Abstract
Alginate is essential for encystment in Azotobacter vinelandii. Transcription of the algD gene, which codes for GDP-mannose dehydrogenase, a key enzyme in the alginate biosynthetic pathway, is initiated at two promoters, one of which, p2, has ςE consensus sequences. AlgU is the A. vinelandii alternative ςE factor. In this study, we constructed an algU mutant (SMU88) which, as expected, is impaired in alginate production, encystment, and transcription of the algD gene from the p2 promoter. Plasmid pJMSAT1, carrying the A. vinelandii algU gene, restored alginate production and encystment to SMU88 and to strain UW136, a naturally occurring algU mutant. Plasmid pSMU865, carrying the A. vinelandii mucABCD genes coding for negative regulators of AlgU activity and previously shown to diminish alginate production in the wild-type strain, ATCC 9046, was shown here to impair encystment and transcription of the algD gene from the p2 algU-dependent promoter. Since nonencysting strain ATCC 9046/pSMU865 produced more alginate than some encysting strains, such as UW136/pJMSAT1, we propose an AlgU role in encystment, independent of the structural role that alginate plays in mature cysts.
Encystment in Azotobacter vinelandii has been studied at the morphological and biochemical levels (9, 10, 22, 27). This differentiation process is induced by growth on n-butanol as the sole carbon source (12); mature cysts develop in 3 to 5 days and are characterized by a central body surrounded by an intine and a thick, laminated exine. Alginate is essential for encystment since it is a component of intine and exine layers (1, 3). The alginate biosynthetic pathways of A. vinelandii and Pseudomonas aeruginosa are very similar (21). The molecular genetics of alginate biosynthesis have been widely studied in P. aeruginosa (for reviews, see references 5 and 17), where the genes encoding the alginate biosynthetic enzymes are organized in an operon starting with algD (4), encoding a GDP-mannose dehydrogenase, which converts GDP-mannose to GDP-mannuronic acid, the substrate for alginate polymerization. The algU mucABCD cluster controls alginate production. AlgU (also known as AlgT) is a ςE homolog (8); it is required for transcription of algD and its own gene, algU (15, 25). The mucA and mucB genes code for negative regulators of AlgU activity; MucA interacts with AlgU as an anti-sigma factor, inhibiting its activity (6, 24, 25, 28).
The A. vinelandii alginate biosynthetic gene cluster is organized in at least three operons, one of which includes the algD gene (3, 13, 18). In the highly mucoid strain A. vinelandii ATCC 9046, algD is transcribed from two promoters: a putative AlgU dependent promoter, p2, and a promoter (p1) showing consensus sequences for the vegetative sigma factor ςD (3). We identified and sequenced the algU and mucABCD genes of A. vinelandii and presented evidence showing that they control alginate biosynthesis in a manner similar to that of the P. aeruginosa homologs (16).
An A. vinelandii strain unable to produce alginate due to a mutation in the algD gene is unable to encyst (3). The naturally occurring A. vinelandii strain UW136 does not produce alginate, due to a mutation in the algU gene, which codes for the AlgU sigma factor (16), and is also unable to encyst (20). The question of whether algU mutations affect encystment exclusively via its effect on alginate biosynthesis or whether AlgU activity is also required for expression of other genes involved in cyst formation was raised. In the present work we present evidence suggesting that AlgU is involved in the formation of mature cysts, independently of its role in alginate production.
Construction and characterization of an ATCC 9046 algU mutant.
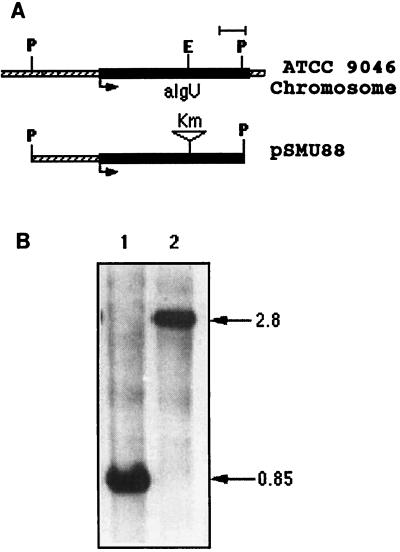
To study the role of AlgU in encystment, we constructed an ATCC 9046 derivative carrying an algU::Km null mutation. A PstI DNA fragment of 828 nucleotides corresponding to algU from A. vinelandii ATCC 9046 (Fig. 1) was cloned into plasmid pBluescript II KS to give pSMU85. A 2-kb SmaI fragment containing a kanamycin resistance gene from plasmid pHP45Ω-Km (7) was inserted into the unique EcoRV site present within the 828-bp fragment of pMSU85 to create an algU::Km mutation between the codons for amino acid residues 115 and 116 of AlgU. The resultant plasmid, pSMU88, which is unable to replicate in A. vinelandii, was introduced into strain ATCC 9046 by transformation. One of the kanamycin-resistant transformants, strain SMU88, was chosen for further analysis. The replacement of the intact algU gene with the algU::Km mutation on the chromosome of the SMU88 mutant was confirmed by Southern blotting (Fig. 1). The algU mutation impaired alginate production and encystment (Table 1). To complement the algU mutation, a 1.6-kb DNA fragment with the algU-mucA region from ATCC 9046, obtained by PCR, was cloned into plasmid pCRII, which confers ampicillin resistance (2). The resultant plasmid, pJMSAT1, which is unable to replicate in A. vinelandii, was transformed into algU mutant strain SMU88. A transformant, SMU88::pJMSAT1, carrying plasmid pJMSAT1 integrated into the chromosome, was selected as a mucoid colony resistant to carbenicillin. Integration of the plasmid was confirmed by Southern blot analysis (data not shown). Strain SMU88::pJMSAT1 was able to encyst (Table 1). We also show that plasmid pDMUM13 carrying the P. aeruginosa algU gene (14), previously shown to restore alginate production to strain UW136 (16), also restored encystment and alginate production to strains SMU88 and UW136 (Table 1). Although the amounts of alginate synthesized by strains SMU88, UW136 complemented with plasmid pDMUM13, and UW136::pJMSAT1 were significantly lower than that of ATCC 9046, they were sufficient for encystment (Table 1).
FIG. 1.
Construction of strain SMU88. (A) Schematic representation of the A. vinelandii algU region and the algU::Km mutation in plasmid pSMU88. Bar, 100 bp. (B) Southern blot hybridization of total genomic DNA digested with PstI endonuclease, with pSMU85 as a probe. Lanes: 1, ATCC 9046; 2, SMU88. Molecular sizes (in kilobases) are indicated on the right.
TABLE 1.
Alginate production and encystmenta
| Strain | Alginate (mg/mg of protein) | Resistance to desiccation (%) |
|---|---|---|
| ATCC 9046 | 2.5 ± 0.50 | 7.2 ± 1.5 |
| ATCC 9046/pSMU865 | 0.46 ± 0.04 | <0.0001 |
| SMU88 | 0.003 ± 0.001 | <0.0001 |
| SMU88::pJMSAT1 | 2.74 ± 0.7 | 7.1 ± 2.5 |
| SMU88/pDMUM13 | 0.170 ± 0.04 | 1.9 ± 0.5 |
| UW136 | 0.004 ± 0.002 | <0.0001 |
| UW136::pJMSAT1 | 0.19 ± 0.02 | 8.8 ± 2.5 |
| UW136/pDMUM13 | 0.16 ± 0.03 | 4.0 ± 1.0 |
Alginate production was determined in cells induced for encystment. Cells grown during 5 days on BS plates with 0.2% n-butanol as the sole carbon source were used to measure alginate production, as described previously (19). Encystment was determined as previously described (3). All values are means of three determinations (± standard deviations, where appropriate).
Effects of plasmid pSMU865 and the algU::Km mutation on transcription from the algD p1 and p2 promoters.
Plasmid pSMU865, carrying the A. vinelandii mucABCD genes, was previously shown to reduce alginate production in strain ATCC 9046 (16). We propose that the effect of pSMU865 on alginate production in ATCC 9046 is due to the negative effect of the mucA and mucB gene products on AlgU activity and therefore on transcription from algDp2. Thus, transcription of the algD gene in strains ATCC 9046/pSMU865 and SMU88 should initiate predominantly from p1, the AlgU-independent promoter. Primer extension experiments were performed on RNA isolated from strains SMU88 and ATCC 9046 in the presence and absence of plasmid pSMU865 and grown in liquid BS medium (11) for 48 h at 30°C. Reactions were performed with a primer extension system (Boehringer), as instructed by the manufacturer. Oligonucleotide primers used were labeled with [γ-32P]dATP (Amersham) at the 5′ end by using polynucleotide kinase and were hybridized to 50 μg of total RNA. After extension with reverse transcriptase, cDNA products were examined by electrophoresis in an 8% polyacrylamide gel. To map transcriptional start points, sequencing reactions were performed on pMSD27 (16) DNA by the dideoxy chain termination method (23) with [γ-32P]dATP and a sequencing kit with the same primers as used for the primer extension reactions.
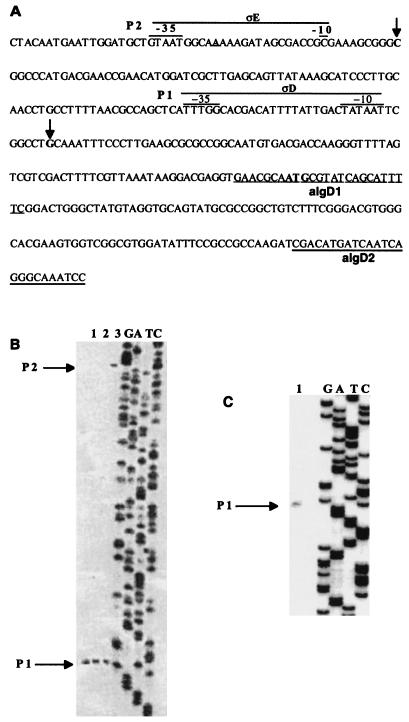
As expected, primer extension products corresponding to p1 and p2 transcripts were observed with ATCC 9046 RNA, but only the primer extension products corresponding to p1 transcripts were observed with RNA from strains SMU88 and ATCC 9046/pSMU865, confirming that p2 is an AlgU-dependent promoter (Fig. 2).
FIG. 2.
Primer extension analysis of algD transcription in strains SMU88 and ATCC 9046, with and without plasmid pSMU865. (A) DNA sequence of the 5′ end of algD. The arrows indicate the start sites of algD transcription, the p1 and p2 promoters are indicated (overbar), and the complementary sequences where oligonucleotides algD1 and algD2 (used for primer extension analysis) were generated are underlined. The algD ATG initiation codon is shown in boldface type. (B and C) Primer extension of the algD gene with oligonucleotide algD1 in strains SMU88 (lane 1) and ATCC 9046 with (lane 2) and without (lane 3) plasmid pSMU865 (B) and with oligonucleotide algD2 in strain SMU88 (C). The algD sequence ladders (GATC) were produced with the oligonucleotides used for primer extension.
In P. aeruginosa, algU mutants are unable to produce alginate, since transcription of the biosynthetic operon starting with algD, as well as some regulatory genes such as algR, is dependent on the presence of an active AlgU sigma factor (15, 26). Similarly, A. vinelandii algU mutants UW136 and SMU88 are also unable to produce alginate; in contrast to P. aeruginosa, however, in A. vinelandii transcription of all other known alginate biosynthetic genes, such as algA and alg8, is not dependent on AlgU (13, 18). As shown here, AlgU is absolutely required for transcription from algDp2, but algD is still transcribed from algDp1 in strain ATCC 9046. Transcription from algDp1 was also detected in strain SMU88, with an oligonucleotide corresponding to the nucleotides coding for amino acids 37 to 44 of AlgD (Fig. 2), indicating that transcription from p1 extends into the algD structural gene. Thus, the fact that an algU mutation abolished alginate production in this strain indicates that in A. vinelandii, transcription of other unidentified regulatory or biosynthetic alginate genes depends on AlgU.
Effect of plasmid pSMU865 on encystment.
We studied encystment in strain ATCC 9046/pSMU865, which produces alginate even though its AlgU activity is not sufficient to initiate transcription from algDp2. Table 1 shows that this strain was unable to form desiccation-resistant cysts, despite the fact that under encysting conditions it produced more alginate than the encysting strains UW136::pJMSAT1, SMU88/pDMUM13, and UW136/pDMUM13. These data strongly suggest that AlgU plays an additional role in the expression of genes involved in cyst formation. The observation that an algU mutation, but not plasmid pSMU865, abrogated alginate production in strain ATCC 9046 leads us to hypothesize that in strain ATCC 9046/pSMU865, a low level of AlgU activity allows some transcription of the unidentified alginate genes mentioned above but not of algD (from p2) and encystment genes.
The putative AlgU requirement for transcription of genes involved in encystment may allow the identification and characterization of such genes.
Ultrastructure analysis of cyst formation.
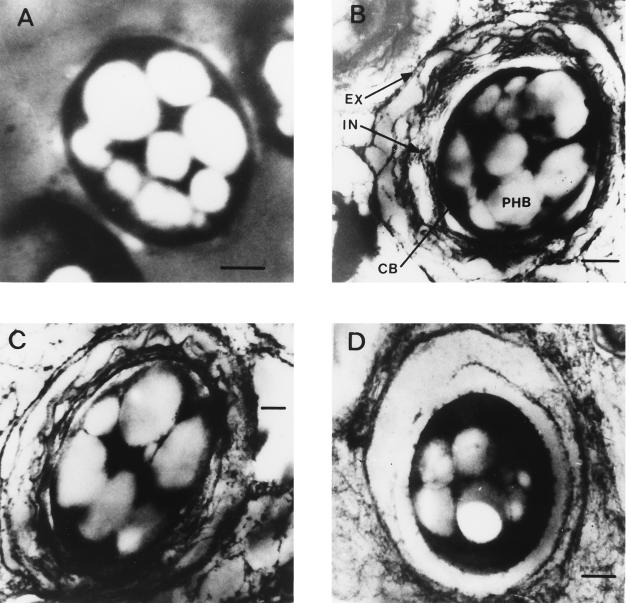
Electron microscopic examination of the cyst structures formed by the mutant strains (Fig. 3) showed that strain SMU88 lacks the intine and exine layers of mature cysts such as those produced by the wild-type strain, ATCC 9046. SMU88/pDMUM13 cysts were similar to those produced by wild-type strain ATCC 9046, consisting of the compacted cell (central body containing poly-β-hydroxybutyrate granules) surrounded by the intine capsule and the exine outer shell. The cyst structures formed by strain ATCC 9046/pSMU865 (Fig. 3D) appear to lack the intine layers. An early electron microscopy study of the development of A. vinelandii cysts (27) revealed that the exine appears in 36 to 48 h, after which the exine thickens and the intine is formed between the exine and the central body; thus, at 36 h the intine appears to be nothing but an empty area. This seems to be the stage at which encystment development is blocked in the absence of AlgU activity.
FIG. 3.
Electron micrographs were produced as described previously (19). Thin sections of A. vinelandii cysts formed by strains SMU88 (A), SMU88/pDMUM13 (B), ATCC 9046 (C), and ATCC 9046/pSMU865 (D) are shown. Abbreviations: EX, exine; IN, intine; CB, central body; PHB, poly-β-hydroxybutyrate. Bars, 0.4 μm.
This study contributes to our understanding of the A. vinelandii differentiation process leading to cyst formation at the molecular level and is the first example of the involvement of an alternative sigma factor in cellular differentiation of a gram-negative bacterium.
Acknowledgments
This work was supported by grant IN212096 from DGAPA UNAM.
We thank L. Servin, F. Bastarrachea, and S. Silver for reviewing the manuscript.
REFERENCES
- 1.Brivonese A C, Sutherland I W. Polymer production by a mucoid strain of Azotobacter vinelandii in batch culture. Appl Microbiol Biotechnol. 1989;30:97–102. [Google Scholar]
- 2.Brosius J. Superpolylinkers in cloning and expression vectors. DNA. 1989;8:759–777. doi: 10.1089/dna.1989.8.759. [DOI] [PubMed] [Google Scholar]
- 3.Campos M-E, Martínez-Salazar J M, Lloret L, Moreno S, Núñez C, Espín G, Soberón-Chávez G. Characterization of the gene coding for GDP-mannose dehydrogenase (algD) from Azotobacter vinelandii. J Bacteriol. 1996;178:1793–1799. doi: 10.1128/jb.178.7.1793-1799.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chitnis C E, Ohman D E. Genetic analysis of the alginate biosynthetic gene cluster of Pseudomonas aeruginosa shows evidence of an operonic structure. Mol Microbiol. 1993;8:583–590. doi: 10.1111/j.1365-2958.1993.tb01602.x. [DOI] [PubMed] [Google Scholar]
- 5.Deretic V, Martin D W, Schurr M J, Mudd M H, Hibler N S, Curcic R, Boucher J C. Regulation of mucoidy in Pseudomonas aeruginosa. Bio/Technology. 1993;11:1133–1136. doi: 10.1038/nbt1093-1133. [DOI] [PubMed] [Google Scholar]
- 6.Deretic V, Schurr M J, Boucher J C, Martin D W. Conversion of Pseudomonas aeruginosa to mucoidy in cystic fibrosis: environmental stress and regulation of bacterial virulence by alternative sigma factors. J Bacteriol. 1994;176:2773–2780. doi: 10.1128/jb.176.10.2773-2780.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fellay R, Frey J, Krisch H. Interposon mutagenesis of soil and water bacteria: a family of DNA fragments designed for in vitro insertional mutagenesis of Gram-negative bacteria. Gene. 1987;52:147–154. doi: 10.1016/0378-1119(87)90041-2. [DOI] [PubMed] [Google Scholar]
- 8.Hershberger C D, Ye R W, Parsek M R, Xie Z D, Chakrabarty A M. The algT (algU) gene of Pseudomonas aeruginosa, a key regulator involved in alginate biosynthesis, encodes an alternative ς factor (ςE) Proc Natl Acad Sci USA. 1995;92:7941–7945. doi: 10.1073/pnas.92.17.7941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hitchins V M, Sadoff H L. Morphogenesis of cysts in Azotobacter vinelandii. J Bacteriol. 1970;104:492–498. doi: 10.1128/jb.104.1.492-498.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hitchins V M, Sadoff H L. Sequential metabolic events during encystment of Azotobacter vinelandii. J Bacteriol. 1973;113:1273–1279. doi: 10.1128/jb.113.3.1273-1279.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kennedy C, Gamal R, Hummphrey R, Ramos J, Brigle K, Dean D. The nifH, nifM and nifN genes of Azotobacter vinelandii: characterization by Tn5 mutagenesis and isolation from pLARF1 gene banks. Mol Gen Genet. 1986;205:318–325. [Google Scholar]
- 12.Lin L P, Sadoff H L. Encystment and polymer production by Azotobacter vinelandii in the presence of β-hydroxybutyrate. J Bacteriol. 1968;98:1335–1341. doi: 10.1128/jb.95.6.2336-2343.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lloret L, Barreto R, León R, Moreno S, Martínez-Salazar J, Espín G, Soberón-Chávez G. Genetic analysis of the transcriptional arrangement of Azotobacter vinelandii alginate biosynthetic genes: identification of two independent promoters. Mol Microbiol. 1996;21:449–457. doi: 10.1111/j.1365-2958.1996.tb02554.x. [DOI] [PubMed] [Google Scholar]
- 14.Martin D W, Holloway D W, Deretic V. Characterization of a locus determining the mucoid status of Pseudomonas aeruginosa: AlgU shows sequence similarities with a Bacillus sigma factor. J Bacteriol. 1993;175:1153–1164. doi: 10.1128/jb.175.4.1153-1164.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin D W, Schurr M J, Yu H, Deretic V. Analysis of promoters controlled by the putative sigma factor AlgU regulating conversion to mucoidy in Pseudomonas aeruginosa: relationship to ςE and stress response. J Bacteriol. 1994;176:6688–6696. doi: 10.1128/jb.176.21.6688-6696.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Martínez-Salazar J M, Moreno S, Nájera R, Boucher J C, Espín G, Soberón-Chávez G, Deretic V. Characterization of the genes coding for the putative sigma factor AlgU and its negative regulators MucA, MucB, MucC, and MucD in Azotobacter vinelandii and evaluation of their roles in alginate biosynthesis. J Bacteriol. 1996;178:1800–1808. doi: 10.1128/jb.178.7.1800-1808.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.May T, Chakrabarty A M. Pseudomonas aeruginosa: genes and enzymes of alginate synthesis. Trends Microbiol. 1994;2:151–157. doi: 10.1016/0966-842x(94)90664-5. [DOI] [PubMed] [Google Scholar]
- 18.Mejía-Ruíz H, Guzmán J, Moreno S, Soberón-Chávez G, Espín G. The Azotobacter vinelandii alg8 and alg44 genes are essential for alginate synthesis and can be transcribed from an algD-independent promoter. Gene. 1997;199:271–277. doi: 10.1016/s0378-1119(97)00380-6. [DOI] [PubMed] [Google Scholar]
- 19.Mejía-Ruíz H, Moreno S, Guzmán J, Nájera R, León R, Soberón-Chávez G, Espín G. Isolation and characterization of an Azotobacter vinelandii algK mutant. FEMS Microbiol Lett. 1997;156:101–106. doi: 10.1111/j.1574-6968.1997.tb12712.x. [DOI] [PubMed] [Google Scholar]
- 20.Page W J. Formation of cyst-like structures by iron-limited Azotobacter vinelandii strain UW during prolonged storage. Can J Microbiol. 1983;29:1110–1118. [Google Scholar]
- 21.Pindar D F, Bucke C. The biosynthesis of alginic acid by Azotobacter vinelandii. Biochem J. 1975;152:617–622. doi: 10.1042/bj1520617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sadoff H L. Encystment and germination in Azotobacter vinelandii. Bacteriol Rev. 1975;39:516–539. doi: 10.1128/br.39.4.516-539.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schurr M J, Martin D W, Mudd M H, Deretic V. Gene cluster controlling conversion to alginate-overproducing phenotype in Pseudomonas aeruginosa: functional analysis in a heterologous host and role in the instability of mucoidy. J Bacteriol. 1994;176:3375–3382. doi: 10.1128/jb.176.11.3375-3382.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schurr M J, Yu H, Martínez-Salazar J M, Boucher J C, Deretic V. Control of AlgU, a member of the ςE-like family of stress sigma factors, by the negative regulators MucA and MucB and Pseudomonas aeruginosa conversion to mucoidy in cystic fibrosis. J Bacteriol. 1996;178:4997–5004. doi: 10.1128/jb.178.16.4997-5004.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wozniak D J, Ohman D E. Transcriptional analysis of the Pseudomonas aeruginosa genes algR, algB, and algD reveals a hierarchy of alginate gene expression which is modulated by algT. J Bacteriol. 1994;176:6007–6014. doi: 10.1128/jb.176.19.6007-6014.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wyss O, Newmann M G, Socolofsky M D. Development and germination of the Azotobacter cyst. J Biophys Biochem Cytol. 1961;10:555–565. doi: 10.1083/jcb.10.4.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xie Z-D, Hershberger C D, Shankar S, Ye R W, Chakrabarty A M. Sigma factor–anti-sigma factor interaction in alginate synthesis: inhibition of AlgT by MucA. J Bacteriol. 1996;178:4990–4996. doi: 10.1128/jb.178.16.4990-4996.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]