Abstract
pbpA, a gene encoding penicillin-binding protein (PBP) 1 of Staphylococcus aureus, was cloned in an Escherichia coli MC1061 transformant which grew on a plate containing 512 μg of vancomycin per ml. This gene encodes a 744-amino-acid sequence which conserves three motifs of PBPs, SXXK, SXN, and KTG. The chromosomal copy of pbpA could be disrupted only when RN4220, a methicillin-sensitive S. aureus strain, had additional copies of pbpA in its episome. Furthermore, these episomal copies of pbpA could not be eliminated by an incompatible plasmid when the chromosomal copy of pbpA was disrupted beforehand. Based on these observations, we concluded that pbpA is essential for the growth of methicillin-sensitive S. aureus.
Staphylococcus aureus, one of the most medically important pathogens, causes both hospital- and community-acquired infections worldwide. Its resistance to multiple drugs, especially to β-lactam antibiotics, is a major therapeutic problem. S. aureus penicillin-binding proteins (PBPs), which synthesize its cell wall, are the targets of β-lactam antibiotics. Methicillin-sensitive S. aureus (MSSA) has four PBPs, PBP1 to -4, and methicillin-resistant S. aureus (MRSA) has an additional PBP (4, 10, 41), PBP2′, which is recognized as a major cause of its high resistance to β-lactam antibiotics. The gene encoding PBP2′, mecA, and several other genes have been shown to contribute to the β-lactam resistance of S. aureus (6, 7, 20, 21, 26, 36). The mecA gene was cloned by selecting a tobramycin-resistant Escherichia coli clone having a genomic fragment of MRSA in which sequences encoding methicillin and tobramycin resistance were linked (27). A gene encoding PBP2 was cloned from a λgt11 library by using antiserum which was raised against excised PBP2 from a sodium dodecyl sulfate (SDS)-polyacrylamide gel (31). The gene encoding PBP4 was cloned by Tn551 insertion mutagenesis in a penicillin-resistant PBP4 overproducer (21). Here we describe another method for cloning the gene encoding PBP1 of S. aureus and genetically show the essentiality of this gene for the growth of MSSA.
Bacterial strains and plasmids.
Bacterial strains and plasmids used in this study are summarized in Table 1. RN4220 (24) and BB255 (6) were kindly provided by R. P. Novick and B. Berger-Bächi, respectively.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Genotype and relevant markersa | Source or reference |
|---|---|---|
| Strains | ||
| E. coli MC1061 | 11 | |
| S. aureus | ||
| NCTC8325 | Type strain of S. aureus | 33 |
| BB255 | NCTC8325, penicillinase plasmid cured | 6 |
| RN4220 | NCTC8325, penicillinase plasmid cured, hly mutant, restriction minus | 24 |
| AW302 | RN4220(pAW11) | This study |
| AW303 | RN4220(pPBPA) | This study |
| AW304 | AW303 pbpA::pΔPBPA | This study |
| AW305 | AW302(pAW12) | This study |
| AW306 | AW303(pAW12) | This study |
| AW307 | AW304(pAW12) | This study |
| Plasmids | ||
| pAW119 | Cloning shuttle vector, pMB1 ori for replication in E. coli, pAMα1 ori for replication in gram-positive organisms, Tetr | This study |
| pAW9 | Suicide vector in S. aureus, p15A ori for replication in E. coli, Tetr | This study |
| pAW10 | Small (3.9-kb) shuttle vector, p15A ori for replication in E. coli, pAMα1 ori for replication in gram-positive organisms, Tetr | This study |
| pAW11 | Cloning shuttle vector, pMB1 ori for replication in E. coli, pAMα1 ori for replication in gram-positive organisms, Emr | This study |
| pAW12 | Cloning shuttle vector, the same origins as pAW11, Kmr | This study |
| pPBPA | pAW11 with a KpnI-KpnI 3.1-kb fragment containing orf2 and pbpA | This study |
| pΔPBPA | pAW9 with a HindIII-HindIII 936-bp fragment containing a middle part of pbpA | This study |
| pORF2 | pAW9 with a Sau3AI-HindIII 993-bp fragment containing intact orf2 | This study |
Tetr, tetracycline resistant; Emr, erythromycin resistant; Kmr, kanamycin resistant; ori, origin.
Transformation of E. coli and S. aureus.
E. coli MC1061 (11) and S. aureus RN4220 were transformed by electroporation by using a Gene Pulser system (Bio-Rad, Richmond, Calif.). Methods for the preparation of MC1061 competent cells and conditions of the transformation were described previously (15). A method for the preparation of RN4220 competent cells is described below. One fresh colony was inoculated into 5 ml of filtrated brain heart infusion (BHI) broth (Difco, Detroit, Mich.) and incubated at 37°C with gentle shaking. The cells were harvested at mid-log phase, i.e., optical density at 550 nm of <0.5, and rinsed twice with a buffer containing 1.1 M sucrose and 2 mM MgCl2 at 4°C. After being concentrated 50 times by the rinse steps, the cells were transformed with DNAs by the application of current from a 25-μF condenser, which was charged to 2.0 kV, to a 1-mm-wide Gene Pulser cuvette (Bio-Rad), which was connected to a 200-Ω resistance in parallel. The cells were immediately mixed with 1 ml of BHI broth containing 1.1 M sucrose and incubated at 37°C for 1 h for the expression of antibiotic resistance markers. Then they were incubated on BHI agar containing appropriate concentrations of antibiotics at 37°C overnight. The DNAs used for electroporation were purified by the CsCl ultracentrifuge.
Antibiotics.
Vancomycin was purchased from Shionogi (Osaka, Japan) and Sigma (St. Louis, Mo.). Tetracycline, erythromycin, and kanamycin were purchased from Sigma and used at concentrations of 2, 1, and 10 μg/ml, respectively, unless otherwise stated.
Construction of a plasmid library.
A total of 5 μg of genomic DNA of NCTC8325 (33) was partially digested by Sau3AI (New England Biolabs, Beverly, Mass.), and fragments ranging from 3 to 5 kb in length were enriched by agarose gel electrophoresis. One hundred nanograms of pAW119 was digested with BamHI (New England Biolabs), treated with calf intestinal alkaline phosphatase (Boehringer Mannheim, Mannheim, Germany), and ligated with the genomic fragments of NCTC8325. These DNAs were used for the transformation of MC1061. At 37°C, the transformants were selected on LB agar (Lennox; Difco) containing 512 μg of vancomycin and 10 μg of tetracycline per ml. After 18 h, the colonies obtained were streaked on duplicate LB plates containing 10 μg of tetracycline per ml; one set of the plates was incubated at 37°C, and the other set was incubated at 42°C for 18 h. Clones which grew at 37°C but which could not grow or which grew very slowly at 42°C were analyzed for their inserts. An introduced plasmid of each clone was digested with HincII (New England Biolabs) or was doubly digested with EcoRI (New England Biolabs) and PstI (New England Biolabs), and the clones were compared to each other by agarose gel electrophoresis. Five clones were selected (see below), and their PBP activities were studied.
PBP assay.
Detailed methods for the PBP assay were described previously (41). In brief, PBPs were labeled with [benzyl-14C]penicillin (Amersham, Buckinghamshire, England), separated by SDS–8% polyacrylamide gel electrophoresis, and detected by fluorography.
pbpA disruption and its analysis.
To obtain pbpA disruptant AW304, RN4220 was first transformed with pPBPA to obtain AW303 and then AW303 was transformed with 2 μg of pΔPBPA and cultured with erythromycin and tetracycline. Plasmids of S. aureus clones were extracted with a Miniprep kit (Perkin-Elmer Applied Biosystems, Foster City, Calif.) according to the manufacturer’s instructions except for adding lysostaphin (Sigma) to resuspension buffer to a final concentration of 100 μg/ml and for incubation at 37°C for 10 min at the resuspension step. For amplification of chromosomal fragments around pbpA, PCR was performed by using primers 5′-TACTGGACATTCTAATGGTC-3′ and 5′-TTCCGACTCTATCACTTGTC-3′, and by using TaKaRa LA Taq (Takara Shuzo Co., Kyoto, Japan). Thermal cycles of 96°C for 20 s, 58°C for 20 s, and 72°C for 10 min were repeated 20 times. For Southern hybridization analysis, the resulting amplified products were transferred to a Hybond-N+ membrane (Amersham) and probed with whole pAW9 DNAs, which were labeled with digoxigenin (DIG) by using DIG-High Prime (Boehringer Mannheim). CDP Star (Boehringer Mannheim) was used for chemiluminescence detection by anti-digoxigenin-AP Fab fragments (Boehringer, Mannheim).
Plasmid incompatibility.
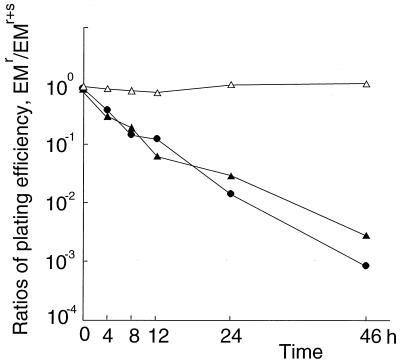
AW305, AW306, and AW307 were cultured at 37°C in 5 ml of LB broth (Lennox; Difco) containing kanamycin for AW305 and AW306 and containing kanamycin and tetracycline for AW307. Every 12 h, each clone was diluted 1,000-fold with the same medium. At times zero and at 4, 8, 12, 24, and 46 h after the first inoculation, AW305 and AW306 were harvested and plated on LB agar (Lennox; Difco) with or without erythromycin. AW307 was harvested at the same times and was plated on LB agar (Lennox; Difco) containing tetracycline with or without erythromycin. After the incubation of each clone at 37°C for 18 h, the number of colonies on each plate was counted, and ratios of plating efficiencies with erythromycin to those without erythromycin were calculated.
Nucleotide sequencing.
DNA sequences were determined by automatic DNA sequencers ABI PRISM 373A and 310 (Perkin-Elmer Applied Biosystems) with a DyePrimer and/or a DyeTerminator cycle sequencing kit (Perkin-Elmer Applied Biosystems).
Cloning of pbpA.
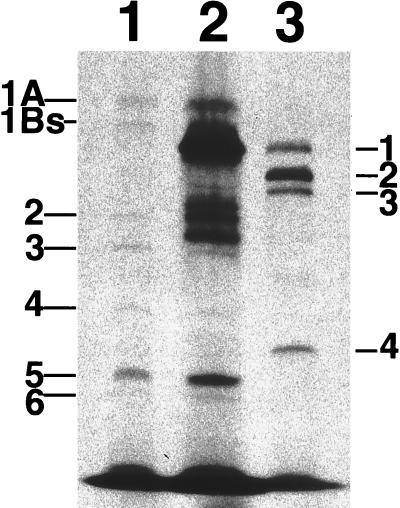
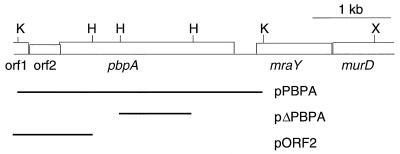
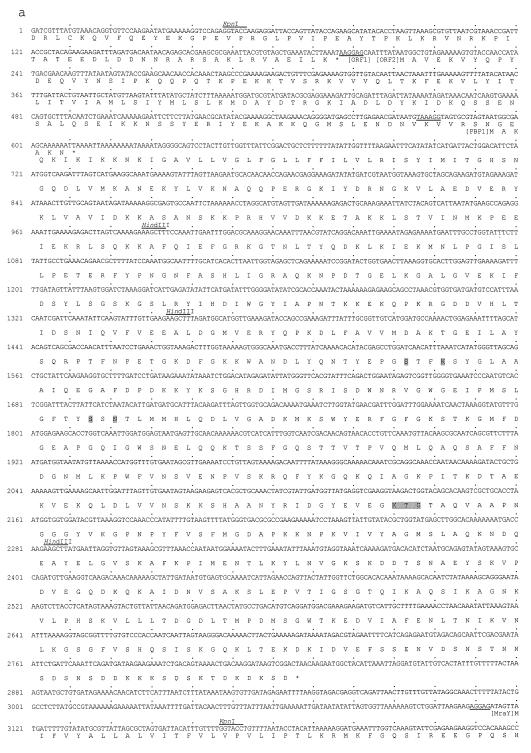
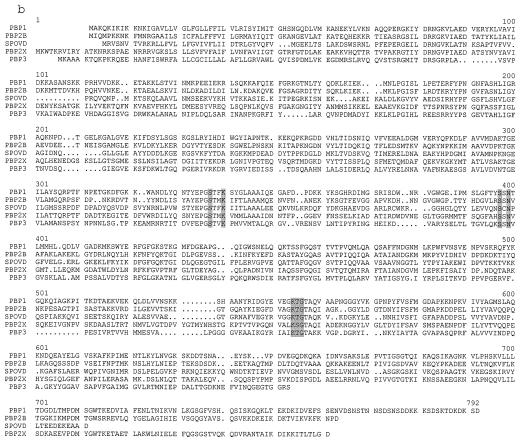
Previously, we found that the plating efficiency of an E. coli MC1061 strain having mecA in its episome was about 1,000 times higher than that of a strain having a control plasmid when they were cultured with 512 μg of vancomycin per ml at 37°C. At 42°C, the strain having mecA grows very slowly, while the growth rate of the control strain at 42°C did not remarkably differ from that at 37°C (unpublished data). Based on these findings, we speculated that the PBP gene(s) of S. aureus might be cloned by positive selection of E. coli MC1061 transformants by using vancomycin at 37°C and by subsequent negative selection at 42°C. As the first screening step, a NCTC8325 genomic library containing about 20,000 transformants of MC1061 was plated on LB agar containing 512 μg of vancomycin and 10 μg of tetracycline per ml. After incubation at 37°C for 18 h, 71 transformants were obtained. Thirty-two transformants could not grow or grow very slowly at 42°C. In 12 of these 32 transformants, five common restriction patterns were observed in inserts of their plasmids (data not shown). To check the penicillin-binding activities of these transformants, we performed a PBP assay for five representative transformants, each of which showed one of the five common restriction patterns of the inserts. As shown in Fig. 1, one transformant, which had a 4.9-kb genomic fragment of NCTC8325, had extra PBP activities (lane 2). These activities were not observed in a control E. coli clone having pAW119 (lane 1). The mobility of the strongest band of this clone was the same as that of PBP1 of S. aureus BB255 (lanes 2 and 3). We determined the nucleotide sequence of the 4.9-kb fragment and found a total of five open reading frames (ORFs); three ORFs are located inside of the fragment and the other two, lacking N-terminal or C-terminal amino acid sequences, are located at each side of the fragment (Fig. 2). A part of the nucleotide sequence of this 4.9-kb fragment and its deduced amino acid sequences are shown in Fig. 3a. The third ORF encodes a 744-amino-acid sequence, which shows a high degree of homology to those of PBP2B of Bacillus subtilis (45) (50.6% similarity and 40.6% identity), SpoVD of B. subtilis (12) (40.8% similarity and 31.5% identity), PBP2X of Streptococcus pneumoniae (25) (37.9% similarity and 32.4% identity), and PBP3 of E. coli (32) (33.2% similarity and 26.3% identity). The amino acid sequence of the product of the third ORF of S. aureus and those of all these PBPs, including that of spore-making protein SpoVD, share the conserved motifs of PBPs, SXXK, SXN, and KT(S)G (Fig. 3b). In addition, the calculated molecular size of the deduced amino acid sequence of the product of the third ORF was 82.7 kDa, which corresponded to the molecular size of PBP1 of S. aureus as estimated by its mobility in a gel used for the PBP assay (41). Based on these observations, we concluded that the gene encoding PBP1 of S. aureus NCTC8325 has been cloned in the 4.9-kb fragment. We propose the name pbpA because its product has the highest molecular weight of any of the PBPs of S. aureus. A pulsed-field gel electrophoresis analysis revealed that this gene is located on the SmaI-B fragment of the NCTC8325 chromosome (35, 42) (data not shown).
FIG. 1.
Fluorography of PBPs labeled with [benzyl-14C]penicillin after separation by SDS–8% polyacrylamide gel electrophoresis. Lane 1, an E. coli MC1061 transformant having pAW119; lane 2, an E. coli MC1061 transformant having a 4.9-kb genomic fragment of S. aureus NCTC8325; lane 3, S. aureus BB255. Designations of PBPs of E. coli and S. aureus are indicated at the left and right sides of the fluorography, respectively. On each lane, 100 μg of proteins of the membrane fraction was applied.
FIG. 2.
A restriction map of a 4.9-kb genomic fragment of S. aureus NCTC8325 containing five ORFs, which are shown by boxes. The name of each ORF is indicated under the map. Each thick line under the map corresponds to an insert of each plasmid, whose name is indicated at the right side of each line. K, KpnI; H, HindIII; X, XbaI.
FIG. 3.
(a) Nucleotide sequence of a part of the 4.9-kb fragment of NCTC8325 shown in Fig. 2, and its deduced amino acid sequence. The putative ribosome binding site of each ORF is underlined. The conserved motifs of PBPs are shown by shading in the amino acid sequence of PBP1. KpnI and HindIII sites used for constructing the plasmids shown in Fig. 2 are indicated above the nucleotide sequence. (b) Amino acid sequence alignment of PBPs of various organisms including PBP1 of S. aureus. Common motifs of PBPs are shown by shadowing PBP1, PBP1 of S. aureus; PBP2B, PBP2B of B. subtilis; SPOVD, SpoVD of B. subtilis; PBP2X, PBP2X of S. pneumoniae; PBP3, PBP3 of E. coli.
Essentiality of pbpA.
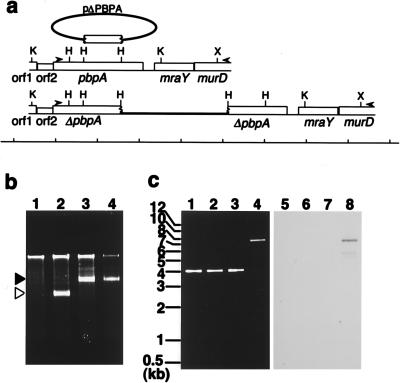
To test the essentiality of pbpA for growth of S. aureus genetically, we tried to disrupt this gene by homologous recombination. For its high level of competency, we adopted RN4220, a restriction-minus derivative of NCTC8325, and optimized conditions for transforming this strain by electroporation. With the conditions described in Materials and Methods, 5 × 105 transformants per μg of pAW10 (3.9 kb in length) can usually be obtained. In addition, we constructed pΔPBPA, which cannot replicate in S. aureus and which has a 936-bp HindIII-HindIII fragment of a middle portion of pbpA as an insert (Fig. 2, 3a, and 4a). Because this fragment lacks codons for both N-terminal and C-terminal sequences of PBP1, it will disrupt pbpA by a single homologous recombination (Fig. 4a). As a control, we constructed pORF2, which is another suicide plasmid having a 993-bp fragment containing complete orf2 (Fig. 2 and 3a) located upstream of pbpA and which will not disrupt orf2 on the chromosome. In the first experiment, we tried to transform RN4220 with 5 μg of pΔPBPA or pORF2. Table 2 shows the results of three independent experiments. A total of 147 transformants were obtained from pORF2, while no transformant could be obtained from pΔPBPA. A Southern hybridization analysis of randomly selected transformants of pORF2 showed that pORF2 was integrated into the chromosomal copy of orf2 by homologous, not by illegitimate, recombination (data not shown). In the next experiment, we tried to transform AW302 and AW303 with pΔPBPA. AW302 had control plasmid pAW11 and had only one copy of pbpA on its chromosome, while AW303 had extra copies of pbpA in its episome (Table 1). In this experiment, transformants were easily obtained from AW303, while no transformant could be obtained from AW302. One of 11 analyzed transformants of AW303, AW304, had a plasmid showing the same electrophoretic mobility as that of pPBPA (Fig. 4b). However, the length of the PCR-amplified fragment from the chromosome around pbpA of AW304 increased from 4.1 to 7.6 kb; the difference corresponds to the length of pΔPBPA (Fig. 4a and c). A whole fragment of pAW9, an original suicide plasmid used to make pΔPBPA (Table 1), hybridized to this 7.6-kb fragment of AW304 (Fig. 4c). These results showed that the chromosomal copy of pbpA of AW304 was disrupted by homologous recombination with pΔPBPA, while the episomal copies of pbpA on pPBPA were unaffected. In the other 10 transformants of AW303, the episomal copies of pbpA on pPBPA were disrupted and the chromosomal copy of pbpA was intact (data not shown). We think that this observed frequency was reasonable, because the copy number of pPBPA, estimated by Southern hybridization analysis, was about 10 (data not shown). In the third experiment, we tried to eliminate pPBPA from AW304 by using incompatible plasmid pAW12, which has the same replication origin as pPBPA but which has a different selection marker (Kmr; Table 1). At first, we transformed AW302, AW303, and AW304 with pAW12 and selected them with both erythromycin and kanamycin to obtain AW305, AW306, and AW307, respectively. In the presence of erythromycin and kanamycin, AW305, AW306, and AW307 can tentatively retain pAW11 and pAW12, pPBPA and pAW12, and pPBPA and pAW12, respectively (34). Then, these clones were cultured without erythromycin. When AW305 and AW306 were cultured with kanamycin alone, the numbers of erythromycin-resistant clones decreased logarithmically (Fig. 5). However, almost all populations of AW307 retained resistance to erythromycin even though they were cultured without erythromycin for 46 h (Fig. 5). This observation means that the extrachromosomal pbpA on pPBPA cannot be eliminated if the chromosomal copy of pbpA is disrupted beforehand. Based on the results of these three experiments (the failure to obtain transformants from RN4220 with pΔPBPA, the success in obtaining a pbpA disruptant in the presence of pPBPA, and the observed retention of pPBPA in a chromosomal pbpA disruptant AW304), we concluded that pbpA of S. aureus (MSSA) is essential for its growth.
FIG. 4.
(a) Restriction maps of the genomic fragment around pbpA before and after integration of suicide plasmid pΔPBPA. The name of each ORF is indicated under the maps, and the size marker is indicated at the bottom. Short vertical lines in the size marker are 1 kb apart. Arrowheads show the locations of the primers used for PCR shown in panel c. Abbreviations of the restriction enzymes are as defined in the legend for Fig. 2. (b) Plasmid profiles of S. aureus clones. The plasmid fraction of each clone was analyzed by agarose gel electrophoresis. Solid and open triangles indicate pPBPA and pAW11, respectively. Lane 1, RN4220; lane 2, AW302; lane 3, AW303; lane 4, AW304. (c) PCR-amplified fragments separated by agarose gel electrophoresis (lanes 1 to 4) and Southern hybridization analysis (lanes 5 to 8) by using pAW9 as a probe. Lanes 1 and 5, RN4220; lanes 2 and 6, AW302; lanes 3 and 7, AW303; lane 4 and 8, AW304. The sizes of size markers are indicated at the left side of the panel.
TABLE 2.
Result of transformation with suicide plasmids
| Suicide plasmid | No. of transformants in expt:
|
||
|---|---|---|---|
| 1 | 2 | 3 | |
| pΔPBPA | 0 | 0 | 0 |
| pORF2 | 54 | 11 | 82 |
FIG. 5.
Elimination of pAW11 and pPBPA by incompatible plasmid pAW12. After being cultured in LB broth with kanamycin at 37°C for the indicated numbers of hours, AW305 (•), AW306 (▴), and AW307 (▵) were plated on LB agar with or without erythromycin. Calculated ratios of plating efficiency with versus without erythromycin were plotted at each harvesting time.
As described in this report, a total of four genes encoding PBP1, PBP2′ (27), PBP2 (31), and PBP4 (21) have been cloned and sequenced from S. aureus. As mentioned above, all genes other than that of PBP2 were cloned without purification of PBPs. To clone pbpA, encoding PBP1 of S. aureus, we used vancomycin for selecting E. coli MC1061 transformants. A population analysis of an MC1061 clone having pPBPA (Table 1, Fig. 2) showed vancomycin resistance similar to that conferred by mecA (data not shown). It remains unknown, however, whether pbpA or orf2 or both on pPBPA are necessary for the E. coli clone to be resistant to vancomycin. The reason for the observed vancomycin resistance is difficult to explain. One hypothesis is that the PBPs of S. aureus may protect the peptidoglycan precursor from an attack by vancomycin at the periplasm of E. coli. Although PBP overproduction has been shown to be related to the glycopeptide resistance of S. aureus (30), it is not probable that the PBPs of S. aureus function in E. coli, and even the translocation of the PBPs of S. aureus from the cytoplasm to the periplasm of E. coli cannot be proven by the PBP assay used in this study (41). The second hypothesis is that the overproduced PBPs of S. aureus may alter the concentrations of various peptidoglycan precursors such as UDP-N-acetylmuramoyl-tri-, tetra-, and pentapeptides in the cytoplasm of E. coli. In enterococci, altered concentrations of these precursors were shown to be related to glycopeptide resistance (2). It is interesting to analyze the remaining four E. coli transformants, which grew with 512 μg of vancomycin per ml and which each had one of the common restriction patterns in the introduced plasmids but did not show detectable extra-PBP activity. They may contain a gene(s) involved in the synthesis or assembly of peptidoglycan precursors of S. aureus. On the other hand, we think that the negative selection at 42°C is less specific than vancomycin selection. This temperature may be toxic to an E. coli transformant in which PBP1 of S. aureus is overproduced (Fig. 2).
The deduced amino acid sequence of pbpA of S. aureus shows high degrees of homology to various PBPs of other organisms (see above). At present, the functions of PBP3, encoded by ftsI, of E. coli have been analyzed more throughly than those of other PBPs. A temperature-sensitive mutant of ftsI is aseptate and filamentous at the restrictive temperature (40), and overproduction of PBP3 in the ftsI mutant results in the suppression of filamentation (16). A study using azthreonam, which inhibits the PBP3 of E. coli specifically, confirms the notion that PBP3 is a septum-making enzyme (17). A carboxy truncation mutant of PBP2B of B. subtilis is reported to be filamentous (45), and no disruptant of the associated gene can be obtained (14). The deletion of PBP2X of S. pneumoniae has been shown to be lethal too (23). It is most probable that PBP1 of S. aureus also contributes to the synthesis of the peptidoglycan septum. Further investigation by immuno- and/or fluorescence staining of PBP1 to show its localization in dividing cells of S. aureus will provide more information about the function(s) of PBP1 as such techniques did for PBP3 of E. coli (44).
Interestingly, four ORFs other than that of pbpA on the cloned 4.9-kb fragment (Fig. 2) also show homologies to the members of cell division gene cluster mra of E. coli (29), in which ftsI is located; deduced amino acid sequences corresponding to orf1, orf2, mraY, and murD of S. aureus (Fig. 2) have homologies to those corresponding to yabC (8, 9) (49.2% similarity to a partial sequence), ftsL (18) (31.8% similarity), mraY (22) (55.0% similarity), and murD (28) (37.3% similarity to a partial sequence) of E. coli, respectively. B. subtilis also has homologous genes in the neighborhood of pbpB, which encode PBP2B; deduced amino acid sequences corresponding to orf1, orf2, mraY, and murD of S. aureus (Fig. 2) have homologies to those corresponding to yllC (45) (73.8% similarity to a partial sequnece), ftsL (45) (39.3% similarity), mraY (13) (67.9% similarity), and murD (13) (55.0% similarity to a partial sequnece) of B. subtilis, respectively. All these genes of E. coli and B. subtilis are thought to be involved in the synthesis of peptidoglycan or its precursors (12–14, 16, 18, 22, 28, 29, 45). In addition, E. coli has other cell wall-synthesizing genes, ftsZ and ddl, within the mra region (8, 9). Our preliminary sequencing data spanning the 14 kb downstream of pbpA confirmed the presence of ftsZ of S. aureus (1) 5.4 kb from pbpA, but we could not find any ORFs homologous to the ddl or van genes of enterococci (3). This observation agrees with that in a recently published report (37), which described a nucleotide sequence spanning 12.1 kb around pbpA.
Several genetic methods have been established to show the essentiality of E. coli genes for its growth. These methods include obtaining a temperature-sensitive mutant (40), conditional expression with an araC promoter (18), and selection of a disruptant with rpsL (19) or sacB (38). A mutation in rpsL confers streptomycin resistance on E. coli, but this resistance is sensitive dominant (19). At first, we tried to apply this selection to S. aureus by using its own rpsL gene (43). This gene, however, does not work as well that of E. coli. So we tried to disrupt pbpA by high-efficiency transformation with a suicide plasmid and by a subsequent single homologous recombination. By combining the data from the transformation and recombination experiments with the result of the plasmid incompatibility experiment, we could genetically show the essentiality of pbpA for the growth of MSSA. Our conclusion agrees with the previously postulated essentiality of PBP1 on the basis of PBP assays (5, 39).
Competitive PBP assays of MRSA have shown that β-lactam antibiotics much more easily bind to PBP1- to -4 than to PBP2′ (41), which leads to a well-accepted notion that PBP2′ can substitute for all other PBPs in the presence of β-lactam antibiotics. This notion, however, has not yet been proven genetically. A recent study by Pinho et al. shows that a Tn551 insertion near the C terminus of PBP2 of a MRSA strain results in a reduction of its methicillin resistance (36). Our study shows that PBP1 disruption is lethal to MSSA, but its essentiality and even its contribution to the methicillin resistance of MRSA remain unknown. By using a phage 80α transduction system, whether PBP2′ can substitute for PBP1 or not is under investigation.
Nucleotide sequence accession numbers.
The DNA sequence data described in this paper have been deposited in the GenBank/EMBL/DDBJ database with accession no. D28879 and AB007500.
Acknowledgments
We thank R. N. Novick and B. Berger-Bächi for supplying S. aureus strain RN4220 and BB255, respectively.
This work was supported by a grant from the Ministry of Health and Welfare.
REFERENCES
- 1.Alessi, D. M., and E. R. Olson. GenBank accession number U06462.
- 2.Arthur M, Depardieu F, Reynolds P, Courvalin P. Quantitative analysis of the metabolism of soluble cytoplasmic peptidoglycan precursors of glycopeptide-resistant enterococci. Mol Microbiol. 1996;21:33–44. doi: 10.1046/j.1365-2958.1996.00617.x. [DOI] [PubMed] [Google Scholar]
- 3.Arthur M, Reynolds P, Courvalin P. Glycopeptide resistance in enterococci. Trends Microbiol. 1996;4:401–407. doi: 10.1016/0966-842X(96)10063-9. [DOI] [PubMed] [Google Scholar]
- 4.Beck W D, Berger-Bächi B, Kayser F H. Additional DNA in methicillin-resistant Staphylococcus aureus and molecular cloning of mec-specific DNA. J Bacteriol. 1986;165:373–378. doi: 10.1128/jb.165.2.373-378.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beise F, Labischinski H, Giesbrecht P. Selective inhibition of penicillin-binding proteins and its effects on growth and architecture of Staphylococcus aureus. FEMS Microbiol Lett. 1988;55:195–202. [Google Scholar]
- 6.Berger-Bächi B, Barberis-Maino L, Strässle A, Kayser F H. FemA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterization. Mol Gen Genet. 1989;219:263–269. doi: 10.1007/BF00261186. [DOI] [PubMed] [Google Scholar]
- 7.Berger-Bächi B, Strässle A, Gustafson J E, Kayser F H. Mapping and characterization of multiple chromosomal factors involved in methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1992;36:1367–1373. doi: 10.1128/aac.36.7.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Blattner, F. R. GenBank accession numbers AE000118 and U00096.
- 9.Blattner F R, Plunkett III G, Bloch C A, Perna N T, Burland V, Riley M, Collado-Vides J, Glasner J D, Rode C K, Mayhew G F, Gregor J, Davis N W, Kirkpatrick H A, Goeden M A, Rose D J, Mauand B, Shao Y. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 10.Brown D F, Reynolds P E. Intrinsic resistance to beta-lactam antibiotics in Staphylococcus aureus. FEBS Lett. 1980;122:275–278. doi: 10.1016/0014-5793(80)80455-8. [DOI] [PubMed] [Google Scholar]
- 11.Casadaban M J, Cohen S N. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J Mol Biol. 1980;138:179–207. doi: 10.1016/0022-2836(80)90283-1. [DOI] [PubMed] [Google Scholar]
- 12.Daniel R A, Drake S, Buchanan C E, Scholle R, Errington J. The Bacillus subtilis spoVD gene encodes a mother-cell-specific penicillin-binding protein required for spore morphogenesis. J Mol Biol. 1994;235:209–220. doi: 10.1016/s0022-2836(05)80027-0. [DOI] [PubMed] [Google Scholar]
- 13.Daniel R A, Errington J. DNA sequence of the murE-murD region of Bacillus subtilis 168. J Gen Microbiol. 1993;139:361–370. doi: 10.1099/00221287-139-2-361. [DOI] [PubMed] [Google Scholar]
- 14.Daniel R A, Williams A M, Errington J. A complex four-gene operon containing essential cell division gene pbpB in Bacillus subtilis. J Bacteriol. 1996;178:2343–2350. doi: 10.1128/jb.178.8.2343-2350.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dower W J, Miller J F, Ragsdale C W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988;16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreira L C, Keck W, Betzner A, Schwarz U J. In vivo cell division gene product interactions in Escherichia coli K-12. J Bacteriol. 1987;169:5776–5781. doi: 10.1128/jb.169.12.5776-5781.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Georgopapadakou N H, Smith S A, Sykes R B. Mode of action of azthreonam. Antimicrob Agents Chemother. 1982;21:950–956. doi: 10.1128/aac.21.6.950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Guzman L M, Barondess J J, Beckwith J. FtsL, an essential cytoplasmic membrane protein involved in cell division in Escherichia coli. J Bacteriol. 1992;174:7716–7728. [PMC free article] [PubMed] [Google Scholar]
- 19.Hashimoto-Gotoh T, Tsujimura A, Kuriyama K, Matsuda S. Construction and characterization of new host-vector systems for the enforcement-cloning method. Gene. 1993;137:211–216. doi: 10.1016/0378-1119(93)90008-q. [DOI] [PubMed] [Google Scholar]
- 20.Henze U, Sidow T, Wecke J, Labischinski H, Berger-Bächi B. Influence of femB on methicillin resistance and peptidoglycan metabolism in Staphylococcus aureus. J Bacteriol. 1993;175:1612–1620. doi: 10.1128/jb.175.6.1612-1620.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henze U U, Berger-Bächi B. Staphylococcus aureus penicillin-binding protein 4 and intrinsic β-lactam resistance. Antimicrob Agents Chemother. 1995;39:2415–2422. doi: 10.1128/aac.39.11.2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ikeda M, Wachi M, Jung H K, Ishino F, Matsuhashi M. The Escherichia coli mraY gene encoding UDP-N-acetylmuramoyl-pentapeptide: undecaprenyl-phosphate phospho-N-acetylmuramoyl-pentapeptide transferase. J Bacteriol. 1991;173:1021–1026. doi: 10.1128/jb.173.3.1021-1026.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kell C M, Sharma U K, Dowson C G, Town C, Balganesh T S, Spratt B G. Deletion analysis of the essentiality of penicillin-binding proteins 1A, 2B and 2X of Streptococcus pneumoniae. FEMS Microbiol Lett. 1993;106:171–175. doi: 10.1111/j.1574-6968.1993.tb05954.x. [DOI] [PubMed] [Google Scholar]
- 24.Kreiswirth B N, Lofdahl S, Betley M J, O’Reilly M, Schlievert P M, Bergdoll M S, Novick R P. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature. 1983;305:709–712. doi: 10.1038/305709a0. [DOI] [PubMed] [Google Scholar]
- 25.Laible G, Hakenbeck R, Sicard M A, Joris B, Ghuysen J M. Nucleotide sequences of the pbpX genes encoding the penicillin-binding proteins 2x from Streptococcus pneumoniae R6 and a cefotaxime-resistant mutant, C506. Mol Microbiol. 1989;3:1337–1348. doi: 10.1111/j.1365-2958.1989.tb00115.x. [DOI] [PubMed] [Google Scholar]
- 26.Maki H, Yamaguchi T, Murakami K. Cloning and characterization of a gene affecting the methicillin resistance level and the autolysis rate in Staphylococcus aureus. J Bacteriol. 1994;176:4993–5000. doi: 10.1128/jb.176.16.4993-5000.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsuhashi M, Song M D, Ishino F, Wachi M, Doi M, Inoue M, Ubukata K, Yamashita N, Konno M. Molecular cloning of the gene of a penicillin-binding protein supposed to cause high resistance to beta-lactam antibiotics in Staphylococcus aureus. J Bacteriol. 1986;167:975–980. doi: 10.1128/jb.167.3.975-980.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mengin-Lecreulx D, Parquet C, Desviat L R, Plá J, Flouret B, Ayala J A, van Heijenoort J. Organization of the murE-murG region of Escherichia coli: identification of the murD gene encoding the d-glutamic-acid-adding enzyme. J Bacteriol. 1989;171:6126–6134. doi: 10.1128/jb.171.11.6126-6134.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miyakawa T, Matsuzawa H, Matsuhashi M, Sugino Y. Cell wall peptidoglycan mutants of Escherichia coli K-12: existence of two clusters of genes, mra and mrb, for cell wall peptidoglycan biosynthesis. J Bacteriol. 1972;112:950–958. doi: 10.1128/jb.112.2.950-958.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moreira B, Boyle-Vavra S, deJonge B L M, Daum R S. Increased production of penicillin-binding protein 2, increased detection of other penicillin-binding proteins, and decreased coagulase activity associated with glycopeptide resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1997;41:1788–1793. doi: 10.1128/aac.41.8.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murakami K, Fujimura T, Doi M. Nucleotide sequence of the structural gene for the penicillin-binding protein 2 of Staphylococcus aureus and the presence of a homologous gene in other staphylococci. FEMS Microbiol Lett. 1994;117:131–136. doi: 10.1111/j.1574-6968.1994.tb06754.x. [DOI] [PubMed] [Google Scholar]
- 32.Nakamura M, Maruyama I N, Soma M, Kato J, Suzuki H, Horota Y. On the process of cellular division in Escherichia coli: nucleotide sequence of the gene for penicillin-binding protein 3. Mol Gen Genet. 1983;191:1–9. doi: 10.1007/BF00330881. [DOI] [PubMed] [Google Scholar]
- 33.Novick R. Properties of a cryptic high-frequency transducing phage in Staphylococcus aureus. Virology. 1967;33:155–166. doi: 10.1016/0042-6822(67)90105-5. [DOI] [PubMed] [Google Scholar]
- 34.Novick R P. Plasmid incompatibility. Microbiol Rev. 1987;51:381–395. doi: 10.1128/mr.51.4.381-395.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Patte P A. Genetic and physical map of Staphylococcus aureus NCTC8325. In: O’Brien S J, editor. Genetic maps. 6th ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1993. pp. 2.106–2.113. [Google Scholar]
- 36.Pinho M G, Ludovice A M, Wu S, De Lencastre H. Massive reduction in methicillin resistance by transposon inactivation of the normal PBP2 in a methicillin-reisistant strain of Staphylococcus aureus. Microb Drug Resist. 1997;3:409–413. doi: 10.1089/mdr.1997.3.409. [DOI] [PubMed] [Google Scholar]
- 37.Pucci M J, Thanassi J A, Discotto L F, Kessler R E, Dougherty T J. Identification and characterization of cell wall-cell division gene clusters in pathogenic gram-positive cocci. J Bacteriol. 1997;179:5632–5635. doi: 10.1128/jb.179.17.5632-5635.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Quandt J, Hynes M F. Versatile suicide vectors which allow direct selection for gene replacement in gram-negative bacteria. Gene. 1993;127:15–21. doi: 10.1016/0378-1119(93)90611-6. [DOI] [PubMed] [Google Scholar]
- 39.Reynolds P E. The essential nature of staphylococcal penicillin-binding proteins. In: Actor P, Daneo-Moore L, Higgins M L, Salton M R J, Shockman G D, editors. Antibiotic inhibition of bacterial cell surface assembly and function. Washington, D.C: American Society for Microbiology; 1988. pp. 343–351. [Google Scholar]
- 40.Suzuki H, Nishimura Y, Hirota Y. On the process of cellular division in Escherichia coli: a series of mutants of E. coli altered in the penicillin-binding proteins. Proc Natl Acad Sci USA. 1978;75:664–668. doi: 10.1073/pnas.75.2.664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Utsui Y, Yokota T. Role of an altered penicillin-binding protein in methicillin- and cephem-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1985;28:397–403. doi: 10.1128/aac.28.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wada A, Ohta H, Kulthanan K, Hiramatsu K. Molecular cloning and mapping of 16S-23S rRNA gene complexes of Staphylococcus aureus. J Bacteriol. 1993;175:7483–7487. doi: 10.1128/jb.175.22.7483-7487.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wada, A., and H. Watanabe. GenBank accession number U20869.
- 44.Weiss D S, Pogliano K, Carson M, Guzman L M, Fraipont C, Nguyen-Disteche M, Losick R, Beckwith J. Localization of the Escherichia coli cell division protein FtsI (PBP3) to the division site and cell pole. Mol Microbiol. 1997;25:671–681. doi: 10.1046/j.1365-2958.1997.5041869.x. [DOI] [PubMed] [Google Scholar]
- 45.Yanouri A, Daniel R A, Errington J, Buchanan C E. Cloning and sequencing of the cell division gene pbpB, which encodes penicillin-binding protein 2B in Bacillus subtilis. J Bacteriol. 1993;175:7604–7616. doi: 10.1128/jb.175.23.7604-7616.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]