Abstract
Purpose:
Dysregulation of viral-like repeat RNAs are a common feature across many malignancies that are linked with immunological response, but the characterization of these in hepatocellular carcinoma (HCC) is understudied. In this study, we performed RNA in situ hybridization (RNA-ISH) of different repeat RNAs, immunohistochemistry (IHC) for immune cell subpopulations, and spatial transcriptomics to understand the relationship of HCC repeat expression, immune response, and clinical outcomes.
Experimental Design:
RNA-ISH for LINE1, HERV-K, HERV-H, and HSATII repeats and IHC for T-cell, Treg, B-cell, macrophage, and immune checkpoint markers were performed on 43 resected HCC specimens. Spatial transcriptomics on tumor and vessel regions of interest was performed on 28 specimens from the same cohort.
Results:
High HERV-K and high LINE1 expression were both associated with worse overall survival. There was a positive correlation between LINE1 expression and FOXP3 T-regulatory cells (r = 0.51 p < 0.001) as well as expression of the TIM3 immune checkpoint (r = 0.34, p = 0.03). Spatial transcriptomic profiling of HERV-K high and LINE-1 high tumors identified elevated expression of multiple genes previously associated with epithelial mesenchymal transition, cellular proliferation, and worse overall prognosis in HCC including SSX1, MAGEC2, and SPINK1.
Conclusion:
Repeat RNAs may serve as useful prognostic biomarkers in HCC and may also serve as novel therapeutic targets. Additional study is needed to understand the mechanisms by which repeat RNAs impact HCC tumorigenesis.
INTRODUCTION
The majority of the human genome is composed of non-coding repeat elements (1, 2) that are normally silenced in adult differentiated tissues, but are de-repressed in the setting of carcinogenesis (3–5). The expression patterns of repeat species, and association with immune cell infiltrates, vary significantly across different cancer types (5–7). These relationships appear to be linked to differences in repeat RNA sequence (8) and secondary structure (9), which both contribute to the activation of innate immune anti-viral responses (9–17). More recently, human endogenous retrovirus (HERV) repeats have been associated with response to immune checkpoint inhibitors (7, 18–20). However, satellite repeats appear to be associated with poor immunological response to tumors and with more invasive phenotypes (7, 21). Altogether, the diverse and expansive repertoire of repeat RNA species in the human genome are linked with differences in interactions with the immune microenvironment.
In this study, we sought to understand the relationship of repeat RNA expression and the immune response in hepatocellular carcinoma (HCC). Given the known viral drivers of HCC via chronic hepatitis B and C viral (HBV and HCV) infection, we hypothesized that endogenous repeat viral-like elements are important in carcinogenesis and the immunological response in HCC. Prior work has demonstrated the worsened prognosis of HCC patients with hypomethylation of the LINE1 repeat (22–25) and evidence of retrotransposition events in viral and non-viral HCC (26). Further, there is mounting evidence of the interplay between HBV and HCV driven HCC oncogenesis with LINE1 (27, 28). Here, we have developed and optimized RNA in situ hybridization (RNA-ISH) for different repeat RNA species for use in formalin-fixed paraffin-embedded tissue and quantified the expression of these viral-like elements in a cohort of resected primary HCC tumors. We found differences in the expression levels of each of these repeat species and correlation with tumor immune infiltrates. Finally, we utilized spatial transcriptomics to define cancer cell and endothelial cell expression patterns linked with repeat expression profiles.
METHODS
Tissue Procurement and Annotation
Patient tumor materials were obtained under Massachusetts General Hospital IRB protocol 2011P001236 and Dana-Farber Harvard Cancer Center IRB protocol 02-240. Archived FFPE samples in tissue microarray (TMA) format from 43 patients who underwent surgical resection or liver transplantation for the treatment of hepatocellular carcinoma between March 2004 and December 2015 were obtained. Clinical and pathologic data was obtained through review of the Massachusetts General Hospital electronic medical record.
RNA In Situ Hybridization (RNA-ISH)
Detection of each of the repeat RNA levels was performed with an automated RNA-ISH assay on the Leica Biosystems BondRx 6.0 auto-stainer platform using the Affymetrix ViewRNA platform. The ViewRNA eZL Detection Kit (Affymetrix) was used on the Bond RX immunohistochemistry and ISH Staining System with BDZ 6.0 software (Leica Biosystems). The Bond RX user-selectable settings for part 2 were as follows: VjewRNA eZ-L Detectjon 1-plex (Red) protocol; ViewRNA Dewax1 Preparation protocol; ViewRNA Enzyme 2 (10); ViewRNA Probe Hybridization 3hrs. With these settings, the RNA unmasking conditions for the FFPE tissue consisted of 10-minute incubation with Proteinase K from the Bond Enzyme Pretreatment Kit at 1:1000 dilution (Leica Biosystems). HERV-K (Cat# DVF1-19321), HSATII (Cat# VA1-10874), LINE1 (Cat# DVA1-19767), and HERV-H (Cat # DVF1-19702) Ez probes were diluted as 1:40 in ViewRNA Probe Diluent (Affymetrix). Post run, slides were rinsed with water, air dried for 30 minutes at room temperature and mounted using Dako Ultramount (Dako, Carpinteria,CA).
Immunohistochemistry
Immunohistochemistry (IHC) was performed on the automated Leica Biosystems BondRx auto-stainer. The antibody clones and conditions are as listed below.
| Marker | Company | Catalog Information | Dilution used | ER Conditions |
|---|---|---|---|---|
| CD3 | Leica Novacastra | Cat #NCL-L-CD3-565 | RTU | 20min ER2 95C |
| CD4 | Leica Novacastra | Cat #NCL-L-CD4-368 | 1:100 | 20min ER2 95C |
| CD8 | Leica Novacastra | Cat #NCL-L-CD8-4B11 | 1:500 | 15-20min ER2 95C |
| FOXP3 | Invitrogen (Thermofisher) | Cat #14-4774-80 | 1:100 | 20min ER2 95C |
| CD20 | Leica Novacastra | Cat #NCL-L-CD20-L26 | 1:100 | 20min ER1 95C |
| CD163 | Leica Novacastra | Cat #NCL-L-CD163 | 1:400 | 20min ER1 95C |
| LAG3 (D2G40) | Cell Signaling | Cat #15372 | 1:200 | 20min ER1 95C |
| TIM3 (D5D5R) | Cell Signaling | Cat #45208 | 1:400 | 20min ER2 95C |
| PD1 (EH33) | Cell Signaling | Cat #43248 | 1:200 | 20min ER2 95C |
| PDL1 (E1L3N) | Cell Signaling | Cat #13684 | 1:200-1:400 | 20min ER2/ER1 95C |
RNA-ISH and IHC Quantification
The RNA-ISH slides were imaged with the Motic EasyScan Infinity Digital Pathology Scanner at 40x magnification. RNA quantification was performed with the Halo Image Analysis Platform by Indica Labs. Individual tissue areas on the TMA corresponding to each patient were annotated. In each tissue area, cellular segmentation was performed by detection of hematoxylin-stained nuclei and the RNA-ISH probe was detected by red chromogen. For HERV-K, HERV-H, and HSATII, the average number of probe copies per cell in the tissue area was quantified. For LINE1, due to the density of stain in the samples with the highest expression, accurate detection of individual red copies was challenging. LINE1 expression was therefore quantified as the probe detected per μm2 tissue area.
The IHC slides were imaged with the Leica Aperio CS-O Digital Pathology Slide Scanner at 40x magnification. Cells were quantified by annotation by an anatomic pathologist (AN) and quantification was performed using the Halo platform (Indica labs) by detecting the chromogen positive area per μm2 of each tissue area on the TMA.
Spatial Transcriptomics Data Generation and Analysis
The GeoMx™ Digital Spatial Profiler (DSP) platform (NanoString Technologies, Seattle, WA) was used to characterize the transcriptional profile of each tumor. Serial sections from the FFPE TMA blocks were prepared. Each slide was stained with photocleavable oligonucleotide probes from the GeoMx™ Cancer Transcriptome Atlas, which contains probes against 18,676 unique mRNAs. The slides were then stained with fluorescent antibodies against arginase (Anti-Arginase 1/ARG1/liver Arginase Antibody [Alexa Fluor 532], Novus NBP1-32731AF532) and CD31 (Anti-CD31 antibody [EPR3094], Abcam (ab76533) conjugated using Alexa Fluor® 647 Conjugation Kit - Lightning-Link from Abcam) to allow for segmentation of tumor cells and endothelial cells, respectively. After staining, 300μm regions of interest (ROIs) were selected. A photomask was applied based on the different emission spectra of the fluorescent markers, allowing the instrument to segment the tissue into distinct tumor and endothelial area of interest (AOIs). Ultraviolet light was then applied to cleave and collect the oligonucleotides from each AOI for sequencing on the Illumina NextSeq 500/550.
The sequencing data was quantile normalized for analysis. Differential expression analysis was performed comparing the differences in transcriptional profile of repeat-high and repeat-low tumors defined as the upper tercile and lower tercile of repeat RNA expression quantified by RNA-ISH. We obtained cell type-enriched spatial transcriptomes for cancer and endothelial cells separately. For each cell type, we performed differential gene expression analyses on the transcriptomes from repeat-high versus repeat-low individuals using Wilcoxon rank-sum test. To determine differentially expressed genes, we used a log fold change threshold of 0.5 and a p-value threshold of 0.05 for FDR-adjusted p-values. Results are visualized in volcano plots and heatmaps.
Statistical Analysis
Survival analyses using Kaplan Meier analysis with log-rank test for significance, and correlation analysis between repeat RNA expression and immune markers performed using Pearson’s correlation were performed with Stata Statistical Software 17.0. Two-sided FDR-adjusted p-values of < 0.05 were deemed statistically significant.
Data Availability Statement
All images and primary data will be made available upon request. Spatial transcriptomic expression matrices will be available upon request.
Code Availability Statement
All code and statistical packages are detailed in the methods above and will be made available upon request. All software for RNA expression and digital image data analysis is described in the methods above and all software will be provided upon request.
RESULTS
Patient characteristics
Characteristics of the patient population are summarized in Table 1. The median age of patients at the time of operation was 59 years old. The majority of patients (79%) had underlying cirrhosis with the most common etiology being chronic HCV infection (44%) followed by chronic HBV infection (14%). Of the 43 patients in the cohort, 33 underwent surgical resection and 10 underwent liver transplantation. The median survival after surgery was 4.2 years (95% CI 2.0 – 7.9 years) and the 5-year survival of the cohort was 46.5%.
Table 1.
Characteristics of study population
| Sex, number (%) | |
| Male | 28 (65%) |
| Female | 15 (35%) |
|
| |
| Age, years | |
| Median | 59.4 |
| Range | 38.6 – 86.6 |
|
| |
| Cirrhosis, number (%) | |
| Absent | 9 (21%) |
| Present | 34 (79%) |
| Ishak Grade | |
| 2 | 3 (7%) |
| 3 | 2 (5%) |
| 4 | 2 (5%) |
| 5 | 8 (19%) |
| 6 | 15 (35%) |
|
| |
| Etiology of Cirrhosis | |
| HCV | 19 (44%) |
| HBV | 6 (14%) |
| Alcohol | 3 (7%) |
| NASH | 3 (7%) |
| Other | 3 (7%) |
|
| |
| Surgery | |
| Resection | 33 (77%) |
| Transplant | 10 (23%) |
|
| |
| Tumor Size, cm | |
| Median | 4.6 |
| Range | 1.2 – 15 |
|
| |
| Differentiation, number (%) | |
| Well | 7 (16%) |
| Moderate-Well | 1 (2%) |
| Moderate | 28 (65%) |
| Moderate-Poor | 3 (7%) |
| Poor | 3 (7%) |
|
| |
| Serum AFP, ng/mL | |
| Median | 45.8 |
| Range | 2 – 37510 |
Repeat RNA expression in hepatocellular carcinoma
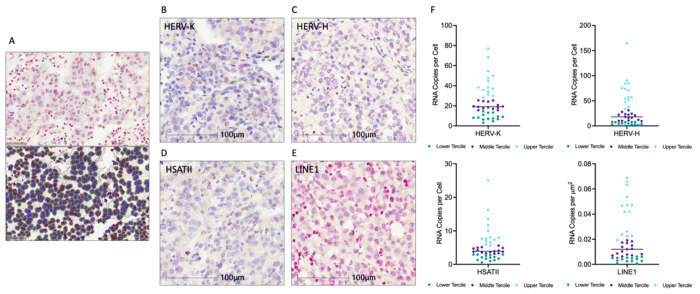
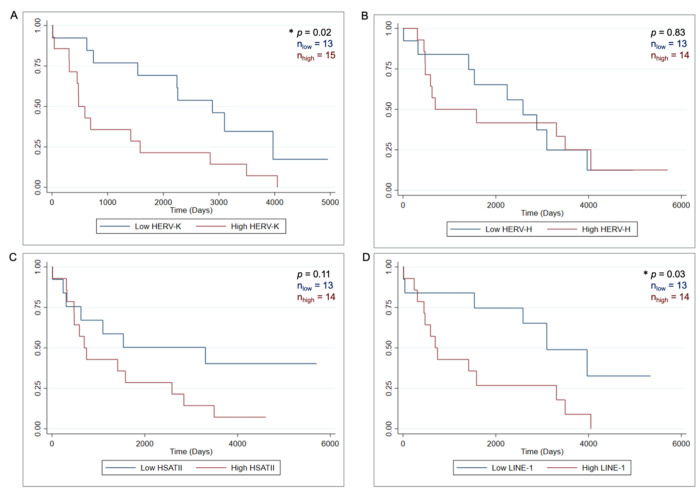
Repeat RNA expression was evaluated using RNA-ISH across our HCC cohort samples. Digital imaging followed by cellular segmentation and RNA-ISH signal quantification using the HALO AI platform (Fig. 1A) was used to quantify expression of HERV-K, HERV-H, LINE1 and HSATII in the HCC tumor tissue as shown in (Fig. 1B–E). Patient samples were divided into terciles based on the expression of each repeat RNA element. Patients with expression in the upper tercile were designated as “high” and samples in the lower tercile were designated as “low” (Fig. 1F). The relationship between expression of each parameter and overall survival (OS) was assessed using Kaplan Meier analysis and log-rank test to determine significance (Fig. 2). High HERV-K expression was associated with worsened overall survival than low HERV-K expression (median OS 1.31 vs. 7.90 years, p = 0.02). Similarly, high LINE1 expression was also associated with worsened overall survival (median OS 1.90 vs. 8.49 years, p = 0.03). There were no significant differences in survival between patients with HERV-H and HSATII high and low tumors. Altogether, these findings indicate that there are repeat expression profiles in HCC with higher HERV-K and LINE1 expression being associated with poor prognosis.
Figure 1. Repeat RNA Expression Quantification in Hepatocellular Carcinoma.
A. RNA-ISH stained slide and corresponding digital cell segmentation for ISH quantification on Halo AI platform. B-E. Representative images of RNA-ISH for HERV-K, HERV-H, HSATII, and LINE1. F. Dot plots of repeat RNA expression, n = 43, median and terciles shown.
Figure 2. Kaplan-Meier survival curves of HCC stratified by repeat-expression level.
A. HERV-K low (n = 13) vs HERV-K high (n = 15) B. HERV-H low (n = 13) vs HERV-H high (n = 14) C. HSATII low (n = 13) vs HSATII high (n = 14) D. LINE1 low (n = 13) vs LINE1 high. P value is log-rank test.
Distinct immune infiltrates associated with different repeat expression profiles
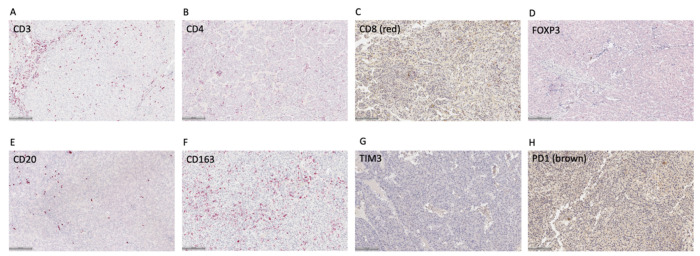
Given prior work demonstrating different immune infiltrates are associated with certain repeat elements, we evaluated for a panel of immune cell markers in our cohort of HCC samples using IHC. This included markers for T cells (CD3, CD4, CD8), regulatory T cells (Tregs; FOXP3), B cells (CD20), and macrophages (CD163). We also evaluated for immune checkpoint molecules including LAG3, TIM3, PD1, and PDL1. We quantified total immune cells expressing each marker over a tumor area defined by gastrointestinal pathologists (AC, VD). Representative IHC images for each of these markers are shown in Fig. 3A–H.
Figure 3. Representative images of immune cell and immune checkpoint immunohistochemistry in HCC samples.
Scale bar = 100 μm
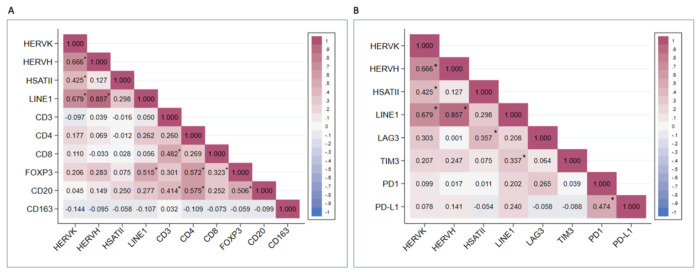
The relationship between expression of each repeat species and the tumor immune infiltrate was assessed using Pearson’s correlation (Fig. 4). We found that the expression of several of the repeat RNA species were correlated (Supplemental Fig. 1). HERV-K expression positively correlated with HERV-H (r = 0.67 p < 0.001), HSATII (r = 0.43, p = 0.005), and LINE1 (r =0.68, p < 0.001). There was also a strong positive correlation between LINE1 and HERV-H expression (r = 0.86, p < 0.001). There was no statistically significant correlation between HSATII and either LINE1 (r = 0.30, p = 0.05) or HERV-H (r = 0.13, p = 0.42). This distinct co-expression pattern of HSATII is consistent with our prior work in colorectal, pancreatic, and ovarian cancer (21). When assessing the relationship between repeat RNA expression and the immune cell infiltrate, there was a positive correlation between LINE1 expression and FOXP3 T-regulatory cells (r = 0.51 p < 0.001). LINE1 also had weakly positive correlations between CD4 (r = 0.26, p = 0.09) and CD20 (r = 0.28, p = 0.08), however these were not statistically significant. HERV-K and HERV-H both had weakly positive correlations with FOXP3, but these were not statistically significant (r = 0.21, p = 0.20 and r = 0.28, p = 0.07 respectively). CD163 and CD8 had minimal correlation with any of the RNA repeats. With regard to immune checkpoint protein expression, HSATII expression positively correlated with LAG3 (r = 0.36, p = 0.02), and LINE1 expression positively correlated with TIM3 (r = 0.34, p = 0.03). LINE1 also positively correlated with LAG3 (r = 0.21; p =0.20), PD1 (r = 0.20; p = .21), and PDL1 (r = 0.24; p = 0.14) however these were not statistically significant relationships. There were no statistically significant relationships between HERV-K or HERV-H and the immune checkpoints.
Figure 4. Correlation matrices of repeat RNAs, immune cells, and immune checkpoints.
A. Pearson’s correlations of repeat expression and T-cells (CD3, CD4, CD8), T-regulatory cells (FOXP3), B-cells (CD20), and macrophages (CD163). B. Pearson’s correlations of repeat expression and expression of immune checkpoints LAG3, TIM3, PD1, and PD-L1. * p < 0.05
Spatial transcriptomic analysis reveals differences in gene expression profiles of tumors and vessels in high repeat HCCs vs low repeat HCCs
To obtain a more comprehensive molecular analysis of repeat RNA patterns with coding genes, we performed spatial transcriptomics in tumor cell and vessel compartments of 28 HCC samples from the same TMAs. Spatial transcriptomic profiling of HERV-K high and LINE1 high tumors revealed several differentially expressed genes in the tumor and vessel compartments (Fig. 5A). In HERV-K high tumors (Fig. 5B–C) there was high expression of SSX1 and SPINK1, genes that have previously been independently associated with poor prognosis in multiple cancers (29, 30). In addition, CEBPA, IRAK1, MAP2K2, GP1, and CD63 were also highly expressed in HERV-K high tumors. In HERV-K low tumors there was significantly higher expression of EPCAM, a well-established epithelial gene, and SOCS2, a suppressor of cytokine-induced JAK/STAT signaling that has been implicated in the regulation of epithelial-mesenchymal transition in cancers (31, 32). In the vessels of HERV-K high tumors (Fig. 5D–E), we noted elevated expression of SSX1 and SPINK1, as well as TUBB, OAZ1, SREBF1, GPX1, CEBPA, SOD2, IDH1, and ATOX1. Altogether, these findings support shared co-expression of SSX1 and SPINK1 in both HCC tumor cells and in the surrounding supporting endothelial cell ecosystem in HERV-K high tumors.
Figure 5. GeoMx Digital Spatial Profiling of Hepatocellular Carcinoma Tumor Samples.
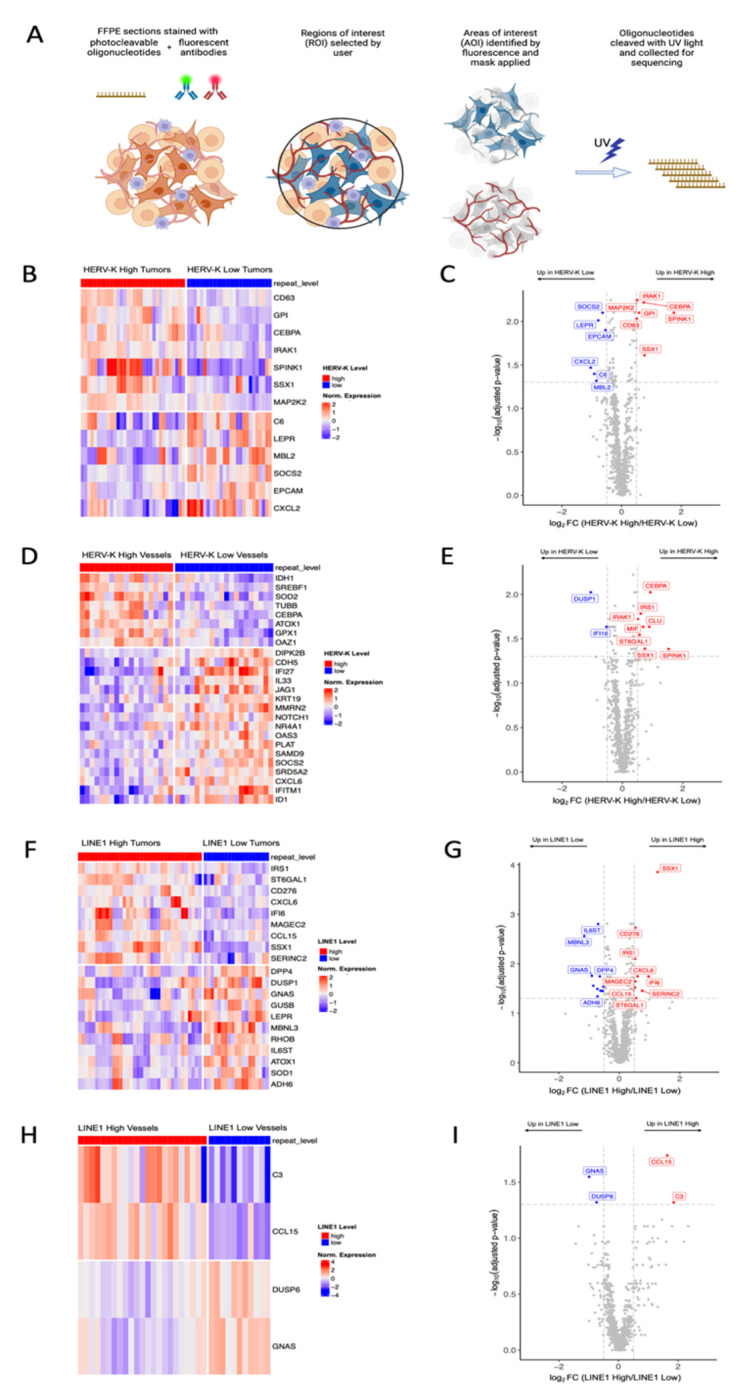
A. Schematic of GeoMx staining workflow. B-C. Heatmap and volcano plot of differentially expressed genes in HERV-K high and HERV-K low tumor AOIs. D-E. Heatmap and volcano plot of differentially expressed genes in HERV-K high and HERV-K low vessel AOIs. F-G. Heatmap and volcano plot of differentially expressed genes in LINE1 high and LINE1 low tumor AOIs. H-I. Heatmap and volcano plot of differentially expressed genes in LINE1 high and LINE1 low vessel AOIs. Differentially expressed genes determined by log fold change threshold of 0.5 and FDR-adjusted p-value threshold of 0.05.
In the tumors with high LINE1 expression (Fig. 5F–G), several genes were highly expressed including SSX1, MAGEC2, CD276, IRS1, CXCL6, IFI6, CCL15, SERINC2, and ST6GAL1. There were fewer differentially expressed genes when comparing LINE1 high and LINE1 low vessels (Fig. 5H–I), but C3 and CCL15 were highly expressed in the vessels of LINE1 high tumors. In summary, LINE1 high tumors shared similar SSX1 enrichment with HERV-K high tumors, but there were additional distinct gene programs in these tumors, suggesting fundamental differences in coding gene co-expression patterns between LINE1 and HERV-K repeat expression.
DISCUSSION
The development and progression of hepatocellular carcinoma is influenced by complex interactions in the tumor microenvironment between tumor cells, immune cells, and the tumor vasculature. In this study, we quantified the expression of LINE1, HSATII, HERV-K, and HERV-H repeat RNA species in HCC tumors. We found differences in overall survival associated with different repeat profiles, and positive correlations between repeat expression and multiple immune-suppressive markers. Patients with LINE1 high tumors and HERV-K high tumors had worse overall survival, consistent with the findings of prior work (22 – 24). To further explore the association between repeat RNA expression and clinical outcomes in HCC, we characterized the immune milieu by immunohistochemistry and spatial transcriptomics to understand the molecular differences in the tumor and vascular compartments of HCC.
We found that LINE1 expression positively correlated with both FOXP3 and TIM3 expression, implying an association between high LINE1 expression and an immune suppressive phenotype. TIM3 is a known negative regulator of the human immune system effector function, with known roles in the development of immune tolerance and the negative regulation of the immune response to chronic viral infection (33–35). Prior work has indicated that TIM3 is preferentially expressed on HCC tumor-derived Treg cells and that TIM3+FOXP3+Treg cells exhibit a particularly suppressive effect on CD8+ T-cells when compared with TIM3-FOXP3+Treg cells (36, 37). High-LINE1 expression has been observed in quiescent T-cells, with LINE1 knockdown resulting in a reduction in TIM3, LAG3, and PD-1 positive cells, suggesting that LINE1 directly contributes to an immune suppressive tumor microenvironment (38).
Spatial transcriptomic analysis revealed several differentially expressed genes in HERV-K high and LINE1 high tumors that have previously been associated with more aggressive tumor behavior and worse prognosis. In HERV-K high tumors and vessels, SPINK1 was highly expressed. SPINK1 overexpression has been noted in several cancers including HCC and has been associated with poor prognosis potentially due to its ability to function as a growth factor via EGFR signaling (30). There was significantly lower expression of EPCAM in HERV-K high tumors which suggests HERV-K low tumors may maintain more epithelial differentiation than HERV-K high tumors. Loss of EPCAM has been identified as an early event in epithelial-mesenchymal transition (39). This is further supported by the increased expression of SOCS2 in HERV-K low tumors. Increased SOCS2 expression has been shown to inhibit EMT, and the downregulation of SOCS2 has been associated with EMT activation, increased metastasis, and worse overall prognosis in HCC and other cancers (31, 32, 40). In both LINE1 high and HERV-K high tumors, members of the Cancer Testis Antigen (CTA) group (SSX1 and MAGEC2) were highly expressed. CTAs are a group of genes normally expressed in the gametes of males. Although the biological functions of CTAs are poorly understood, in HCC overexpression of CTAs has been associated with epithelial-mesenchymal transition and cancer progression (29, 41). High SSX1 and MAGEC2 specifically have both previously been associated with poor overall survival across numerous solid-organ and hematologic malignancies (42). High CTA and high repeat RNA expression may both be indicators of DNA hypomethylation and overall genomic instability, which have been linked with a worse prognosis in HCC. Given these findings, LINE1 and HERV-K may be useful prognostic biomarkers for more aggressive forms of HCC.
In addition to their potential as prognostic biomarkers, our findings indicate that repeat RNA expression profiles maybe useful in identifying patients that could benefit from novel therapeutic options. With response rates to immunotherapies in advanced HCC ranging from 15% in nivolumab monotherapy to about 30% in nivolumab-ipilimumab combination therapy and atezolizumab-bevacizumab combination therapies, additional progress is needed to identify novel therapeutic targets to augment treatment response. Given their limited expression in normal tissues and high expression in many cancers, CTAs have been explored as potential novel targets for immunotherapy in the form of cancer vaccines (43). Similarly, CD276 (also known as B7-H3) which was highly expressed in LINE1 high tumors, has limited expression in normal tissues but high expression in cancer tissues, and CAR-T cells targeting B7-H3 protein have demonstrated immune-mediated anti-tumor effects in pre-clinical models (44).
These highly expressed repeat RNAs may also represent potential novel therapeutic targets. Of the repeat RNA species evaluated in this study, high expression of the two that encode for protein products – HERV-K and LINE1 – were associated with worse overall survival. HERV-K is considered the most transcriptionally active of the endogenous retroviruses, encoding for proviral Gag, Pro, Pol, and Env proteins (45). Specific targeting of HERV-K proteins with monoclonal antibodies (46), HERV-K reactive cytotoxic T-cells derived from patient serum (47) (48), and CAR T-cells (49) have demonstrated anti-tumor effects in preclinical models. LINE1 encodes for ORF1 an RNA-binding protein and ORF2 which has endonuclease and reverse transcriptase activity. These protein products of LINE1 may similarly be targetable in HCC.
The RNA-ISH staining and quantification in this study was performed on FFPE sections and quantified using digital pathology analysis, techniques that can be readily translated to clinical applications. Given the differences in HCC clinical outcomes based on different repeat RNA expression profiles, repeat RNAs may be useful biomarkers for predicting prognosis and identifying potential therapeutic targets in HCC patients at risk for poor clinical outcomes.
The limitations of this study include the small sample size and the retrospective nature of the analysis. The tissue samples were operative samples from patients who underwent surgical resection or transplantation for management of their HCC, therefore patients with unresectable or metastatic disease who may have different tumor biology were not represented in the cohort. Finally, while differing tumor immune profiles were identified with correlational analyses, additional mechanistic experiments are needed to further assess any potential causal relationships.
In summary, our study is the first to characterize different repeat elements in HCC. We identified potentially novel repeat RNA prognostic biomarkers, associations between certain repeat RNAs and immune and additional microenvironmental features, providing novel mechanistic insight into the relationship of repeats with the coding transcriptome. Subsequent mechanistic studies will strengthen these immune and vascular microenvironmental features and generate further motivation for additional therapeutic interventions targeting the repeat transcriptome.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Angelique Gilbert and Danielle Bestoso for laboratory and administrative support. We are thankful for the patients that provided tumor material for this project.
This work was supported by research funding from the National Institutes of Health R01CA240924 (DTT), U01CA228963 (DTT), 3R01CA240924-04S1 (AC), and ACD-Biotechne (DTT).
Footnotes
COMPETING INTERESTS STATEMENT
DTT has received consulting fees from ROME Therapeutics, Sonata Therapeutics, and Tekla Capital. DTT is a founder and has equity in ROME Therapeutics, PanTher Therapeutics and TellBio, Inc., which is not related to this work. DTT is on the advisory board for ImproveBio, Inc. DTT has received honorariums from Moderna and Ikena Oncology that are not related to this work. DTT receives research support from ACD-Biotechne, AVA LifeScience GmbH, and Incyte Pharmaceuticals, which was not used in this work. DTT’s interests were reviewed and are managed by Massachusetts General Hospital and Mass General Brigham in accordance with their conflict-of-interest policies.
REFERENCES
- 1.Britten RJ, Kohne DE. Repeated sequences in DNA. Hundreds of thousands of copies of DNA sequences have been incorporated into the genomes of higher organisms. Science. 1968;161(3841):529–40. [DOI] [PubMed] [Google Scholar]
- 2.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. [DOI] [PubMed] [Google Scholar]
- 3.Ting DT, Lipson D, Paul S, Brannigan BW, Akhavanfard S, Coffman EJ, et al. Aberrant overexpression of satellite repeats in pancreatic and other epithelial cancers. Science. 2011;331(6017):593–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rodic N, Sharma R, Sharma R, Zampella J, Dai L, Taylor MS, et al. Long interspersed element-1 protein expression is a hallmark of many human cancers. The American journal of pathology. 2014;184(5):1280–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N. Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell. 2015;160(1-2):48–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Desai N, Sajed D, Arora KS, Solovyov A, Rajurkar M, Bledsoe JR, et al. Diverse repetitive element RNA expression defines epigenetic and immunologic features of colon cancer. JCI insight. 2017;2(3):e91078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Solovyov A, Vabret N, Arora KS, Snyder A, Funt SA, Bajorin DF, et al. Global Cancer Transcriptome Quantifies Repeat Element Polarization between Immunotherapy Responsive and T Cell Suppressive Classes. Cell Rep. 2018;23(2):512–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tanne A, Muniz LR, Puzio-Kuter A, Leonova KI, Gudkov AV, Ting DT, et al. Distinguishing the immunostimulatory properties of noncoding RNAs expressed in cancer cells. Proceedings of the National Academy of Sciences of the United States of America. 2015;112(49):15154–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mehdipour P, Marhon SA, Ettayebi I, Chakravarthy A, Hosseini A, Wang Y, et al. Epigenetic therapy induces transcription of inverted SINEs and ADAR1 dependency. Nature. 2020;588(7836):169–73. [DOI] [PubMed] [Google Scholar]
- 10.Leonova KI, Brodsky L, Lipchick B, Pal M, Novototskaya L, Chenchik AA, et al. p53 cooperates with DNA methylation and a suicidal interferon response to maintain epigenetic silencing of repeats and noncoding RNAs. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(1):E89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chiappinelli KB, Strissel PL, Desrichard A, Li H, Henke C, Akman B, et al. Inhibiting DNA Methylation Causes an Interferon Response in Cancer via dsRNA Including Endogenous Retroviruses. Cell. 2015;162(5):974–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roulois D, Loo Yau H, Singhania R, Wang Y, Danesh A, Shen SY, et al. DNA-Demethylating Agents Target Colorectal Cancer Cells by Inducing Viral Mimicry by Endogenous Transcripts. Cell. 2015;162(5):961–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moufarrij S, Srivastava A, Gomez S, Hadley M, Palmer E, Austin PT, et al. Combining DNMT and HDAC6 inhibitors increases anti-tumor immune signaling and decreases tumor burden in ovarian cancer. Sci Rep. 2020;10(1):3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stone ML, Chiappinelli KB, Li H, Murphy LM, Travers ME, Topper MJ, et al. Epigenetic therapy activates type I interferon signaling in murine ovarian cancer to reduce immunosuppression and tumor burden. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(51):E10981–E90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheng W, LaFleur MW, Nguyen TH, Chen S, Chakravarthy A, Conway JR, et al. LSD1 Ablation Stimulates Anti-tumor Immunity and Enables Checkpoint Blockade. Cell. 2018;174(3):549–63 e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cañadas I, Thummalapalli R, Kim JW, Kitajima S, Jenkins RW, Christensen CL, et al. Tumor innate immunity primed by specific interferon-stimulated endogenous retroviruses. Nat Med. 2018;24(8):1143–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Griffin GK, Wu J, Iracheta-Vellve A, Patti JC, Hsu J, Davis T, et al. Epigenetic silencing by SETDB1 suppresses tumour intrinsic immunogenicity. Nature. 2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Panda A, de Cubas AA, Stein M, Riedlinger G, Kra J, Mayer T, et al. Endogenous retrovirus expression is associated with response to immune checkpoint blockade in clear cell renal cell carcinoma. JCI insight. 2018;3(16). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parikh AR, Szabolcs A, Allen JN, Clark JW, Wo JY, Raabe M, et al. Radiation therapy enhances immunotherapy response in microsatellite stable colorectal and pancreatic adenocarcinoma in a phase II trial. Nat Cancer. 2021;2(11):1124–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith CC, Beckermann KE, Bortone DS, De Cubas AA, Bixby LM, Lee SJ, et al. Endogenous retroviral signatures predict immunotherapy response in clear cell renal cell carcinoma. J Clin Invest. 2018;128(11):4804–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Porter RL, Sun S, Flores MN, Berzolla E, You E, Phillips IE, et al. Satellite repeat RNA expression in epithelial ovarian cancer associates with a tumor immunosuppressive phenotype. J Clin Invest. 2022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harada K, Baba Y, Ishimoto T, Chikamoto A, Kosumi K, Hayashi H, et al. LINE-1 methylation level and patient prognosis in a database of 208 hepatocellular carcinomas. Ann Surg Oncol. 2015;22(4):1280–7. [DOI] [PubMed] [Google Scholar]
- 23.Zhu C, Utsunomiya T, Ikemoto T, Yamada S, Morine Y, Imura S, et al. Hypomethylation of long interspersed nuclear element-1 (LINE-1) is associated with poor prognosis via activation of c-MET in hepatocellular carcinoma. Ann Surg Oncol. 2014;21 Suppl 4:S729–35. [DOI] [PubMed] [Google Scholar]
- 24.Anwar SL, Hasemeier B, Schipper E, Vogel A, Kreipe H, Lehmann U. LINE-1 hypomethylation in human hepatocellular carcinomas correlates with shorter overall survival and CIMP phenotype. PLoS One. 2019;14(5):e0216374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng Y, Hlady RA, Joyce BT, Robertson KD, He C, Nannini DR, et al. DNA methylation of individual repetitive elements in hepatitis C virus infection-induced hepatocellular carcinoma. Clin Epigenetics. 2019;11(1):145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schauer SN, Carreira PE, Shukla R, Gerhardt DJ, Gerdes P, Sanchez-Luque FJ, et al. L1 retrotransposition is a common feature of mammalian hepatocarcinogenesis. Genome Res. 2018;28(5):639–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sudhindar PD, Wainwright D, Saha S, Howarth R, McCain M, Bury Y, et al. HCV Activates Somatic L1 Retrotransposition-A Potential Hepatocarcinogenesis Pathway. Cancers (Basel). 2021;13(20). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lau CC, Sun T, Ching AK, He M, Li JW, Wong AM, et al. Viral-human chimeric transcript predisposes risk to liver cancer development and progression. Cancer Cell. 2014;25(3):335–49. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y, Xu X, Ni X, Pan J, Chen M, Lin Y, et al. Gene-based cancer-testis antigens as prognostic indicators in hepatocellular carcinoma. Heliyon. 2023;9(3):e13269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Räsänen K, Itkonen O, Koistinen H, Stenman UH. Emerging Roles of SPINK1 in Cancer. Clin Chem. 2016;62(3):449–57. [DOI] [PubMed] [Google Scholar]
- 31.Zhou Y, Zhang Z, Wang N, Chen J, Zhang X, Guo M, et al. Suppressor of cytokine signalling-2 limits IGF1R-mediated regulation of epithelial–mesenchymal transition in lung adenocarcinoma. Cell Death & Disease. 2018;9(4):429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lv Y, Xie X, Zou G, Kong M, Yang J, Chen J, Xiang B. miR-181b-5p/SOCS2/JAK2/STAT5 axis facilitates the metastasis of hepatoblastoma. Precis Clin Med. 2023;6(4):pbad027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wolf Y, Anderson AC, Kuchroo VK. TIM3 comes of age as an inhibitory receptor. Nat Rev Immunol. 2020;20(3):173–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jin H-T, Anderson AC, Tan WG, West EE, Ha S-J, Araki K, et al. Cooperation of Tim-3 and PD-1 in CD8 T-cell exhaustion during chronic viral infection. Proc Natl Acad Sci USA. 2010;107(33):14733–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jones RB, Ndhlovu LC, Barbour JD, Sheth PM, Jha AR, Long BR, et al. Tim-3 expression defines a novel population of dysfunctional T cells with highly elevated frequencies in progressive HIV-1 infection. J Exp Med. 2008;205(12):2763–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yan J, Zhang Y, Zhang J-P, Liang J, Li L, Zheng L. Tim-3 expression defines regulatory T cells in human tumors. PLoS ONE. 2013;8(3):e58006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sakuishi K, Ngiow SF, Sullivan JM, Teng MWL, Kuchroo VK, Smyth MJ, Anderson AC. TIM3(+)FOXP3(+) regulatory T cells are tissue-specific promoters of T-cell dysfunction in cancer. Oncoimmunology. 2013;2(4):e23849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marasca F, Sinha S, Vadalà R, Polimeni B, Ranzani V, Paraboschi EM, et al. LINE1 are spliced in non-canonical transcript variants to regulate T cell quiescence and exhaustion. Nat Genet. 2022;54(2):180–93. [DOI] [PubMed] [Google Scholar]
- 39.Pastushenko I, Blanpain C. EMT Transition States during Tumor Progression and Metastasis. Trends in Cell Biology. 2019;29(3):212–26. [DOI] [PubMed] [Google Scholar]
- 40.Qiu X, Zheng J, Guo X, Gao X, Liu H, Tu Y, Zhang Y. Reduced expression of SOCS2 and SOCS6 in hepatocellular carcinoma correlates with aggressive tumor progression and poor prognosis. Molecular and Cellular Biochemistry. 2013;378(1):99–106. [DOI] [PubMed] [Google Scholar]
- 41.Gu X, Mao Y, Shi C, Ye W, Hou N, Xu L, et al. MAGEC2 Correlates With Unfavorable Prognosis And Promotes Tumor Development In HCC Via Epithelial-Mesenchymal Transition. Onco Targets Ther. 2019;12:7843–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.D’Angelo A, Kilili H, Chapman R, Generali D, Tinhofer I, Luminari S, et al. Immune-related pan-cancer gene expression signatures of patient survival revealed by NanoString-based analyses. PLoS ONE. 2023;18(1):e0280364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Duperret EK, Liu S, Paik M, Trautz A, Stoltz R, Liu X, et al. A Designer Cross-reactive DNA Immunotherapeutic Vaccine that Targets Multiple MAGE-A Family Members Simultaneously for Cancer Therapy. Clinical Cancer Research. 2018;24(23):6015–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Du H, Hirabayashi K, Ahn S, Kren NP, Montgomery SA, Wang X, et al. Antitumor Responses in the Absence of Toxicity in Solid Tumors by Targeting B7-H3 via Chimeric Antigen Receptor T Cells. Cancer Cell. 2019;35(2):221–37.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tönjes RR, Löwer R, Boller K, Denner J, Hasenmaier B, Kirsch H, et al. HERV-K: the biologically most active human endogenous retrovirus family. J Acquir Immune Defic Syndr Hum Retrovirol. 1996;13 Suppl 1:S261–7. [DOI] [PubMed] [Google Scholar]
- 46.Wang-Johanning F, Rycaj K, Plummer JB, Li M, Yin B, Frerich K, et al. Immunotherapeutic Potential of Anti-Human Endogenous Retrovirus-K Envelope Protein Antibodies in Targeting Breast Tumors. JNCI: Journal of the National Cancer Institute. 2012;104(3):189–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rycaj K, Plummer JB, Yin B, Li M, Garza J, Radvanyi L, et al. Cytotoxicity of Human Endogenous Retrovirus K-Specific T Cells toward Autologous Ovarian Cancer Cells. Clinical Cancer Research. 2015;21(2):471–83. [DOI] [PubMed] [Google Scholar]
- 48.Kang BH, Momin N, Moynihan KD, Silva M, Li Y, Irvine DJ, Wittrup KD. Immunotherapy-induced antibodies to endogenous retroviral envelope glycoprotein confer tumor protection in mice. PLoS ONE. 2021;16(4):e0248903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Krishnamurthy J, Rabinovich BA, Mi T, Switzer KC, Olivares S, Maiti SN, et al. Genetic Engineering of T Cells to Target HERV-K, an Ancient Retrovirus on Melanoma. Clinical Cancer Research. 2015;21(14):3241–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All images and primary data will be made available upon request. Spatial transcriptomic expression matrices will be available upon request.