Abstract
Over the last decade, more data has revealed that increased surface expression of the “don’t eat me” CD47 protein on cancer cells plays a role in immune evasion and tumor progression, with CD47 blockade emerging as a new therapy in immuno-oncology. CD47 is critical in regulating cell homeostasis and clearance, as binding of CD47 to the inhibitory receptor SIRPα can prevent phagocytosis and macrophage-mediated cell clearance. The purpose of this study was to examine the role of the CD47-SIRPα signal in platelet homeostasis and clearance. Therapeutic reagents targeting the CD47-SIRPα axis are very promising for treatment of hematologic malignancies and solid tumors, but lead to transient anemia or thrombocytopenia in a subset of patients. We found that platelet homeostatic clearance is regulated through the CD47-SIRPα axis and that therapeutic blockade to disrupt this interaction in mice and in humans has a significant impact on platelet levels. Furthermore, we identified genetic variations at the SIRPA locus that impact platelet levels in humans such that higher SIRPA gene expression is associated with higher platelet levels. SIRPA expression at either end of the normal range may affect clinical outcomes of treatment with anti-CD47 therapy.
Introduction
CD47, also known as integrin-associated protein (IAP), is a transmembrane glycoprotein expressed on hematopoietic and non-hematopoietic cells that, when bound to the highly polymorphic immune inhibitory receptor Signal Regulatory Protein Alpha (SIRPα), reduces phagocytosis and macrophage-mediated clearance of cells. Effectively, macrophages expressing SIRPα receive a ‘don’t eat me’ signal upon binding to CD47, as reviewed by Barclay and Ven den Berg (2014) 1. The potency of the ‘don’t eat me’ signal depends on several factors, including the relative level of surface CD47 2. In the case of pre-cancerous cells, the up-regulation of CD47 contributes to immune evasion of potentially harmful cells. Cancer cells upregulate CD47 expression to evade immune clearance and the level of expression may correlate with poor clinical outcomes in some cancers 3. In contrast, CD47 expression declines with cell age, notably serving as a biological timer for red blood cell (RBC) clearance, whereby loss of CD47 expression on mature red blood cells marks aged erythrocytes heading for clearance 4. Because of its role in regulating cell clearance, CD47 has been targeted in immunotherapeutic treatment approaches for hematologic malignancies and solid tumors 5–7. Inhibition of CD47’s anti-phagocytic signaling is a promising avenue for cancer immunotherapy 5. The CD47-SIRPα axis has also been implicated in the regulation of homeostatic levels of different immune cell populations. Previous studies demonstrated that CD47 blockade in vivo resulted in enhanced erythrophagocytosis leading to transient anemia 6. Furthermore, in a mouse model of induced colitis, mice lacking SIRPα or CD47 also developed anemia, followed by compensatory extramedullary erythropoiesis leading to splenomegaly 8. Interestingly, platelet levels have also been shown to be regulated by the CD47-SIRPα axis 9. With several human clinical trials evaluating the blockade of the CD47-SIRPα axis completed or in progress, there is considerable focus on preventing treatment-induced anemia (Table 1). In many of these trials, transient thrombocytopenia has also been noted in some patients. However, it is not known why some individuals develop anemia or thrombocytopenia, while others do not.
Table 1.
Pharmaceutical intervention with anti-CD47, anti-SIRPα, and thrombocytopenia in clinical trials.
| Strategy to minimize anemia | Reagent (company) | Thrombocytopenia in clinical trials: |
|---|---|---|
| Priming dose to ramp up red blood cell production | Magrolimab (Gilead) | 55% (https://ascopubs.org/doi/full/10.1200/JCO.22.01794) |
| Intratumoral injection | TTI-621 (Trillium) | Observed transient decline in platelet levels (when injected IV) Thrombocytopenia (grade 3 or above) as treatment related adverse event in 20% of patients. (https://aacrjournals.org/clincancerres/article/27/8/2190/672069/Phase-I-Study-of-the-CD47-Blocker-TTI-621-in) |
| Glycosylation specific antibody binding | Lemzoparlimab/TJC4 (I-mab) | In combination with Azacitidine (AZA) treatment, there was decreased platelet count (60%). EMSO Sept. 10, 2022 PDF |
| Antibody binding only CD47 in complex with integrin B1 | AO-176 (Arch Oncology) | Thrombocytopenia as treatment related adverse event in 9/27 (33%) patients (https://ascopubs.org/doi/10.1200/JCO.2021.39.15_suppl.2516). |
| Inert FC | ALX148 (ALX Oncology) | Thrombocytopenia as treatment related adverse event in 10% of patients. (https://ascopubs.org/doi/10.1200/JCO.2020.38.15_suppl.3056) |
| Antibody SIRPα binding to prevent Fcγ receptor recognition | GS-0189 (Gilead) | Thrombocytopenia not observed as a treatment related adverse event in patients. (https://www.ncbi.nlm.nih.gov/pmc/articles/PMC10188468/) |
| High affinity binding to V1 SIRPα allele | BI 765063 (Boehringer Ingelheim) | No related anemia of thrombocytopenia observed. (https://ascopubs.org/doi/abs/10.1200/JCO.2021.39.15_suppl.2623) |
Previously, genetic association studies have provided insight into the pathophysiology of a variety of immune cell traits 10. These earlier studies have identified specific genetic variants that affect platelet and erythrocyte traits in humans. Similarly, variant-to-expression analyses have provided insight into how genetic variants contribute to changes in RNA levels11 and protein expression 12, with possible implications for disease mechanisms. Because of the known connection between CD47/SIRPα and RBC homeostasis and this axis’ association with platelets 5,9, we sought to understand the effect of in vivo CD47 blockade on platelet clearance and the broader association between SIRPA polymorphisms and platelet phenotypes.
Materials and Methods
For clarity of nomenclature, we use SIRPA for the gene name and SIRPα for the protein name throughout the text.
In vivo CD47 Blockade.
All experiments were carried out in accordance with ethical care guidelines set by the Stanford University Administrative Panel on Laboratory Animal Care (APLAC). Female mice were used for all studies. Investigators were not blinded for animal studies.
C57BL/6 mice were injected intraperitoneally with a priming dose of 100 μg, followed by 500μg treatment doses of anti-CD47 MIAP410 (Bio X Cell) antibody, IgG isotype control or PBS every 48 hours for 2 weeks. After euthanasia, cardiac puncture was performed and spleens were collected. Blood was collected in diagnostic tubes containing EDTA and submitted to the veterinary diagnostic laboratory at Stanford for CBC. Spleens were preserved in 4% paraformaldehyde for 12 hours at 4 degrees overnight, then transferred to 70% ethanol for submission to HistoWiz. They were cut at 5 μm thickness and stained for hematoxylin and eosin (H&E) and Ter119.
Meta-analysis of earlier GWAS
We performed a meta-analysis in three cohorts with total N=709,883. These cohorts include meta-analysis of previously published summary statistics from the UK Biobank (N=444,382, accessed through https://alkesgroup.broadinstitute.org/LDSCORE/all_sumstats/), Biobank Japan (N=108,208) 13 and Blood Cell Consortium (N=157,293) 14. We used fixed effects meta-analysis as implemented in META v1.7 15.
FinnGen
FinnGen is a large-scale research study that aims to genotype 500,000 Finnish participants recruited from hospitals as well as prospective and retrospective epidemiological and disease-based cohorts. These data are combined with longitudinal registries that record phenotypes and health events (including ICD-based diagnosis) over the entire lifespan including the National Hospital Discharge Registry (inpatient and outpatient), Causes of Death Registry, the National Infectious Diseases Registry, Cancer Registry, Primary Health Care Registry (outpatient) and Medication Reimbursement Registry.
This study used data from FinnGen Data Freeze 9, which comprises 377,277 individuals. For the purposes of our research, we used registered sleep medication purchases (which constitute a prescription-only medication in Finland) dating from 1995. The primary indication for sleep medication prescription in Finland is sleep problems, and of these, non-benzodiazepine drugs are used for insomnia treatment 16.
FinnGen ethics statement
Patients and control subjects in FinnGen provided informed consent for biobank research, based on the Finnish Biobank Act. Alternatively, separate research cohorts, collected before the Finnish Biobank Act came into effect (in September 2013) and before start of FinnGen (August 2017), were collected based on study-specific consents and later transferred to the Finnish biobanks after approval by Fimea (Finnish Medicines Agency), the National Supervisory Authority for Welfare and Health. Recruitment protocols followed the biobank protocols approved by Fimea. The Coordinating Ethics Committee of the Hospital District of Helsinki and Uusimaa (HUS) statement number for the FinnGen study is Nr HUS/990/2017.
The FinnGen study is approved by the Finnish Institute for Health and Welfare (permit numbers: THL/2031/6.02.00/2017, THL/1101/5.05.00/2017, THL/341/6.02.00/2018, THL/2222/6.02.00/2018, THL/283/6.02.00/2019, THL/1721/5.05.00/2019 and THL/1524/5.05.00/2020), Digital and population data service agency (permit numbers: VRK43431/2017–3, VRK/6909/2018–3, VRK/4415/2019–3), the Social Insurance Institution (permit numbers: KELA 58/522/2017, KELA 131/522/2018, KELA 70/522/2019, KELA 98/522/2019, KELA 134/522/2019, KELA 138/522/2019, KELA 2/522/2020, KELA 16/522/2020), Findata permit numbers THL/2364/14.02/2020, THL/4055/14.06.00/2020,,THL/3433/14.06.00/2020, THL/4432/14.06/2020, HL/5189/14.06/2020, THL/5894/14.06.00/2020, THL/6619/14.06.00/2020, THL/209/14.06.00/2021, THL/688/14.06.00/2021, THL/1284/14.06.00/2021, THL/1965/14.06.00/2021, THL/5546/14.02.00/2020 and Statistics Finland (permit numbers: TK-53–1041-17 and TK/143/07.03.00/2020 (earlier TK-53–90-20)).
The Biobank Access Decisions for FinnGen samples and data utilized in FinnGen Data Freeze 7 include: THL Biobank BB2017_55, BB2017_111, BB2018_19, BB_2018_34, BB_2018_67, BB2018_71, BB2019_7, BB2019_8, BB2019_26, BB2020_1, Finnish Red Cross Blood Service Biobank 7.12.2017, Helsinki Biobank HUS/359/2017, Auria Biobank AB17–5154 and amendment #1 (August 17 2020), Biobank Borealis of Northern Finland_2017_1013, Biobank of Eastern Finland 1186/2018 and amendment 22 § /2020, Finnish Clinical Biobank Tampere MH0004 and amendments (21.02.2020 & 06.10.2020), Central Finland Biobank 1–2017, and Terveystalo Biobank STB 2018001.
PheWAS
PheWAS analysis using FinnGen release 9 data for individual endpoints as predefined by the FinnGen project for coagulation and ischemic stroke. We also used Open Targets Genetics platform (https://genetics.opentargets.org/) 17 to explore association of SIRPA risk variants with other traits and biomarker GWAS.
Mendelian Randomization
The meta-platelets summary statistics for effect size, standard error and P-values were used to find significant lead variants (P-value <5×10−8) using FUMA platform 18. Lead variants were subsequently used in a two-sample bidirectional Mendelian Randomization (MR) analysis. We estimated the effect from platelets on stroke and coronary artery disease using previous GWAS as identified from the GWAS catalog (June 2020 version, see Supplementary information for specific catalog IDs) 19. The MR was performed with the twosamplemr R package 20. Significant causality was explored based on Inverse Variance Weighting (IVW) statistical method (P-value < 0.05), and effect estimate was compared with estimates from MR Egger, IVW, Weighted Median, Simple Mode and Weighted Mode methods. Pleiotropy was assessed using MR Egger intercept.
Human Subjects
For platelet RNA studies, study approval was provided by the Stanford University Institutional Review Board (#18329). We collected blood from patients with myeloproliferative neoplasms, MPNs (specifically essential thrombocythemia, ET in this study) enrolled in the Stanford University and Stanford Cancer Institute Hematology Tissue Bank between December 2017– 2020. All MPN ET patient peripheral blood samples were obtained under written informed consent and were fully anonymized. All relevant ethical regulations were followed.
Eligibility criteria included age ≥18 years and Stanford MPN clinic diagnosis of essential thrombocythemia (defined using the consensus criteria at the time of this study). For healthy controls, blood was collected from eleven asymptomatic adult donors selected at random from the Stanford Blood Center. All donors were asked for consent for genetic research. For both ET patients and healthy controls, blood was collected into acid citrate-dextrose (ACD, 3.2%) sterile yellow-top tubes (Becton, Dickinson and Co.) and platelets were isolated by established 21,22,23,24 purification protocols. Blood was processed within 4 h of collection for all samples. The time from whole blood collection to platelet isolation was similar between healthy donors and MPN patients. For next generation RNA-sequencing (RNA-seq), 1×109 isolated platelets lysed in Trizol were processed following established procedures 21,22,25 for RNA isolation (RNA integrity numbers >7.0), extraction and library preparation. Twelve pooled samples with individual indices were run on an Illumina HiSeq 4000 (Patterned flow cell with Hiseq4000 SBS v3 chemistry) as 2 X 75bp paired end sequencing with a coverage goal of 40M reads/sample. Resultant platelet transcriptomic data was library-size-corrected, variance-stabilized, and log2-transformed using the R package DESeq2 26.
Results
Thrombocytopenia emerges as a common adverse event across distinct CD47 blockade reagents, despite varying approaches aimed at minimizing anemia resulting from CD47-mediated red blood cell clearance (Table 1). Due to the role of the CD47-SIRPα axis in cell clearance and platelet homeostasis, we sought to investigate the possibility that blocking CD47 could directly trigger thrombocytopenia.
CD47 blockade in vivo induces murine splenomegaly, splenic red pulp expansion from erythropoiesis, and increased megakaryocytes.
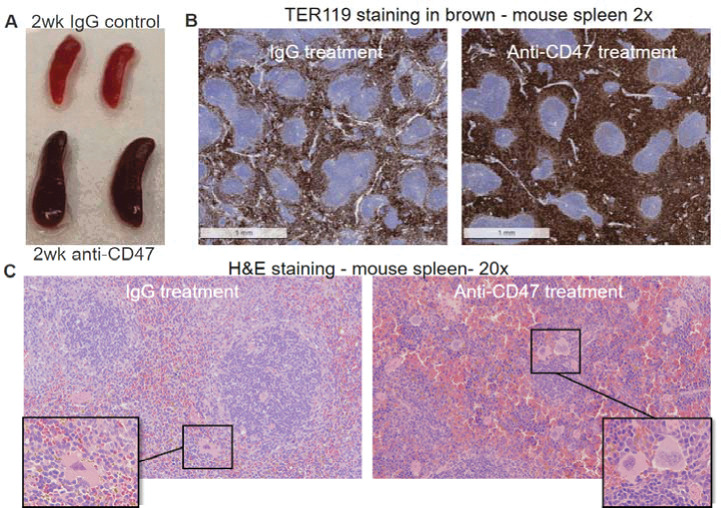
Prior studies have shown that sustained CD47 blockade in vivo is most effective and survivable after an initial dose of a smaller quantity of the antibody is given, termed the priming dose. This priming dose triggers compensatory erythropoiesis and prevents fatal anemia. To investigate the effects of CD47 blockade in a murine model, we injected C57BL/6 mice with a priming dose of either PBS, IgG isotype control, or anti-mouse CD47 antibody, MIAP410, two days before commencing treatment. We then injected PBS, IgG, or MIAP410 every two days for two weeks. Upon dissection, splenomegaly was grossly apparent in specimens who received anti-CD47 mAb treatment (Fig. 1A). Histological examination showed expansion of red pulp due to extramedullary erythropoiesis (Fig. 1B). Furthermore, megakaryocyte infiltration into the spleen was observed (Fig. 1C), suggesting that the murine spleen becomes a site of increased platelet production in the setting of CD47 blockade.
Figure 1. CD47 blockade induces splenomegaly, splenic red pulp expansion from erythropoiesis, and increased megakaryocytes.
In vivo, CD47 blockade leads to splenomegaly with extramedullary erythropoiesis. Mice were treated with MIAP410 (clone-3 anti-CD47) for 2 weeks. (A) Spleens were removed and photographed with visible increase in size and pigment of spleens from animals who received the CD47 blockade treatment (bottom) compared to the IgG control treated animals (top). (B) Spleen sections 5uM thick were stained with anti-TER119 demonstrating expansion of erythroid cells in the red pulp of the spleen. (C) Hematoxylin and eosin (H&E) staining of interfollicular spaces of spleens demonstrates that the white pulp of the spleen is diminished in anti-CD47-treated mice, with the space now devoted to extramedullary erythropoiesis and platelet production, with significant increase in megakaryocytes evident in anti-CD47-treated mice (representative megakaryocytes shown in the boxes). (H&E) images at 200 μM resolution.
Anti-CD47 treatment increases platelet clearance.
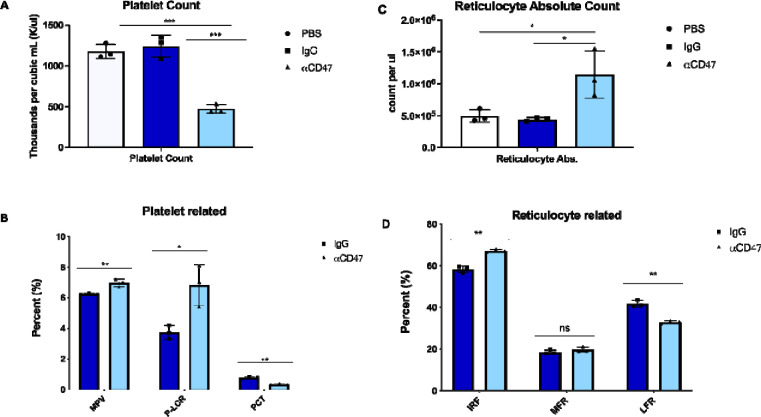
Platelets use CD47 to evade phagocytosis by macrophages 9. To understand the effects of CD47 blockade on platelet homeostasis with greater precision, we collected mouse blood via cardiac puncture at the time of euthanasia and submitted those samples for complete blood count (CBC). Mice treated with MIAP410 had approximately three-fold fewer platelets than controls treated with either PBS or IgG isotype control (p = 0.0003, p = 0.0008, Fig. 2A). In addition to a reduction in the absolute number of platelets, the remaining platelets were approximately 12% larger in volume than controls (p = 0.0079, Fig. 2B MPV). There is an inverse relationship between MPV and platelet count (Fig. 2A and 2B), which has also been observed in human clinical data 27. P-LCR, which represents the percentage of platelets larger than 12fL, increased in treated mice (Fig. 2B P-LCR). Absolute reticulocyte count was also significantly increased in the treated mice (p = 0.0147, Fig. 2C), which demonstrates the hematopoietic system’s concerted efforts to compensate for the anemic and thrombocytopenic effects of anti-CD47 treatment. Based on maturation stage, reticulocytes can be classified into subtypes, where high fluorescence reticulocytes (HFR) and medium fluorescence reticulocytes (MFR) are immature and low fluorescence reticulocytes (LFR) are mature 28. In the treated mice, immature reticulocyte fraction (IRF) significantly increased (p = 0.0011, Fig. 2D), and LFR decreased (p = 0.0011, Fig. 2D), showing the effect of CD47 blockade with reticulocyte maturation.
Figure 2. Platelets are reduced in number but increases in volume upon CD47 blockade in vivo.
Complete blood counts were run after 2wks of anti-CD47 blockade in C57/B6 mice. A) Platelet count shown in thousands per cubic mL. B) Platelet related percentages shown as indicated, MPV: mean platelet volume (average platelet size), P-LCR: platelet large cell ratio, PCT: volume occupied by platelets in blood as percentage C) Reticulocyte absolute count shown per microliter. D) Reticulocyte related percentages shown as indicated, IRF: immature reticulocyte fraction, MFR: middle-fluorescence reticulocytes, LFR: low-fluorescence reticulocytes. CBC counts were run on two separate comparisons of CD47 blockade, one representative experiment is shown n=3 mice per group, significance of unpaired T-test comparisons shown as (*p ≤.05, **p ≤.01, ***p ≤.005).
Variation at SIRPA affects platelet levels in humans
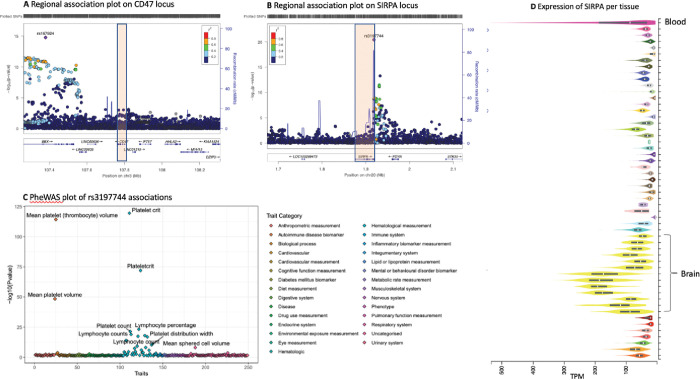
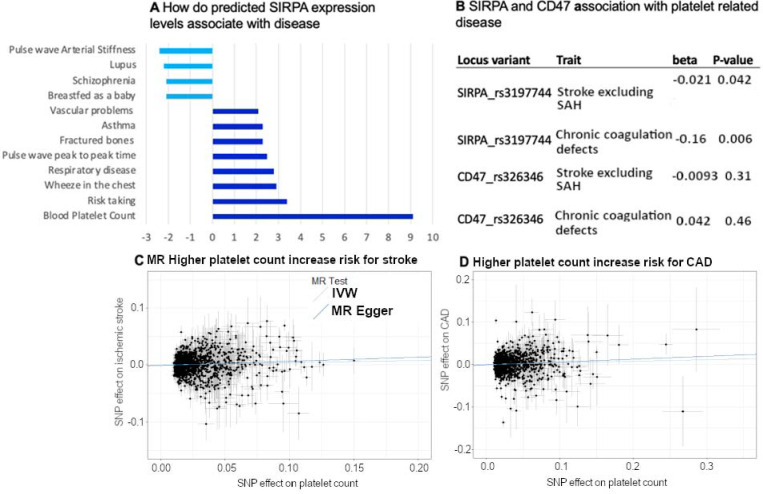
Previous genome-wide association studies have implied that platelet levels are modulated by common genetic risk factors, including even SIRPA 29. We therefore performed a meta-analysis of N=752,910 individuals using publicly available GWAS evidence for 645 independent loci (Supplementary information). The significant loci included BBX-CD47 and SIRPA. Specifically, we discovered GWAS associations at the 3’ end of CD47, close to the BBX gene (rs167924, beta = −0.018, p= 7.40e-23; Fig. 3A). In addition, we discovered that variants in the 3’UTR of SIRPA (rs3197744 beta = −0.023, p = 9.88e-32, Fig. 3B) are associated with the amount of platelets in those individuals.
Figure 3. Variants at SIRPA are associated with platelet levels.
(A) Variation at 3’ to CD47 at BBX locus associated with platelet levels. (B) Variant rs3197744 at SIRPA locus associates with amount of platelets. (C) Examining associations across currently available GWAS results (PheWAS) over 2500 independent studies at rs3197744 at SIRPA locus shows specific association with platelet and immune cell measurements. (D) Expression profile of SIRPA across human tissues shows highest expression in the brain and in blood.
We explored the phenotypic associations of these variants with stroke and coagulation traits, as well as other publicly available traits. We observed that the variants at SIRPA had specific associations with blood cell traits overall. The association with platelets was strongest, which has also been observed in previous GWAS 29(Fig. 3C, and Supplementary information). The expression profile of SIRPA RNA across human tissues observed the highest expression levels in whole blood and the brain (Fig. 3D), as would be expected from the high SIRPα protein expression observed on neurons and myeloid cells. Overall, these findings show a contribution from variants in the SIRPA 3’UTR and at BBX-CD47 loci to platelet levels, in alignment with our functional studies.
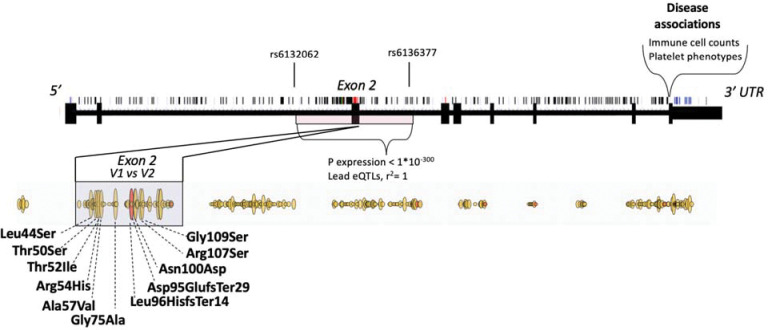
Genetic variation affecting structure, expression and disease associations on SIRPA locus.
SIRPA is highly polymorphic, encompassing several intronic and coding variants. Genetic variation at the SIRPA locus has previously focused on exploring coding variation at the second exon, where missense variants can be divided into two common haplotypes that are referred to as V1 and V2 (Fig. 4). These variants are part of a haplotype from rs6132062 to rs6136377 that spans several regulatory elements from intron one to two and has the largest impact on SIRPA gene expression in blood 30. In addition, the regulatory variants affecting SIRPA gene expression are also in high linkage disequilibrium (LD, r2=1) with the V1 haplotype, so that V1 expression also correlates with the amount of overall SIRPA expression. In contrast, nearly all disease and trait associations with the SIRPA locus are located in the 3’UTR and are not in LD with the eQTL signal, or with V1 or V2 haplotypes. Consequently, the SNPs that are associated with platelet levels are also located at the 3’UTR of SIRPA.
Figure 4. Schematic representation of genetic variation affecting structure, expression and disease associations on SIRPA locus.
Single nucleotide polymorphisms are marked on top of the gene structure. Missense variants are highlighted in red as part of the gene structure and outlined yellow below the gene structure. Frameshift/insertions or deletions are highlighted with red as part of the missense variants. Haplotype from rs6132062 to rs6136377 encompassing exon 2 has the largest impact on SIRPA gene expression in blood. In addition, exon 2 contains missense variants that can be divided into two common haplotypes that are referred to as V1 and V2. V1 allele is shown first in the graph. V1 is in linkage disequilibrium (r2=1) with the SIRPA eQTLs (rs6132062 and rs6136377) having impact therefore both on expression and on coding sequence. Disease associations with SIRPA are located in the 3’UTR including the variants that associate with platelet levels.
SIRPA expression and broader context of platelets in human disease
We expect these genetic variants to affect the number of platelets through one of two mechanisms. First, polymorphisms can lead to changes in amino acids and have a direct effect on protein function. Second, polymorphisms may occur at transcriptional regulatory regions, where they lead to changes in the amount or properties of the transcript, and thus affect the gene product or protein function. This is often cell type-specific and controlled by tissue-specific transcriptional regulators. Such differences can be explored through expression quantitative trait loci (eQTLs), location on active chromatin and tissue specificity, or transcription-wide analysis (TWAS). Indeed, when we examined SIRPA expression in humans and assessed its association with the amount of platelets using TWAS 31, we saw a significant positive correlation with SIRPA gene expression and the amount of platelets (Z=9.3, p= Best TWAS P=1.1e-19, Fig. 5A).
Figure 5. SIRPA expression and broader context of platelets in human disease.
(A) TWAS analysis of SIRPA expression with human phenotypes. X-axis shows effect on expression by Z-statistics. (B) Association of leading SIRPA and CD47 SNPs with stroke excluding subarachnoid hemorrhage (SAH) and coagulation traits that have been implicated in platelet function. (C) Analysis of causality using Mendelian randomization from platelet levels to stroke, (D) and to coronary artery disease.
To understand the contribution from coding variants at the SIRPA locus, we explored the earlier computed burden analyses from the Genebass project (https://genebass.org/), which comprises analysis of variants in 281,852 individuals with exome sequence data from the UK Biobank. Burden analysis of missense variants at SIRPA showed an association of missense variants with a higher percentage of platelets in the blood as measured with plateletcrit (p = 0.014, Fig. 5A), but a nominal association with a subset of other platelet and reticulocyte traits (Supplementary information).
We then assessed whether SIRPA variants that contribute to platelet levels associate with other phenotypes known to be related to platelet levels. Indeed, the platelet-associated SNPs on SIRPA are nominally associated with coagulation and with ischemic stroke phenotypes in FinnGen (Fig. 5B). We also tested the causality behind higher platelet levels and ischemic stroke and coronary artery disease using genetic instruments and a Mendelian randomization approach. We observed a causal effect from higher platelet levels in higher risk for ischemic stroke and coronary artery disease (Fig. 5C and D).
SIRPA and CD47 are inversely correlated, and are independently associated with increased platelet count in ET patients
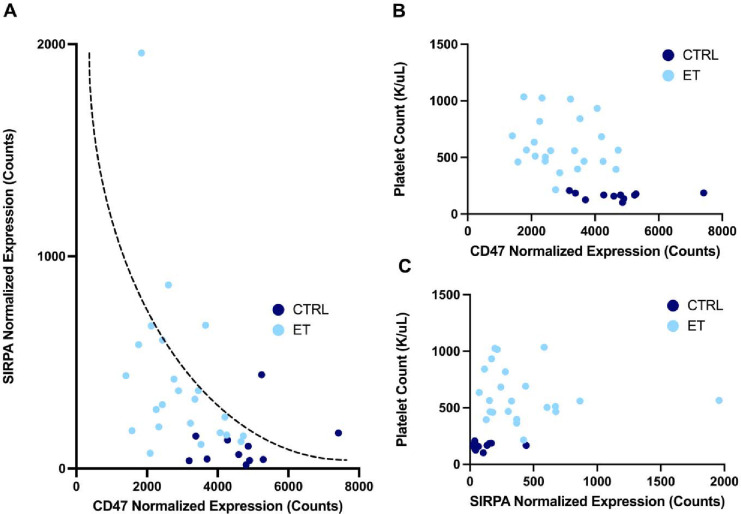
We further evaluated the CD47-SIRPα axis in a second disease phenotype, essential thrombocythemia (ET), which is also associated with increased platelet counts 32. Here, we measured platelet transcriptomic expression of CD47 and SIRPα as a function of platelet counts in patients with ET (n=23) compared with that of healthy donors (n=11). CD47 and SIRPα platelet RNA expression levels were inversely correlated (Fig. 6A), with increasingly higher expression of SIRPA in ET patients compared to healthy donors. Therefore, as a function of platelet counts, CD47 expression levels were also inversely correlated with higher platelet counts in ET patients versus healthy donors, and vice versa for SIRPα expression levels (Fig. 6B and C).
Figure 6. SIRPA and CD47 are inversely correlated, and are independently associated with platelet count in ET patients.
(A) Correlation of platelet normalized gene expression between CD47 and SIRPA for patients with essential thrombocythemia and healthy donors. Closed circles navy or light blue indicate sample type respectively of healthy donors as controls (CTRL, n=11) and patients with essential thrombocythemia (ET, n=23). Inverse correlation is indicated by a dotted curve as a guide to the eye. Platelet gene expression as assessed by normalized counts of (B) SIRPA and (C) CD47 as a function of patient platelet counts (K/uL) at the time of specimen banking and platelet isolation. Closed circles navy or light blue indicate sample type respectively of healthy donors as controls (CTRL, n=11) and patients with essential thrombocythemia (ET, n=23).
Discussion
We have shown that platelet homeostasis and clearance is regulated through the CD47-SIRPα axis and that therapeutic blockade to disrupt this interaction has a significant impact on platelet levels. Our findings summarize and validate previous observations from clinical studies, blockade assays from animal studies, and genetic evidence.
The CD47-SIRPα axis has an established role in platelet hematopoiesis, but the impact of the highly polymorphic SIRPA gene on population level heterogeneity of outcomes from blocking the CD47-SIRPa axis are not known. Yamao et al. (2002) 33 showed that modulation of SIRPα expression and signaling can increase platelet clearance and lead to insufficient compensatory megakaryocytopoiesis and platelet production. They used mice lacking the intracellular domain of SIRPα and showed increased splenic megakaryocytes and thrombocytopenia, similar to what we observed after blockade of CD47 in WT mice. CD47-deficient mice on a C57BL/6 background have also been shown to produce platelets with considerably shorter half-lives and consequently exhibit thrombocytopenia, with a dose-response effect between total CD47 knockouts, heterozygotes and wild-type mice 9,34. CD47−/− platelets transferred into wild-type mice underwent increased and accelerated clearance, and blockade of SIRPα increases phagocytosis of wild-type platelets to the level of CD47−/− platelets. Thus, deficiencies in either CD47 or SIRPα individually appear to be sufficient to drive up platelet clearance. This suggests that both, but not one or the other, are necessary for platelet survival and homeostasis. Our in vivo studies using CD47 blockade recapitulate these results, showing increased megakaryocytes in the spleen of mice treated with an antibody against the SIRPα-binding domain of CD47, and showing simultaneously reduced platelet count and increased reticulocyte count in the circulation. While the platelet numbers are reduced, individual platelets are hypertrophic, which raises the possibility of a compensatory increase not only in megakaryocyte number but also in the rate of megakaryocyte-platelet cleavage, resulting in larger platelet fragments. Clinical trials of anti-CD47 biologics have implied the existence of a similar mechanism in humans, though few studies exist that elucidate the details.
We hypothesized a connection between the observed clinical trial data and murine experiments with genome-wide association studies (GWAS), where we have identified genetic variants at the SIRPA locus 3’UTR that associate with platelet levels 29. To understand the association and possible underlying mechanisms, we performed a meta-analysis of publicly available cohorts. In our analysis, we first identified regulatory variants at intronic and untranslated regions of SIRPA. While the primary region showing ‘platelet effect’ is seen at the 3’UTR of SIRPA, our analysis also shows that higher SIRPA expression in humans is associated with higher platelet levels. Furthermore, the well-established V1-haplotype of non-synonymous genetic variants at exon 2 of SIRPA correlates with higher SIRPA expression. We identified three partially independent genetic mechanisms: a) 3’UTR association, b) intronic eQTLs, and c) non-synonymous coding variants that contribute at the SIRPA locus and translate to higher platelet levels.
Ultimately, the clinical relevance of platelet biology centers on individuals’ risk of thromboembolic events like stroke and CAD. Our analysis demonstrates a connection between SIRPA variants, platelet levels and risk of stroke and CAD. It is known that there exists a role for CD47 in atherosclerosis, where it has been shown that ApoE−/− mice on high fat diets develop atherosclerosis partly due to upregulation of CD47 on plaque smooth muscle cells 35. Anti-CD47 antibodies can help prevent atherosclerosis in monotherapy 36, and in combination with anti-TNFα can reduce preformed plaques, a process in which plaque macrophages remove necrotic cores by efferocytosis as well as pathogenic clonal smooth muscle cells by programmed cell removal (PrCR) 36,37. Since the CD47- SIRPα axis is known to contribute to atherosclerosis, this prompts the idea that genetic polymorphisms within the axis can impact the potential of axis modulation in stroke and CAD.
To further demonstrate the link between CD47 and SIRPα expression levels and platelet count. We analyzed the CD47- SIRPα axis in the context of ET. ET patients with increased platelet levels exhibit elevated SIRPα platelet RNA expression levels in comparison to healthy patients, and the SIRPα expression levels are inversely correlated to CD47 levels. Disruption of the CD47- SIRPα axis seems to directly impact platelet levels, suggesting that genetic variation within the axis may have a similar effect.
Taken together, our findings bring up a novel aspect of CD47 blockade in the context of genetic polymorphisms of SIRPA that can impact platelet biology. Future studies should investigate the extent to which these polymorphisms could potentially help identify patients at greater risk of developing platelet-related diseases, including thrombocytopenia, ET, CAD, and stroke, with treatments that interfere with this signaling pathway and should probe further the functional impact of these differences in platelet levels and characteristics.
Supplementary Material
Key points.
Platelet homeostasis is regulated through the CD47-SIRPα axis and therapeutic blockade to disrupt this interaction impacts platelet levels
Common genetic variants at SIRPA locus associate with platelet levels
Acknowledgments
We want to acknowledge the participants and investigators of the FinnGen study. The authors wish to thank members of the Weissman Lab at the Institute for Stem Cell Biology & Regenerative Medicine at Stanford University School of Medicine and Histowiz for their processing of our histological samples. We also thank Dr. Jason Gotlib at Stanford University, MPN ET patients at the Stanford Cancer Institute, and the healthy donors at the Stanford Blood Center for their contribution to the platelet transcriptome research that enabled further study of select candidate genes in this work.
FUNDING
Research reported in this publication was supported by the Fairbairn family foundation; the Robert J. Kleberg, Jr., and Helen C. Kleberg Foundation; the Virginia and D. K. Ludwig Fund for Cancer Research; M.C.T. and Y.Y.Y. were supported by Stanford Immunology training grant 5T32AI007290, and M.C.T. was also supported by the NIH NRSA 1 F32 AI124558-01 award. M.C.T, P.S.H, and A.S. are also funded by the NIH/NIAID R01 AI178713 award.
A.K. was funded by US National Institutes of Health grant (1K08HG010061-01A1 and 3UL1TR001085-04S1) and the MPN Research Foundation Challenge Grant.
This work was supported by NIH/National Cancer Institute Outstanding Investigator Award R35CA220434 (to I.L.W.); NIH/NIAID R01 AI143889 (to I.L.W.); and the D.K. Ludwig Fund for Cancer Research.
FinnGen is funded by two grants from Business Finland (HUS 4685/31/2016 and UH 4386/31/2016) and the following industry partners: AbbVie Inc., AstraZeneca UK Ltd, Biogen MA Inc., Bristol Myers Squibb (and Celgene Corporation & Celgene International II Sàrl), Genentech Inc., Merck Sharp & Dohme Corp, Pfizer Inc., GlaxoSmithKline Intellectual Property Development Ltd., Sanofi US Services Inc., Maze Therapeutics Inc., Janssen Biotech Inc, Novartis AG, and Boehringer Ingelheim. Following biobanks are acknowledged for delivering biobank samples to FinnGen: Auria Biobank (www.auria.fi/biopankki), THL Biobank (www.thl.fi/biobank), Helsinki Biobank (www.helsinginbiopankki.fi), Biobank Borealis of Northern Finland (https://www.ppshp.fi/Tutkimus-ja-opetus/Biopankki/Pages/Biobank-Borealis-briefly-in-English.aspx), Finnish Clinical Biobank Tampere (www.tays.fi/en-US/Research_and_development/Finnish_Clinical_Biobank_Tampere), Biobank of Eastern Finland (www.ita-suomenbiopankki.fi/en), Central Finland Biobank (www.ksshp.fi/fi-FI/Potilaalle/Biopankki), Finnish Red Cross Blood Service Biobank (www.veripalvelu.fi/verenluovutus/biopankkitoiminta) and Terveystalo Biobank (www.terveystalo.com/fi/Yritystietoa/Terveystalo-Biopankki/Biopankki/). All Finnish Biobanks are members of BBMRI.fi infrastructure (www.bbmri.fi). Finnish Biobank Cooperative -FINBB (https://finbb.fi/) is the coordinator of BBMRI-ERIC operations in Finland. The Finnish biobank data can be accessed through the Fingenious® services (https://site.fingenious.fi/en/) managed by FINBB. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict of interest declaration
H.M.O., M.C.T., Y.Y.Y, and I.L.W. are co-inventors on pct/us2019/050650 which is related to this work. M.C.T., Y.Y.Y, and I.L.W. are co-inventors on PCT/US2020/015905 related to this work. M.C.T. and I.L.W. are co-inventors on a patent application (63/107,295) related to this work. M.C.T., M.S. and I.L.W. are co-inventors on a patent application (17/425,224) related to this work. I.L.W. is an inventor on U.S. patent 2019/0092873 A1 CD47, Targeted Therapies for the Treatment of Infectious Disease. I.L.W. is a cofounder, director, and stockholder in FortySeven Inc., a public company that was involved in CD47-based immunotherapy of cancer during this study but was acquired by Gilead. At the time of this submission, I.L.W. has no formal relationship with Gilead, and is engaged in co-founding a company dealing with atherosclerosis and CD47.
References
- 1.Barclay AN, Van den Berg TK. The interaction between signal regulatory protein alpha (SIRPα) and CD47: structure, function, and therapeutic target. Annu Rev Immunol. 2014;32:25–50. doi: 10.1146/annurev-immunol-032713-120142. Epub 2013 Nov 6. [DOI] [PubMed] [Google Scholar]
- 2.Chao MP, Takimoto CH, Feng DD, McKenna K, Gip P, Liu J, Volkmer JP, Weissman IL, Majeti R. Therapeutic Targeting of the Macrophage Immune Checkpoint CD47 in Myeloid Malignancies. Front Oncol. 2020. Jan 22;9:1380. doi: 10.3389/fonc.2019.01380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Takimoto CH, Chao MP, Gibbs C, et al. The Macrophage ‘Do not eat me’ signal, CD47, is a clinically validated cancer immunotherapy target. Ann Oncol. 2019;30(3):486–489. [DOI] [PubMed] [Google Scholar]
- 4.Oldenborg PA, Zheleznyak A, Fang YF, Lagenaur CF, Gresham HD, Lindberg FP. Role of CD47 as a marker of self on red blood cells. Science. 2000;288(5473):2051–2054. [DOI] [PubMed] [Google Scholar]
- 5.Advani R, Flinn I, Popplewell L, et al. CD47 Blockade by Hu5F9-G4 and Rituximab in Non-Hodgkin’s Lymphoma. N Engl J Med. 2018;379(18):1711–1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Willingham SB, Volkmer JP, Gentles AJ, et al. The CD47-signal regulatory protein alpha (SIRPa) interaction is a therapeutic target for human solid tumors. Proc Natl Acad Sci U S A. 2012;109(17):6662–6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Majeti R, Chao MP, Alizadeh AA, et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138(2):286–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bian Z, Shi L, Guo YL, et al. Cd47-Sirpalpha interaction and IL-10 constrain inflammation-induced macrophage phagocytosis of healthy self-cells. Proc Natl Acad Sci U S A. 2016;113(37):E5434–5443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Olsson M, Bruhns P, Frazier WA, Ravetch JV, Oldenborg PA. Platelet homeostasis is regulated by platelet expression of CD47 under normal conditions and in passive immune thrombocytopenia. Blood. 2005;105(9):3577–3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Astle WJ, Elding H, Jiang T, et al. The Allelic Landscape of Human Blood Cell Trait Variation and Links to Common Complex Disease. Cell. 2016;167(5):1415–1429 e1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Consortium GT. The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Battle A, Khan Z, Wang SH, et al. Genomic variation. Impact of regulatory variation from RNA to protein. Science. 2015;347(6222):664–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nagai A, Hirata M, Kamatani Y, et al. Overview of the BioBank Japan Project: Study design and profile. J Epidemiol. 2017;27(3S):S2–S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eicher JD, Chami N, Kacprowski T, et al. Platelet-Related Variants Identified by Exomechip Meta-analysis in 157,293 Individuals. Am J Hum Genet. 2016;99(1):40–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liu JZ, Tozzi F, Waterworth DM, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42(5):436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lie JD, Tu KN, Shen DD, Wong BM. Pharmacological Treatment of Insomnia. P T. 2015. Nov;40(11):759–71. [PMC free article] [PubMed] [Google Scholar]
- 17.Ghoussaini M, Mountjoy E, Carmona M, et al. Open Targets Genetics: systematic identification of trait-associated genes using large-scale genetics and functional genomics. Nucleic Acids Res. 2021;49(D1):D1311–D1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Watanabe K, Taskesen E, van Bochoven A, Posthuma D. Functional mapping and annotation of genetic associations with FUMA. Nat Commun. 2017;8(1):1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.MacArthur J, Bowler E, Cerezo M, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017;45(D1):D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13(11):e1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Campbell RA, Franks Z, Bhatnagar A, et al. Granzyme A in Human Platelets Regulates the Synthesis of Proinflammatory Cytokines by Monocytes in Aging. J Immunol. 2018;200(1):295–304. doi: 10.4049/jimmunol.1700885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rowley JW, Oler AJ, Tolley ND, et al. Genome-wide RNA-seq analysis of human and mouse platelet transcriptomes [published correction appears in Blood. 2014 Jun 12;123(24):3843]. Blood. 2011;118(14):e101–e111. doi: 10.1182/blood-2011-03-339705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Middleton EA, Rowley JW, Campbell RA, et al. Sepsis alters the transcriptional and translational landscape of human and murine platelets. Blood. 2019;134(12):911–923. doi: 10.1182/blood.2019000067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Amisten S. A rapid and efficient platelet purification protocol for platelet gene expression studies. Methods Mol Biol. 2012;788:155–172. doi: 10.1007/978-1-61779-307-3_12 [DOI] [PubMed] [Google Scholar]
- 25.Thomas S, Krishnan A. Platelet Heterogeneity in Myeloproliferative Neoplasms. Arteriosclerosis, Thrombosis, and Vascular Biology. 2021;41(11):2661–2670. doi: 10.1161/ATVBAHA.121.316373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bessman JD, Williams LJ, Gilmer PR Jr. Mean platelet volume. The inverse relation of platelet size and count in normal subjects, and an artifact of other particles. Am J Clin Pathol. 1981. Sep;76(3):289–93. doi: 10.1093/ajcp/76.3.289. [DOI] [PubMed] [Google Scholar]
- 28.Urbina A., & Palomino F. In vitro kinetics of reticulocyte subtypes: Maturation after red blood cell storage in additive solution-1 (AS-1). Hematology, Transfusion and Cell Therapy, 2018;40(2);143–150. 10.1016/j.htct.2017.12.002 [DOI] [Google Scholar]
- 29.Eicher JD, Chami N, Kacprowski T, Nomura A, Chen MH, Yanek LR, Tajuddin SM, Schick UM, Slater AJ, Pankratz N, Polfus L, Schurmann C, Giri A, Brody JA, Lange LA, Manichaikul A, Hill WD, Pazoki R, Elliot P, Evangelou E, Tzoulaki I, Gao H, Vergnaud AC, Mathias RA, Becker DM, Becker LC, Burt A, Crosslin DR, Lyytikäinen LP, Nikus K, Hernesniemi J, Kähönen M, Raitoharju E, Mononen N, Raitakari OT, Lehtimäki T, Cushman M, Zakai NA, Nickerson DA, Raffield LM, Quarells R, Willer CJ, Peloso GM, Abecasis GR, Liu DJ; Global Lipids Genetics Consortium; Deloukas P, Samani NJ, Schunkert H, Erdmann J; CARDIoGRAM Exome Consortium; Myocardial Infarction Genetics Consortium; Fornage M, Richard M, Tardif JC, Rioux JD, Dube MP, de Denus S, Lu Y, Bottinger EP, Loos RJ, Smith AV, Harris TB, Launer LJ, Gudnason V, Velez Edwards DR, Torstenson ES, Liu Y, Tracy RP, Rotter JI, Rich SS, Highland HM, Boerwinkle E, Li J, Lange E, Wilson JG, Mihailov E, Mägi R, Hirschhorn J, Metspalu A, Esko T, Vacchi-Suzzi C, Nalls MA, Zonderman AB, Evans MK, Engström G, Orho-Melander M, Melander O, O’Donoghue ML, Waterworth DM, Wallentin L, White HD, Floyd JS, Bartz TM, Rice KM, Psaty BM, Starr JM, Liewald DC, Hayward C, Deary IJ, Greinacher A, Völker U, Thiele T, Völzke H, van Rooij FJ, Uitterlinden AG, Franco OH, Dehghan A, Edwards TL, Ganesh SK, Kathiresan S, Faraday N, Auer PL, Reiner AP, Lettre G, Johnson AD. Platelet-Related Variants Identified by Exomechip Meta-analysis in 157,293 Individuals. Am J Hum Genet. 2016. Jul 7;99(1):40–55. doi: 10.1016/j.ajhg.2016.05.005. Epub 2016 Jun 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Võsa U, Claringbould A, Westra H-J, et al. Unraveling the polygenic architecture of complex traits using blood eQTL metaanalysis. bioRxiv. 2018:447367. [Google Scholar]
- 31.Gusev A, Ko A, Shi H, et al. Integrative approaches for large-scale transcriptome-wide association studies. Nat Genet. 2016;48(3):245–252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Panova-Noeva M, Marchetti M, Spronk HM, et al. Platelet-induced thrombin generation by the calibrated automated thrombogram assay is increased in patients with essential thrombocythemia and polycythemia vera. Am J Hematol. 2011;86(4):337–342. doi: 10.1002/ajh.21974 [DOI] [PubMed] [Google Scholar]
- 33.Yamao T, Noguchi T, Takeuchi O, et al. Negative regulation of platelet clearance and of the macrophage phagocytic response by the transmembrane glycoprotein SHPS-1. J Biol Chem. 2002;277(42):39833–39839. [DOI] [PubMed] [Google Scholar]
- 34.Laboratory TJ. Mouse Genome Database (MGD) at the Mouse Genome Informatics website, Bar Harbor, Maine. World Wide Web Vol. 2020: The Jackson Laboratory. [Google Scholar]
- 35.Proefferocytic Therapy Promotes Transforming Growth Factor-β Signaling and Prevents Aneurysm Formation. Circulation. 2018. Feb 13;137(7):750–753. doi: 10.1161/CIRCULATIONAHA.117.030389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lagasse E, Weissman IL. bcl-2 inhibits apoptosis of neutrophils but not their engulfment by macrophages. J Exp Med. 1994;179(3):1047–1052. doi: 10.1084/jem.179.3.1047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chao MP, Majeti R, Weissman IL. Programmed cell removal: a new obstacle in the road to developing cancer. Nat Rev Cancer. 2011;12(1):58–67. Published 2011 Dec 8. doi: 10.1038/nrc3171 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.