivirus infection (Fig. 1B). When ACE2 was absent, rVSV-S failed to infect HEK293T cells (Fig. 1 C and D), despite their high expression of α5β1.
Fig. 1.
Integrin α5β1 does not contribute to the cell entry mediated by SARS-CoV-2 spike. (A) Surface expression of α5 and ACE2 in HEK293T-ACE2 stable cells with or without α5 knockout. (B) The HEK293T-ACE2, HEK293T-ACE2-α5-KO, or HEK293T-ACE2-α5-KO overexpressing α5 were infected for 48 h with VSV-G, SARS-S, or SARS2-S-pseudotyped lentiviruses. The cells were harvested for luciferase activity measurement using Bright-Glo Luciferase Assay System. Data are mean ± SD from four independent repeats. (C) HEK293T cells transfected with or without ACE2 were infected by recombinant VSV virus carrying SARS2-S and EGFP for 24 h. Representative live-cell images from three independent repeats with three images randomly taken for each well were acquired using AMG EVOS fluorescence microscope with Plan Fluor 2× objective lens (numerical aperture of 0.06), equipped with Sony 1cx285AQ color charge-coupled device camera (CCD). (D) Quantification of rVSV-S virus infection based on cell images in panel (C). Data are mean ± SD from seven randomly taken images of three independent repeats. (E and F) Flow cytometry and immunoblot analysis of Vero E6 cells with and without α5 knockout. (G) Representative images of Vero WT or Vero α5-KO cells infected by rVSV-S or rVSV-G virus for 6 h. The images were acquired using EVOS M7000 imaging system with Plan Fluor 4× objective lens (numerical aperture of 0.13). Ten to Twenty images were randomly taken for each repeat in three independent repeats. (H) Quantification of virus infection results of panel (G). The infection was measured as number of EGFP-positive objects, mean EGFP area, or total EGFP area. Data are mean ± SD. Unpaired two-tailed t test. (I) HEK293T-ACE2 or HEK293T-ACE2-α5-KO cells were infected for 6 h with rVSV-S in the absence or presence of 0.1 mM ATN-161 or pooled sera of COVID-19 vaccinated donors. The infection was measured by flow cytometry. Data are mean ± SD from five independent repeats.
To validate these findings, we performed additional experiments using Vero E6 cells that express high levels of endogenous ACE2. The α5 in Vero E6 cells was knocked out using CRISPR/cas9, as demonstrated by flow cytometry and western blot analysis (Fig. 1 E and F). Following infection with rVSV-S or the control rVSV-G virus for 6 h, no discernible differences in the number of infected cells were observed between wild-type Vero E6 and Vero E6 α5-KO cells (Fig. 1 G and H). These results indicate that α5β1 does not contribute to the S-mediated cell entry of SARS-CoV-2.
We examined the effect of ATN-161 on rVSV-S infection of HEK293T-ACE2 cells. Interestingly, the inhibitor exhibited no inhibition effect on rVSV-S infection in either wild-type or α5-KO HEK293T-ACE2 cells, whereas pooled sera from COVID-19 vaccinated donors completely blocked virus infection (Fig. 1I).
Integrin α5β1 Contributes to S-Mediated Cell–Cell Fusion.
The expression of the S protein on cell surface induces cell–cell fusion (32), which facilitates the spread of virus among cells (33). To assess the extent of cell–cell fusion following rVSV-S infection in Vero E6 cells, we measured the mean or total EGFP areas. The results showed that loss of α5 expression in Vero E6 cells significantly reduced cell–cell fusion induced by the rVSV-S virus, while it had no effect on cell–cell fusion induced by the rVSV-G virus (Fig. 1 G and H).
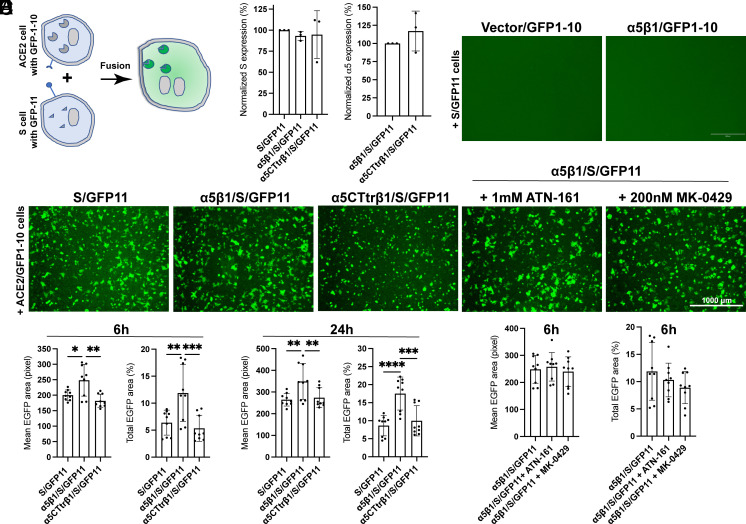
To further investigate a potential role of α5β1 in S-mediated cell–cell fusion, we used a well-established split GFP assay (32). The large GFP1-10 and small GFP11 fragments were expressed separately in HEK293T-α5-KO cells along with ACE2 and SARS2-S, respectively. The fusion of ACE2 and S cells leads to the reconstitution of GFP fluorescence (Fig. 2A). Coexpression of S and GFP11, with or without α5β1, showed comparable levels of S surface expression (Fig. 2B). In the absence of ACE2, no cell–cell fusion was observed (Fig. 2C). When ACE2 and S cells were cocultured for 6 h, substantial cell–cell fusion occurred, as indicated by GFP fluorescence (Fig. 2D). Quantitative analysis of cell–cell fusion, based on mean or total GFP area, showed that coexpression of S and α5β1 significantly increased cell–cell fusion at both 6 h and 24 h (Fig. 2E). To examine whether the signaling function of α5 is involved in this process, we truncated the cytoplasmic tail (CT) of α5 at the conserved GFFKR motif. Remarkably, coexpression of α5-CTtr/β1 with the S protein failed to enhance cell–cell fusion (Fig. 2E), despite α5-CTtr/β1 and α5/β1 exhibiting comparable expression levels (Fig. 2B).
Fig. 2.
Integrin α5β1 contributes to cell–cell fusion mediated by SARS-CoV-2 spike. (A) Diagram of cell–cell fusion assay measured by split GFP assay. (B) Surface expression of SARS2-S and α5 in HEK293T-α5-KO cells. Cells were transfected with indicated constructs for 48 h and subjected to flow cytometry before the cell–cell fusion assay. Data are mean ± SD from four independent repeats. (C) α5β1 alone in the absence of ACE2 does not mediate cell–cell fusion induced by SARS2-S. Representative images of three independent repeats. (D) Representative images of HEK293T-α5-KO cells transfected with ACE2 plus GFP1-10 cocultured for 6 h with HEK293T-α5-KO cells transfected with S/GFP11, α5β1/S/GFP11, or α5CTtrβ1/S/GFP11. The α5β1/S/GFP11 cells were also pretreated with ATN-161 or MK-0429 before coculturing with the ACE2/GFP1-10 cells. Cells were imaged using AMG EVOS fluorescence microscope with Plan Fluor 4× objective lens (numerical aperture of 0.13), equipped with Sony 1cx285AQ color CCD camera. (E and F) Quantification of cell–cell fusion at 6 h and 24 h after coculturing. Three images were randomly taken for each experiment group in three independent repeats. The mean or total EGFP area was measured using CellProfiler software. Data are mean ± SD. One-Way ANOVA Tukey’s multiple comparisons test. *P < 0.05; **P < 0.01; ***P < 0.001; and ****P < 0.0001.
Additionally, we found that ATN-161 and the high-affinity RGD-mimetic inhibitor MK-0429 had no effect on S-mediated cell–cell fusion in the presence of α5β1 (Fig. 2F). These results demonstrate the role of α5β1 in promoting S-mediated cell–cell fusion, which does not depend on the RGD-binding function but rather requires the participation of α5 CT.
Integrin α5β1 Interacts with the S Protein of SARS-CoV-2 Independently of the RGD Motif.
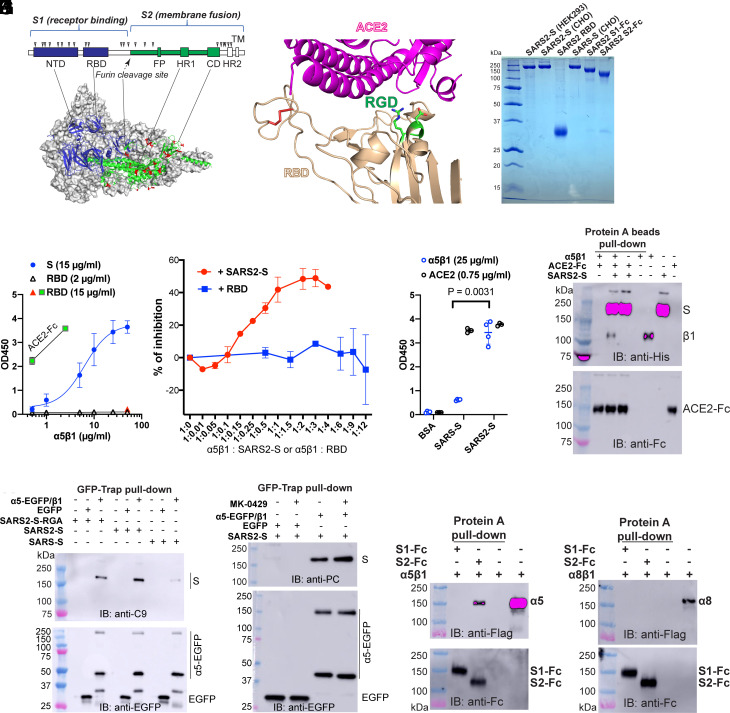
The S protein is composed of S1 and S2 subunits (Fig. 3A), with the S1 subunit containing the receptor binding domain (RBD) responsible for ACE2 binding, and the S2 subunit containing the machinery for membrane fusion (Fig. 3A). The structures of S homotrimer (Fig. 3A) and the complex of RBD bound with ACE2 have been determined (Fig. 3B) (34, 35). The putative integrin binding RGD motif is located in an α-helix of the RBD at the ACE2 binding interface (Fig. 3B). To investigate the interaction between α5β1 and S protein, we used purified ectodomains of SARS2-S and SARS-S, the RBD of SARS2-S, and Fc-tagged S1 and S2 subunits of SARS2-S (Fig. 3C). All proteins were verified to be free of aggregates using size-exclusion chromatography (SEC). enzyme-linked immunosorbent assay (ELISA) was employed to detect the direct interaction of SARS2-S and α5β1 on the plate coated with equal molar concentrations of SARS2-S and RBD. A concentration-dependent binding of α5β1 to SARS2-S was detected by anti-α5 mAb VC5 (Fig. 3D). However, while purified ACE2 bound well to RBD, no binding was detected between α5β1 and RBD coated at 2 or 15 μg/mL (Fig. 3D). Consistently, only soluble SARS2-S but not the RBD inhibited the binding of α5β1 with immobilized SARS2-S (Fig. 3E). Additionally, our ELISA result showed that α5β1 bound to SARS2-S significantly better than SARS-S (Fig. 3F), whereas ACE2 bound equally well to both proteins (Fig. 3F). Furthermore, our pull-down assay using protein A beads showed that α5β1 associated with SARS2-S even in the presence of an abundance of ACE2-Fc (Fig. 3G), suggesting that α5β1 and ACE2 can bind simultaneously to SARS2-S. This result contradicts the RGD-dependent binding model of α5β1 with SARS2-S since the location of the RGD motif on the RBD implies that the binding of α5β1 and ACE2 to the RBD is mutually exclusive (Fig. 3B).
Fig. 3.
Integrin α5β1 interacts with SARS2-S protein independent of the RGD motif. (A) The domain organization and cryo-EM structure of SARS2-S (PDB 6XR8). NTD: N-terminal domain; RBD: receptor binding domain; FP: fusion peptide; HR: heptad repeat; CD: C-terminal domain; TM: transmembrane. The putative N-glycan sites are marked by triangles. The surface exposed residues that are different between SARS-S and SARS2-S S2 subunits are shown as red sticks. (B) Crystal structure of SARS2-S RBD in complex with ACE2 (PDB 6LZG). The RGD motif is shown as green sticks. Disulfide bonds are red sticks. (C) Reducing SDS-PAGE of purified spike proteins. The recombinant stabilized SARS2-S or SARS-S ectodomain was expressed as secreted form in HEK293 or CHO cells. The SARS2-S RBD domain was expressed in HEK293T cells as a secreted form. The SARS2-S S1 and S2 subunits were expressed as secreted form with a C-terminal human IgG Fc tag in HEK293T cells. (D) Interaction of α5β1 with SARS2-S detected by ELISA. The plate was coated with equal molar concentration of S (15 μg/mL) and RBD (2 μg/mL), or RBD at a higher concentration (15 μg/mL). The binding of α5β1 was detected by mAb VC5. Binding of ACE2-Fc at 0.5 and 2.5 μg/ml to RBD was as a control. Data are mean ± SD from three or four independent repeats. (E) Competition ELISA. α5β1 was preincubated with either SARS2-S or RBD at different molar ratio before adding to SARS2-S-coated ELISA plate. Data are mean ± SD from three independent repeats. (F) ELISA plate was coated with 15 μg/mL of SARS-S, SARS2-S, or bovine serum albumin (BSA). Binding of α5β1 at 25 μg/mL or ACE2-Fc at 0.75 μg/mL was measured. Data are mean ± SD from four independent repeats. One-Way ANOVA Tukey’s multiple comparisons test. (G) α5β1 coimmunoprecipitation with SARS2-S and ACE2. Purified α5β1 ectodomain (6×His tag on β1) was mixed with purified ACE2-Fc in the presence or absence of purified 6×His-tagged stabilized SARS2-S ectodomain and pulled down with protein A beads. The samples were first immunoblotted with anti-His and then reblotted with anti-Fc. (H) Pull-down assay of full-length α5β1 and S proteins. HEK293T-α5-KO cells were transfected with α5-EGFP/β1 or EGFP plus C-terminal C9-tagged full-length SARS-S, SARS2-S, or SARS2-S with RGD to RGA mutation. The cell lysates were immunoprecipitated with GFP-Trap beads. The precipitated samples were immunoblotted with anti-C9 and reblotted with anti-EGFP under reducing condition. The intact α5-EGFP and α5 light chain tagged with EGFP were detected. (I) The RGD-mimetic α5β1 inhibitor MK-0429 (IC50 12 nM) had no effect on α5β1 binding with SARS2-S. HEK293T-α5-KO cells were transfected with α5-EGFP/β1 or EGFP plus C-terminal PC-tagged full-length SARS2-S. The cells were treated with MK-0429 at 10 μM for 1 h before pull-down with GFP-Trap beads. (J and K) The purified Flag-tagged α5β1 (J) or α8β1 (K) was incubated with SARS2-S S1-Fc or S2-Fc and precipitated by protein A beads. The precipitated samples were immunoblotted with anti-Flag and anti-Fc under reducing condition.
The pentapeptide ATN-161 (Ac-PHSCN-NH2) has been widely used as an α5β1 antagonist. It was designed based on the synergy sequence PHSRN of fibronectin domain 9 (Fn9) with an R to C substitution (SI Appendix, Fig. S1A) (36). A structural study of α5β1 bound with the Fn7-10 fragment revealed the high-resolution interaction between the synergy site and α5 subunit (37), emphasizing the critical role of the R residue within the PHSRN sequence in interacting with α5 (SI Appendix, Fig. S1A). The R to C substitution in ATN-161 is likely to reduce its interaction with α5 if it binds to the synergy binding site. Our ELISA results showed no significant inhibitory effect of ATN-161 on the interaction between α5β1 and SARS2-S (SI Appendix, Fig. S1B).
Using a pull-down assay with GFP-Trap beads, we observed that α5-EGFP/β1 precipitated the full-length SARS2-S but not SARS-S in HEK293T-α5-KO cells (Fig. 3H and SI Appendix, Fig. S2A). Mutating the RGD motif to RGA had no effect on the association of full-length SARS2-S with α5-EGFP/β1 (Fig. 3H). Furthermore, the RGD-mimetic inhibitor MK-0429 did not impede the pull-down of SARS2-S with α5-EGFP/β1 (Fig. 3I). We also introduced Ala mutations for two conserved Ser residues (S132 and S134) in the β1 subunit, which disrupt RGD binding to α5β1 by interfering with metal ion binding (SI Appendix, Fig. S2B). Our pull-down assay showed that the β1-SSAA mutation had no impact on the association of α5-EGFP/β1 with full-length SARS2-S (SI Appendix, Fig. S2C). Additionally, truncation of the α5 CT did not affect the pull-down of full-length SARS2-S with α5-CTtr/β1-GFP (SI Appendix, Fig. S2D). We also found that α5β1 could associate with purified or surface-expressed S proteins of Delta and Omicron variants of SARS-CoV-2 (SI Appendix, Fig. S2 E and F). These results collectively demonstrate an RGD-independent interaction between the S protein of SARS-CoV-2 and α5β1.
We further investigated the interaction between α5β1 and SARS2-S using purified Fc-tagged S1 and S2 subunits of SARS2-S along with the Flag-tagged α5β1 ectodomain (SI Appendix, Fig. S2G). The pull-down results using protein A beads revealed the association of α5β1 with S2-Fc but not S1-Fc (Fig. 3J and SI Appendix, Fig. S2H). As a control, neither S1-Fc nor S2-Fc captured the Flag-tagged α8β1 (Fig. 3K). As expected, only S1-Fc but not S2-Fc bound the Flag-tagged ACE2 (SI Appendix, Fig. S2I). Additionally, no interaction between α5β1 and the S2-Fc of SARS-S was detected (SI Appendix, Fig. S2J). These data provide evidence for a direct interaction between SARS2-S and integrin α5β1, which is independent of the RGD-containing RBD but relies on the S2 subunit of SARS2-S.
The SARS2-S and α5β1 Interaction Involves α5β1 Homo-Oligomerization.
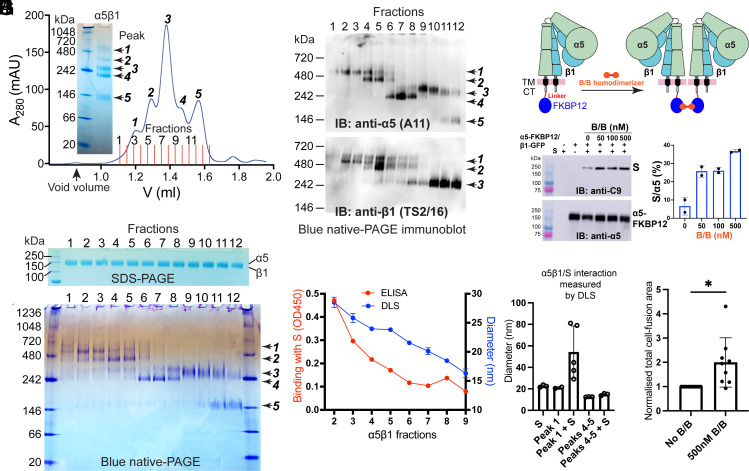
During the purification of the α5β1 ectodomain using high-performance microscale SEC, we observed five elution peaks (Fig. 4A), likely corresponding to five distinct populations migrating differently under native condition on blue native-PAGE (Fig. 4A). Although the 12 factions collected based on five SEC peaks were indistinguishable on reducing SDS-PAGE under denatured condition (Fig. 4B), they exhibited different migration patterns on blue native-PAGE (Fig. 4C). These protein bands migrating differently on blue native-PAGE were confirmed to be α5β1 proteins as determined by anti-α5 and anti-β1 western blotting (Fig. 4D).
Fig. 4.
The SARS2-S and α5β1 interaction involves α5β1 homo-oligomerization. (A) Blue native-PAGE and microscale size exclusion chromatography (SEC) of affinity-purified α5β1 ectodomain. Twelve fractions were collected for the 5 SEC peaks. (B–D) Reducing SDS-PAGE, blue native-PAGE, and immunoblot of the 12 α5β1 SEC fractions in A. (E) Hydrodynamic radii of the α5β1 fractions measured by dynamic light scattering (DLS) and their binding with stabilized SARS2-S ectodomain measured by ELISA. (F) DLS analysis of α5β1 SEC fractions corresponding to peak 1 and peaks 4 to 5 in the absence or presence of SARS2-S protein. Data are mean ± SD from five independent measurements. (G) Model of FKBP12-mediated homodimerization of α5β1. FKBP12 was tagged to the cytoplasmic tail (CT) of α5. Binding of the bivalent FKBP12 ligand B/B homodimerizer induces dimerization of α5β1. (H and I) B/B increased the association of α5-FKBP12/β1-GFP determined by pull-down assay. HEK293T-α5-KO cells were transfected with α5-FKBP12/β1-GFP plus C9-tagged full-length SARS2-S for 48 h. The cells were treated with or without B/B at different concentrations for 1 h before pull-down with GFP-Trap beads. The pull-down samples were subjected to immunoblot with anti-C9 and anti-α5 antibodies. The immunoblot was quantified by S signal as a percentage of α5 signal. Data are mean ± SD from two independent repeats. (J) Cell–cell fusion assay. HEK293T-α5-KO cells transfected with α5-FKBP12/β1 and full-length SARS2-S plus GFP11 were treated with or without B/B and cocultured for 6 h with HEK293T-α5-KO cells transfected ACE2/GFP1-10. The cell fusion was measured as total EGFP area and normalized to the area without B/B. Data are mean ± SD from three independent repeats with three randomly taken images for each repeat. Unpaired two-tailed t test. *P < 0.05.
We then used dynamic light scattering (DLS) to measure the hydrodynamic radii of the SEC fractions, revealing a positive correlation between particle sizes (in diameter) and the SEC retention volumes (in fraction numbers) (Fig. 4E). Intriguingly, our ELISA results revealed that SARS2-S specifically bound to α5β1 fractions with larger diameters but not those with smaller diameter (Fig. 4E). This observation was further validated by DLS assay, which indicated the formation of a complex between SARS2-S with α5β1 fractions from peak-1 but not peaks 4 to 5 (Fig. 4F).
Given that the molecular weight (~500 kDa) and diameter (~25 nm) of α5β1 peak-1 fractions closely resemble those of an α5β1 homodimer, we hypothesized that α5β1 homodimerization might be involved in its interaction with SARS2-S. To test this hypothesis, we introduced a FKBP12 tag to the CT of full-length α5, which undergoes homodimerization upon binding the bivalent ligand B/B homodimerizer (Fig. 4G). Our pull-down results showed that the presence of B/B markedly enhanced the association between full-length SARS2-S and α5-FKBP12/β1-GFP in HEK293T-α5-KO cells (Fig. 4 H and I). Furthermore, B/B also significantly increased cell–cell fusion between HEK293T-α5-KO cells expressing α5-FKBP12/β1 plus SARS2-S and the cells expressing ACE2 (Fig. 4J). These results strongly suggest the involvement of α5β1 homodimerization in the interaction with SARS2-S.
The S Protein of SARS-CoV-2 Induces Inflammatory Responses Mediated by α5β1 and cAMP Regulation in Endothelial Cells.
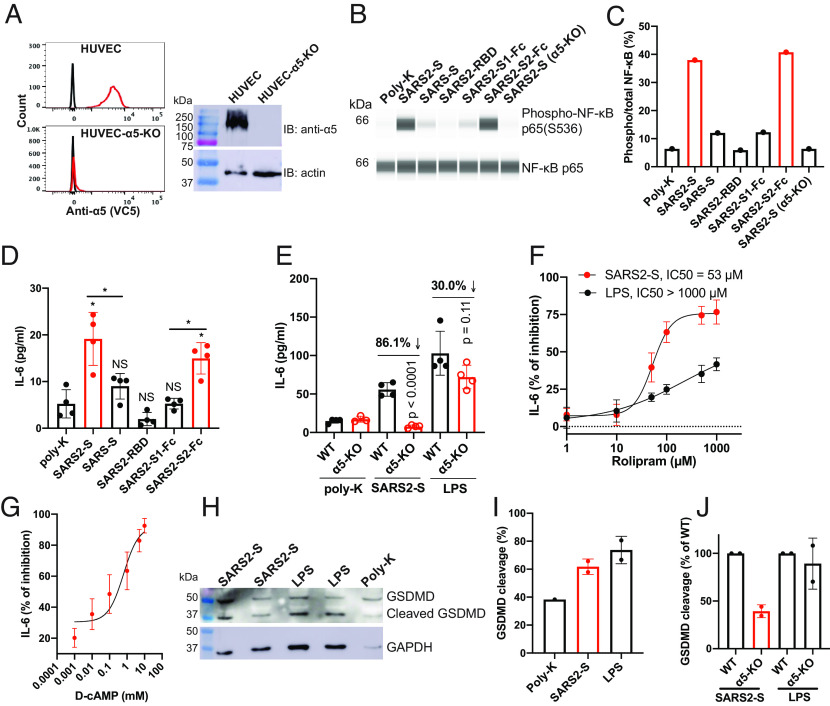
To investigate the potential cellular response resulting from the interaction between SARS2-S and α5β1, we used an immortalized human umbilical vein endothelial cell line (HUVEC) as a model system. We employed CRISPR/Cas9 to knock out the endogenous α5 expression in HUVEC, as demonstrated by flow cytometry and immunoblot analysis (Fig. 5A). Given that α5β1 has been implicated in endothelial inflammation (22, 38), we evaluated NF-κB activation and IL-6 secretion in HUVEC cells attached to plastic surface coated with purified S proteins. Compared to the poly-lysine control, we observed a significant increase in NF-κB phosphorylation in cells adhered to SARS2-S (Fig. 5 B and C and SI Appendix, Fig. S2 A–C). Remarkably, cells adhered to SARS-S or the RBD and S1 subunit of SARS2-S showed little to no NF-κB phosphorylation (Fig. 5 B and C and SI Appendix, Fig. S2 A–C). However, robust NF-κB phosphorylation was observed in cells adhered to the S2 subunit of SARS2-S (Fig. 5 B and C and SI Appendix, Fig. S2 A–C). The loss of α5 expression significantly reduced SARS2-S-induced NF-κB activation (Fig. 5 B and C and SI Appendix, Fig. S2 A–C). Consistent with the NF-κB activation, both SARS2-S and its S2 subunit significantly increased IL-6 secretion in HUVEC cells, whereas SARS-S and the RBD and S1 subunit of SARS2-S had no such effect (Fig. 5D). α5 knock-out significantly diminished SARS2-S-induced IL-6 release, but it had much less effect on LPS-induced IL-6 release (Fig. 5E). These findings demonstrate that SARS2-S, specifically through its S2 subunit, induces an inflammatory response in endothelial cells that is dependent on α5β1.
Fig. 5.
SARS2-S induces inflammatory signaling through α5β1 integrin in HUVECs. (A) Loss of α5 expression in HUVEC-α5-KO cells. The depletion of α5 expression in HUVEC cells shown by flow cytometry (Left) and immunoblot (Right). (B) NF-κB activation in HUVEC cells attached to immobilized S proteins. HUVEC or HUVEC-α5-KO cells were seeded into a plate coated with 10 μg/mL indicated proteins for 2 h. Total cells were collected for Jess automatic western blot. (C) Quantification of immunoblot data of panel (B). One represented data of four repeats is shown. (D) SARS2-S-induced IL-6 release of HUVEC cells. HUVEC cells were seeded into a plate coated with 10 μg/mL indicated proteins for 2 h. The IL-6 concentrations in the supernatants were measured by ELISA. Data are mean ± SD from four independent repeats. One-Way ANOVA Tukey’s multiple comparisons test. *P < 0.05; ns P > 0.05. (E) Loss of α5 significantly reduced SARS2-S- but not LPS-induced IL-6 release in HUVEC cells. Two-tailed unpaired Welch’s t test. Data are mean ± SD from four independent repeats. (F) Dose-dependent inhibition by rolipram of SARS2-S-induced IL-6 release in HUVEC cells. HUVEC cells were treated with different concentrations of rolipram before seeding into a plate coated with SARS2-S or LPS. Data are mean ± SD from three or four independent repeats. (G) Dose-dependent inhibition by Dibutyryl-cAMP (D-cAMP) of SARS2-S-induced IL-6 release in HUVEC cells. Data are mean ± SD from three or four independent repeats. (H and I) SARS2-S- or LPS-induced cleavage of GSDMD in HUVEC detected by immunoblot in total cell lysates (H). The cleavage of GSDMD was presented as a percent of cleaved GSDMD to total GSDMD (I). (J) Loss of α5 greatly reduced SARS2-S- but not LPS-induced GSDMD cleavage in HUVEC cells. For (I) and (J), Data are mean ± SD from two independent repeats.
Previous studies have demonstrated that α5 integrin, through its cytoplasmic interaction with phosphodiesterase-4 (PDE4) isoform D5, plays a role in promoting inflammatory signaling in endothelial cells (38). To investigate the involvement of PDE4 in the SARS2-S-induced inflammatory response, we used a selective PDE4 inhibitor called rolipram (39). A dose–response assay showed that rolipram efficiently blocked SARS2-S-induced IL-6 release, while its impact on LPS-induced IL-6 release was comparably less pronounced (Fig. 5F). Active PDE4 is known to modulate inflammation by degrading cAMP (40), and a decrease in cAMP levels triggers the activation of inflammasome (41). Consistent with this scenario, the SARS2-S-induced IL-6 release was inhibited by elevating intracellular cAMP levels using cell membrane permeable Dibutyryl-cAMP (D-cAMP) (Fig. 5G). Additionally, SARS2-S stimulation led to the cleavage of gasdermin D (GSDMD) (Fig. 5 H and I), a hallmark of inflammasome activation. Knockout of α5 integrin markedly attenuated SARS2-S-indcued GSDMD cleavage, while its impact on LPS-induced GSDMD cleavage was minimal (Fig. 5J). These results indicate that the inflammatory response induced by the interaction between SARS2-S and α5β1 involves the α5-PDE4D-cAMP pathway.
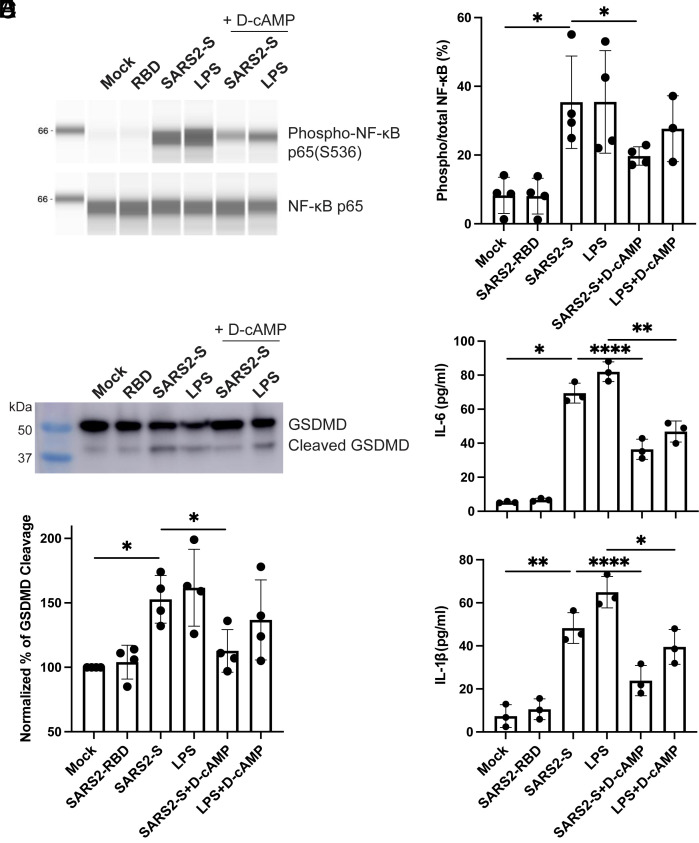
To further validate these findings, we used HULEC-5a, a human-lung-derived microvascular endothelial cell line, which provides a more relevant model for studying lung microvascular injury associated with SARS-CoV-2 infection (42). We exposed HULEC-5a cells to soluble SARS2-S, RBD, or LPS in the absence or presence of D-cAMP. Consistent with our observations in HUVECs, treatment with SARS2-S, but not its RBD, led to NF-κB activation in HULEC-5a cells, and this response was attenuated by D-cAMP (Fig. 6 A and B). Moreover, SARS2-S, but not its RBD, triggered the cleavage of GSDMD (Fig. 6 C and D), which was also inhibited by D-cAMP (Fig. 6 C and D). Additionally, SARS2-S, but not its RBD, induced the secretion of IL-6 and IL-1β, which were effectively blocked by D-cAMP (Fig. 6E). Both IL-1β release and GSDMD cleavage are well-recognized indicators of inflammasome activation (43). As a control, D-cAMP reduced the NF-κB activation, GSDMD cleavage, IL-6 and IL-1β release induced by LPS (Fig. 6 A–E), consistent with the well-established anti-inflammatory function of cAMP.
Fig. 6.
SARS2-S induces inflammatory response in HULECs. (A and B) Soluble SARS2-S-induced NF-κB activation and inhibition by D-cAMP. HULEC-5a cells in suspension were incubated with 10 μg/mL SARS2-S, 10 μg/mL RBD, or 5 μg/mL LPS in the presence or absence of 1 mM D-cAMP for 2 h. The total cell lysates were subjected to Jess automatic western blot for phosphor- and total NF-κB. The blots were quantified based on intensity. (C and D) Soluble SARS2-S-induced GSDMD cleavage and inhibition by D-cAMP. HULEC-5a cells in suspension were treated as in (A and B) for 2 h. The total cell lysates were subjected to western blot for GSDMD. The cleavage of GSDMD was quantified based on intensity. (E) Soluble SARS2-S-induced release of IL-6 and IL-1β and the inhibition by D-cAMP. HULEC-5a cells in suspension were treated as in (A and B) for 2 h. The cell supernatants were subjected to ELISA for measuring the concentrations of IL-6 and IL-1β. Data are mean ± SD from three or four independent repeats. One-way ANOVA Tukey’s multiple comparisons test. *P < 0.05; **P < 0.01; and ****P < 0.0001.
Discussion
Whether integrins function as cell entry receptors for SARS-CoV-2 remains a topic of debate. Our argument is that SARS-CoV-2 cannot effectively infect cells in the absence of ACE2 by using integrins as receptors. Considering the widespread expression of integrins such as α5β1 and αVβ3 in our body, if SARS-CoV-2 were able to infect ACE2-null cells using integrins as receptors, it could lead to more severe disease progression. It is worth noting that mouse and human integrins often share similar ligand-binding properties. However, mice without human ACE2 are not susceptible to SARS-CoV-2 infection (44–47). Our data based on pseudovirus and replication-competent rVSV-S along with α5-KO did not support the involvement of α5β1 in the S-mediated cell entry. However, we did find that α5β1 contributes to S-mediated cell–cell fusion to some extent, which involves the participation of α5 CT. The rVSV-S virus has served as a widely accepted model for the study of SARS2-S-mediated cell entry and inhibition, demonstrating a strong correlation with the authentic SARS-CoV-2 virus (48–50). However, distinctions between VSV and SARS-CoV-2 virions, such as variations in density, distribution, stability, and potential mutations of the S proteins on the viral envelope, can potentially impact the cell fusion function of the S protein. The limitation of using rVSV-S in our study was partially mitigated by using the S protein that expressed on cell surface to mimic the cell fusion between virus-infected cells, showing consistent results with rVSV-S. Nevertheless, we recognize the need for further comprehensive studies to compare our findings with those obtained from authentic SARS-CoV-2 virus.
The RGD-based interaction between integrins and SARS2-S is another subject of debate, with conflicting findings from different research groups. For example, one study reported a surprising high-affinity binding between SARS2-S S1 and α5β1 (12), whereas others found no binding at all (18, 19). Similarly, one study detected an interaction between SARS2-S RBD and αIIbβ3 (17), while another study showed no binding (18). Interestingly, a different study reported similar levels of binding between SARS2-S RBD and both RGD-recognizing (α5β1) and non-RGD-recognizing (α4β1, α4β7, and αLβ2) integrins (14). The reasons for these inconsistencies among studies are not yet clear. Unlike the RGD motifs found in well-characterized integrin ligands, which are typically located in a flexible loop known as RGD finger (51, 52), such as in fibronectin (SI Appendix, Fig. S1A), the RGD motif in SARS2-S resides within a rigid α-helical structure that is unlikely to undergo conformational changes. This makes the backbone and sidechain orientation of the RGD unfavorable for integrin binding (Fig. 3B), as indicated by a molecular dynamics simulation study (53). Our data obtained from ELISA, pull-down, competition, mutagenesis, and functional assays did not support the RGD-dependent interaction between SARS2-S and α5β1. Instead, we found that the interaction may depend on the S2 subunit of SARS2-S. Furthermore, our detailed analysis of purified α5β1 suggested the involvement of α5β1 homo-oligomerization in binding to SARS2-S. Clearly, further structural studies are necessary to gain a more comprehensive understanding of the interaction between SARS2-S and α5β1, as well as other integrins.
We did not observe any inhibition effect of ATN-161 on the interaction between α5β1 and SARS2-S, nor on S-mediated virus infection and cell fusion. Our analysis of the cryo-EM structure of α5β1 in complex with Fn7-10 raised concerns about the mechanism by which ATN-161 works as an inhibitor in functional studies of α5β1. Despite being designed based on the synergistic integrin binding site on Fn9 (SI Appendix, Fig. S1A), the R to C mutation in ATN-161 may significantly reduce its binding affinity with α5β1. One study suggested that ATN-161 might bind to the N terminus of β1, but no supporting data were provided (54). Another study proposed that it interacts with the RBD of SARS2-S and blocks ACE2 binding (55). Molecular docking and molecular dynamics simulation studies have also suggested the binding of ATN-161 on the main protease of SARS-CoV-2 (56). Thus, the exact mechanism by which ATN-161 interacts with α5β1 remains unknown. Caution should be taken when considering the use of ATN-161 as a potential α5β1 antagonist.
SARS2-S has been reported to induce an inflammatory response via TLRs in macrophages. However, there are inconsistent and conflicting data regarding whether TLRs, or which TLRs (TLR2 or TLR4), interact with the SARS2-S (57–60). Integrin α5β1 has been implicated in regulating inflammatory response through its interaction with physiological or pathogen ligands (38, 61, 62). We used HUVEC as a model given the established inflammatory role of α5β1 in this cell line (38) and the absence of surface TLR2 and TLR4 expression in the resting state (63). We found that both immobilized and soluble SARS2-S induced an inflammatory response in endothelial cells, as demonstrated by NF-κB activation and cytokine release. This finding is consistent with another study (22). In agreement with our protein interaction results, the α5β1-dependent inflammatory responses were induced by the SARS2-S or its S2 subunit but not its RBD or S1 subunit. In contrast, SARS-S did not induce NF-κB activation or IL-6 release under our experimental conditions. This mechanism differs from the inflammation mediated by TLRs, which do not discriminate between SARS-CoV and SARS-CoV-2 (8, 64). It is conceivable that SARS-CoV-2 induces inflammation through processes not previously observed in SARS-CoV.
The involvement of PDE4D activation in α5β1-mediated endothelial inflammatory response has been established by both in vitro and in vivo studies (38, 65, 66). Upon activation, PDE4D hydrolyses cAMP, leading to reduced protein kinase A (PKA) activity (40). The reduction in PKA activity subsequently triggers NF-κB activation and the production of proinflammatory cytokines such as IL-6. Additionally, decreased intracellular cAMP levels activate the NLRP3 inflammasome, resulting in the maturation and secretion of IL-1β (41). Our findings that SARS2-S, via α5 integrin, induces GSDMD cleavage and IL-1β release, a downstream response of inflammasome activation (43), align with this scenario. Previous studies have demonstrated the ability of SARS2-S to induce NLRP3 inflammasome activation in macrophages (67, 68). Besides endothelial cells, α5 integrin and PDE4D are widely expressed in various other cell types, including immune cells such as macrophages and neutrophils, lung epithelial cells, and neurons. These cell types are known to play roles in COVID-19 pathogenesis (2, 5). Elevated levels of IL-6 and IL-1β and inflammasome activation have been associated with the severity of COVID-19 (3, 69).
Our data support the involvement of the inflammasome and GSDMD in COVID-19 (4), mediated by the SARS2-S-induced α5-PDE4D-cAMP cascade. PDE4 inhibition has been validated as an anti-inflammatory strategy and may hold potential as a treatment for COVID-19 (70, 71). While our data suggest the important role of cAMP regulation by the SARS2-S/α5β1 signaling in the inflammatory response, further investigation into the involvement of additional α5β1-associated signaling pathways is required. It also remains to be determined whether the SARS2-S/α5β1 interaction and signaling follow the conventional integrin activation model that involves large-scale conformational regulation. Our study provides a molecular basis for targeting the S-α5β1 interaction and its downstream pathway as potential therapeutic approaches for COVID-19.
Materials and Methods
The detailed materials and methods are available in SI Appendix. DNA constructs were either obtained from Addgene or generated through standard molecular cloning techniques. We created α5 integrin knockout cell lines using CRISPR/Cas9 technology. Recombinant proteins were either obtained from BEI Resources or produced in HEK293 cells. Virus infection and cell–cell fusion assays were conducted following established protocols. The interaction between the S proteins and α5β1 was assessed through ELISA, DLS, and pull-down assays using standard procedures. NF-κB activation, cytokine release, and GSDMD cleavage were analyzed using ELISA and western blotting. Statistical analysis was carried out on at least three individual datasets and analyzed with GraphPad Prism software.
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We thank Patrick Wilson for providing plasmids for spike protein expression. We thank Gary Whittaker for providing plasmids for pseudovirus production. We thank Eric Michalski in the VBRI Core lab and Cathy Paddock in Peter Newman’s lab for providing HUVEC cell lines. We thank Kartik Chandran for providing rVSV-G virus. We thank Sean Whelan for providing rVSV-S virus. We thank Jing Li in Timothy Springer’s lab for providing α5β1 protein. We thank Peter Newman and Gilbert White for the critical reading of the manuscript. This work was supported in whole or in part by NIH grants R01HL131836 (J.Z.) and R01GM137143 (J.Z.).
Author contributions
H.Z. and J.Z. designed research; H.Z., Z.W., H.T.T.N., A.J.W., Q.L., and A.L. performed research; H.Z., Z.W., H.T.T.N., A.J.W., and J.Z. analyzed data; and J.Z. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or SI Appendix. For additional information and resource requests, please contact Dr. Jieqing Zhu at Jieqing.Zhu@versiti.org.
Supporting Information
References
- 1.Zhou P., et al. , A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 579, 270–273 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Matheson N. J., Lehner P. J., How does SARS-CoV-2 cause COVID-19? Science 369, 510–511 (2020). [DOI] [PubMed] [Google Scholar]
- 3.Ragab D., Salah Eldin H., Taeimah M., Khattab R., Salem R., The COVID-19 cytokine storm; what we know so far. Front Immunol. 11, 1446 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vora S. M., Lieberman J., Wu H., Inflammasome activation at the crux of severe COVID-19. Nat. Rev. Immunol. 21, 694–703 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merad M., Martin J. C., Pathological inflammation in patients with COVID-19: A key role for monocytes and macrophages. Nat. Rev. Immunol. 20, 355–362 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pons S., Fodil S., Azoulay E., Zafrani L., The vascular endothelium: The cornerstone of organ dysfunction in severe SARS-CoV-2 infection. Crit Care 24, 353 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meinhardt J., et al. , Olfactory transmucosal SARS-CoV-2 invasion as a port of central nervous system entry in individuals with COVID-19. Nat. Neurosci. 24, 168–175 (2021). [DOI] [PubMed] [Google Scholar]
- 8.Khanmohammadi S., Rezaei N., Role of Toll-like receptors in the pathogenesis of COVID-19. J. Med. Virol. 93, 2735–2739 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hoffmann M., et al. , SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181, 271–280.e278 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beddingfield B. J., et al. , The integrin binding peptide, ATN-161, as a novel therapy for SARS-CoV-2 infection. JACC Basic Transl. Sci. 6, 1–8 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simons P., et al. , Integrin activation is an essential component of SARS-CoV-2 infection. Sci. Rep. 11, 20398 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu J., Lu F., Chen Y., Plow E., Qin J., Integrin mediates cell entry of the SARS-CoV-2 virus independent of cellular receptor ACE2. J. Biol. Chem. 298, 101710 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bugatti A., et al. , SARS-CoV-2 infects human ACE2-negative endothelial cells through an alpha(v)beta(3) integrin-mediated endocytosis even in the presence of vaccine-elicited neutralizing antibodies. Viruses 14, 705 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang M., et al. , Lymphocyte integrins mediate entry and dysregulation of T cells by SARS-CoV-2. Signal Transduct Target Ther. 8, 84 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kliche J., Kuss H., Ali M., Ivarsson Y., Cytoplasmic short linear motifs in ACE2 and integrin beta(3) link SARS-CoV-2 host cell receptors to mediators of endocytosis and autophagy. Sci. Signal 14, eabf1117 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meszaros B., et al. , Short linear motif candidates in the cell entry system used by SARS-CoV-2 and their potential therapeutic implications. Sci. Signal 14, eabd0334 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma X., et al. , SARS-CoV-2 RBD and its variants can induce platelet activation and clearance: Implications for antibody therapy and vaccinations against COVID-19. Research (Wash D C) 6, 0124 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhn C. C., et al. , Direct Cryo-ET observation of platelet deformation induced by SARS-CoV-2 spike protein. Nat. Commun. 14, 620 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Norris E. G., Pan X. S., Hocking D. C., Receptor-binding domain of SARS-CoV-2 is a functional alphav-integrin agonist. J. Biol. Chem. 299, 102922 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biering S. B., et al. , SARS-CoV-2 Spike triggers barrier dysfunction and vascular leak via integrins and TGF-beta signaling. Nat. Commun. 13, 7630 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nader D., Kerrigan S. W., Molecular cross-talk between integrins and cadherins leads to a loss of vascular barrier integrity during SARS-CoV-2 infection. Viruses 14, 891 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Robles J. P., et al. , The spike protein of SARS-CoV-2 induces endothelial inflammation through integrin alpha5beta1 and NF-kappaB signaling. J. Biol. Chem. 298, 101695 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nader D., Fletcher N., Curley G. F., Kerrigan S. W., SARS-CoV-2 uses major endothelial integrin alphavbeta3 to cause vascular dysregulation in-vitro during COVID-19. PloS One 16, e0253347 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hynes R. O., Integrins: Bi-directional, allosteric, signalling machines. Cell 110, 673–687 (2002). [DOI] [PubMed] [Google Scholar]
- 25.Hussein H. A., et al. , Beyond RGD: Virus interactions with integrins. Arch Virol. 160, 2669–2681 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Maginnis M. S., Virus-receptor interactions: The key to cellular invasion. J. Mol. Biol. 430, 2590–2611 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stewart P. L., Nemerow G. R., Cell integrins: Commonly used receptors for diverse viral pathogens. Trends Microbiol. 15, 500–507 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Beaudoin C. A., Hamaia S. W., Huang C. L., Blundell T. L., Jackson A. P., Can the SARS-CoV-2 spike protein bind integrins independent of the RGD sequence? Front Cell Infect Microbiol. 11, 765300 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sigrist C. J., Bridge A., Le Mercier P., A potential role for integrins in host cell entry by SARS-CoV-2. Antiviral Res. 177, 104759 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Park E. J., et al. , The spike glycoprotein of SARS-CoV-2 binds to beta1 integrins expressed on the surface of lung epithelial cells. Viruses 13, 645 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amruta N., et al. , In vivo protection from SARS-CoV-2 infection by ATN-161 in k18-hACE2 transgenic mice. Life Sci. 284, 119881 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Buchrieser J., et al. , Syncytia formation by SARS-CoV-2-infected cells. Embo J. 40, e107405 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zeng C., et al. , SARS-CoV-2 spreads through cell-to-cell transmission. Proc. Natl. Acad. Sci. U.S.A. 119, e2111400119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai Y., et al. , Distinct conformational states of SARS-CoV-2 spike protein. Science 369, 1586–1592 (2020), 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang Q., et al. , Structural and functional basis of SARS-CoV-2 entry by using human ACE2. Cell 181, 894–904.e899 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Livant D. L., et al. , Anti-invasive, antitumorigenic, and antimetastatic activities of the PHSCN sequence in prostate carcinoma. Cancer Res. 60, 309–320 (2000). [PubMed] [Google Scholar]
- 37.Schumacher S., et al. , Structural insights into integrin alpha5beta1 opening by fibronectin ligand. Sci. Adv. 7, eabe9716 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yun S., et al. , Interaction between integrin alpha5 and PDE4D regulates endothelial inflammatory signalling. Nat. Cell Biol. 18, 1043–1053 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacKenzie S. J., Houslay M. D., Action of rolipram on specific PDE4 cAMP phosphodiesterase isoforms and on the phosphorylation of cAMP-response-element-binding protein (CREB) and p38 mitogen-activated protein (MAP) kinase in U937 monocytic cells. Biochem. J. 347, 571–578 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tavares L. P., et al. , Blame the signaling: Role of cAMP for the resolution of inflammation. Pharmacol. Res. 159, 105030 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Lee G. S., et al. , The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca2+ and cAMP. Nature 492, 123–127 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang M., et al. , Biomimetic human disease model of SARS-CoV-2-induced lung injury and immune responses on organ chip system. Adv. Sci. (Weinh) 8, 2002928 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chan A. H., Schroder K., Inflammasome signaling and regulation of interleukin-1 family cytokines. J. Exp. Med. 217, e20190314 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bao L., et al. , The pathogenicity of SARS-CoV-2 in hACE2 transgenic mice. Nature 583, 830–833 (2020). [DOI] [PubMed] [Google Scholar]
- 45.Sun S. H., et al. , A mouse model of SARS-CoV-2 infection and pathogenesis. Cell Host Microbe 28, 124–133.e124 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang R. D., et al. , Pathogenesis of SARS-CoV-2 in transgenic mice expressing human angiotensin-converting enzyme 2. Cell 182, 50–58.e58 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sun J., et al. , Generation of a broadly useful model for COVID-19 pathogenesis, vaccination, and treatment. Cell 182, 734–743.e735 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kreutzberger A. J. B., et al. , SARS-CoV-2 requires acidic pH to infect cells. Proc. Natl. Acad. Sci. U.S.A. 119, e2209514119 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Case J. B., et al. , Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2. Cell Host Microbe 28, 475–485.e475 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dieterle M. E., et al. , A replication-competent vesicular stomatitis virus for studies of SARS-CoV-2 spike-mediated cell entry and its inhibition. Cell Host Microbe 28, 486–496.e486 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhu J., Zhu J., Springer T. A., Complete integrin headpiece opening in eight steps. J. Cell Biol. 201, 1053–1068 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Dong X., et al. , Force interacts with macromolecular structure in activation of TGF-beta. Nature 542, 55–59 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Othman H., et al. , SARS-CoV-2 spike protein unlikely to bind to integrins via the Arg-Gly-Asp (RGD) motif of the receptor binding domain: Evidence from structural analysis and microscale accelerated molecular dynamics. Front Mol. Biosci. 9, 834857 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stoeltzing O., et al. , Inhibition of integrin alpha5beta1 function with a small peptide (ATN-161) plus continuous 5-FU infusion reduces colorectal liver metastases and improves survival in mice. Int. J. Cancer J. Int. du Cancer 104, 496–503 (2003). [DOI] [PubMed] [Google Scholar]
- 55.Rabbani G., Ahn S. N., Kwon H., Ahmad K., Choi I., Penta-peptide ATN-161 based neutralization mechanism of SARS-CoV-2 spike protein. Biochem. Biophys. Rep. 28, 101170 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Silva R. S., Souza L. M. P., Costa R. K. M., Souza F. R., Pimentel A. S., Absolute binding free energies of the antiviral peptide ATN-161 with protein targets of SARS-CoV-2. J. Biomol. Struct Dyn 41, 10546–10557 (2022), 10.1080/07391102.2022.2154848. [DOI] [PubMed] [Google Scholar]
- 57.Shirato K., Kizaki T., SARS-CoV-2 spike protein S1 subunit induces pro-inflammatory responses via toll-like receptor 4 signaling in murine and human macrophages. Heliyon 7, e06187 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zheng M., et al. , TLR2 senses the SARS-CoV-2 envelope protein to produce inflammatory cytokines. Nat. Immunol. 22, 829–838 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Khan S., et al. , SARS-CoV-2 spike protein induces inflammation via TLR2-dependent activation of the NF-kappaB pathway. Elife 10, e68563 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao Y., et al. , SARS-CoV-2 spike protein interacts with and activates TLR41. Cell Res. 31, 818–820 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jun H. K., Lee S. H., Lee H. R., Choi B. K., Integrin alpha5beta1 activates the NLRP3 inflammasome by direct interaction with a bacterial surface protein. Immunity 36, 755–768 (2012). [DOI] [PubMed] [Google Scholar]
- 62.Thinwa J., Segovia J. A., Bose S., Dube P. H., Integrin-mediated first signal for inflammasome activation in intestinal epithelial cells. J. Immunol. 193, 1373–1382 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shuang C., Wong M. H., Schulte D. J., Arditi M., Michelsen K. S., Differential expression of Toll-like receptor 2 (TLR2) and responses to TLR2 ligands between human and murine vascular endothelial cells. J. Endotoxin Res. 13, 281–296 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Sariol A., Perlman S., SARS-CoV-2 takes its Toll. Nat. Immunol. 22, 801–802 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yun S., et al. , Integrin alpha5beta1 regulates PP2A complex assembly through PDE4D in atherosclerosis. J. Clin. Invest. 129, 4863–4874 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Budatha M., et al. , Inhibiting integrin alpha5 cytoplasmic domain signaling reduces atherosclerosis and promotes arteriogenesis. J. Am. Heart Assoc. 7, e007501 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Theobald S. J., et al. , Long-lived macrophage reprogramming drives spike protein-mediated inflammasome activation in COVID-19. EMBO Mol. Med. 13, e14150 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Eisfeld H. S., et al. , Viral glycoproteins induce NLRP3 inflammasome activation and pyroptosis in macrophages. Viruses 13, 2076 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rodrigues T. S., et al. , Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J. Exp. Med. 218, e20201707 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dastidar S. G., Rajagopal D., Ray A., Therapeutic benefit of PDE4 inhibitors in inflammatory diseases. Curr. Opin. Invest. Drugs 8, 364–372 (2007). [PubMed] [Google Scholar]
- 71.Dalamaga M., Karampela I., Mantzoros C. S., Commentary: Phosphodiesterase 4 inhibitors as potential adjunct treatment targeting the cytokine storm in COVID-19. Metabolism 109, 154282 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
All study data are included in the article and/or SI Appendix. For additional information and resource requests, please contact Dr. Jieqing Zhu at Jieqing.Zhu@versiti.org.