Abstract
Artemisinins have been a cornerstone of malaria control, but resistance in Plasmodium falciparum, due to mutations in the Kelch13 (K13) protein, threaten these advances. Artemisinin exposure results in a dynamic transcriptional response across multiple pathways, but most work has focused on ring stages and ex vivo transcriptional analysis. We applied single cell RNAseq to two unsynchronized coisogenic parasite lines (K13C580 and K13580Y) over 6 hrs after a pulse exposure to dihydroartemisinin (DHA). Transcription was altered across all stages, with the greatest occurring at the trophozoite and ring stage in both lines. This response involved the arrest of metabolic processes, support for a dormancy phenomenon upon treatment, and the enhancement of protein trafficking and the unfolded protein response. While similar, the response was consistent across stages in K13580Y, with enhanced parasite survival to drug induced stress. Increased surface protein expression was seen in K13580Y parasites at baseline and upon drug exposure, highlighted by the increased expression of PfEMP1 and GARP, a potential therapeutic target. Antibody targeting GARP maintained anti-parasitic efficacy in K13580Y parasites. This work provides single cell insight of gene transcription across all life cycle stages revealing transcriptional changes that could initiate a dormancy state and mediate survival upon treatment.
Keywords: Plasmodium falciparum, drug resistance, artemisinin, dihydroartemisinin, Kelch13, Single-cell, RNA-seq, Transcriptome
INTRODUCTION
Malaria remains one of the most common infectious diseases in the world and one of the greatest global health concerns, with an estimated 247 million reported malaria cases and 619,000 malaria-related deaths in 2022 of which 76% were in children under five years (1). Plasmodium falciparum (Pf) accounts for 99.7% of estimated malaria cases and virtually all the deaths in Africa (1). The World Health Organization recommends artemisinin-based combination therapies (ACTs) for the treatment of uncomplicated falciparum malaria. These combinations rely on the fast-acting properties of the artemisinin derivative to rapidly clear parasites with the longer-acting partner drugs clearing residual parasites, reducing the frequent observation of recrudescent infections when artemisinin drugs are used alone (2). Effective treatment and management of P. falciparum is now endangered by emerging partial resistance to artemisinin with multiple validated mutations in Africa, primarily in Rwanda, Uganda, Ethiopia, and Tanzania (3–9). The emergence of artemisinin partial resistance may precede development of partner drug resistance, jeopardizing the considerable gains made against malaria over the past decade. Given the vast majority of disease burden is in Africa, a public health disaster could arise if artemisinin partial resistance spreads across the continent (10, 11).
Resistance to artemisinin (ART) derivatives first emerged in western Cambodia (12). Clinical artemisinin partial resistance (ART-R) is expressed as a reduced rate of parasite clearance (>5 hr parasite half-life), residual parasitemia 3 days after treatment, or increase rate of recrudescence (13). This resistance biologically manifests as a decreased susceptibility of ring-stage parasites to ART and is highly attributed to nonsynonymous mutations in the ß-propeller domain of the Kelch13 (K13) protein (7, 14, 15). The identification of K13 (PF3D7_1343700) gene mutations as a marker of resistance has provided the impetus for a comprehensive series of studies that track and analyze the distribution of K13 mutations in malaria-endemic regions (7, 16). Over 100 different K13 mutations have been identified (17), but thirteen are considered validated resistance markers and 10 are considered candidate resistance markers (18). The K13580Y mutation is the most dominant in Southeast Asia (19–21). In Eastern Africa, the mutations K13561H,675V,469Y,622I have emerged and are now found in multiple countries (22). The role of K13 mutations in conferring artemisinin partial resistance has been investigated through selection experiments and gene editing (15, 23). While the degree of resistance is modulated by the genetic background (24), K13 mutants exhibit about tenfold higher residual viability than their genetically matched wild-type parasites when exposed to dihydroartemisinin (DHA) at the early ring stage (15), sparking interest in K13’s role in artemisinin treatment response.
K13 protein function remains to be fully understood. It consists of 726 amino acids and exhibits sequence similarity to a class of Kelch/BTB/POZ ubiquitination adaptors (14, 25). K13 protein functions within the endocytosis pathway required for hemoglobin uptake from the host cell (23), which is critical for parasite survival and activation of the endoperoxide bridge of artemisinin (26). Resistant K13 mutations in the propeller are thought to partially reduce normal function, which in turn is thought to diminish hemoglobin endocytosis. Reduced hemoglobin uptake limits the availability of hemoglobin-derived Fe2+-heme necessary for drug activation to kill the rings via alkylation of proximal proteins, leading to reduced ART killing (25, 26).
In this study, we explored the single cell transcriptomic profile of K13580Y (CamWT_C580Y, MRA-1251) and K13C580 (CamWT, MRA-1250) P. falciparum coisogenic lines following DHA exposure using a well based single cell platform, SeqWell (27, 28). The well based single cell platform Smart-seq2 has successfully been used in the past to study Plasmodium berghei, allowing for detection of cells across the complete life cycle, including the extracellular merozoite stage (29). Single cell transcriptomic profile enabled us to measure transcriptional effects of DHA treatment effect on malaria cell cycle stages. We observed parasite stage specific gene expression profiles and also potential vulnerabilities of the parasite’s response to DHA treatment, such as elevated GARP surface protein expression, that can be a logical therapeutic target for effective combination therapies in the face of ART-R.
RESULTS
Stage-specific differential expression between untreated K13580Y and K13C580.
To study the transcriptomic profiles of P. falciparum cells in response to DHA exposure, we isolated coisogenic non-synchronized K13C580 and K13580Y parasites after 48hrs in culture and profiled the single cell transcriptome at different timepoints of treatment with DHA (Figure 1 and 2). The transcriptional differences between K13580Y and K13C580 were determined in the absence of the drug by comparing the gene expression profile of the K13580Y untreated control to K13C580 untreated control (Figure 3). The genetic difference between K13580Y and K13C580 was verified using molecular inversion probes targeting multiple drug-resistant markers (Supplemental Methods, Supplementary Data 1). The coisogenic parasites’ identities were consistent with previous genotyping, only differing in K13 (Figure 2C). K13 gene expression was observed to be highly expressed in the ring stages of both the K13C580 and K13580Y parasites compared to all the other stages (Figure 3). Despite the fact that the K13580Y mutation is not known to affect transcription efficiency of the gene, K13 gene expression was significantly lower (log2FC=−0.92, p-value=2.13e-38) in the K13580Y compared to K13C580 (Figure 3, Supplemental Table S1). This lower K13 expression was observed in the rings, and merozoite stages of K13580Y (Figure 3). With the K13580Y mutation partially affecting hemoglobin uptake, elevated expression of PF3D7_1121600 (EXP1) has been linked to the initiation of the compensatory process involved in minimizing the fitness costs associated with the partial loss of function associated with the K13 mutation (30). We observed elevated expression of EXP1 (log2FC=1.25, p-value=6.5E-136) in the K13580Y relative to K13C580 rings.
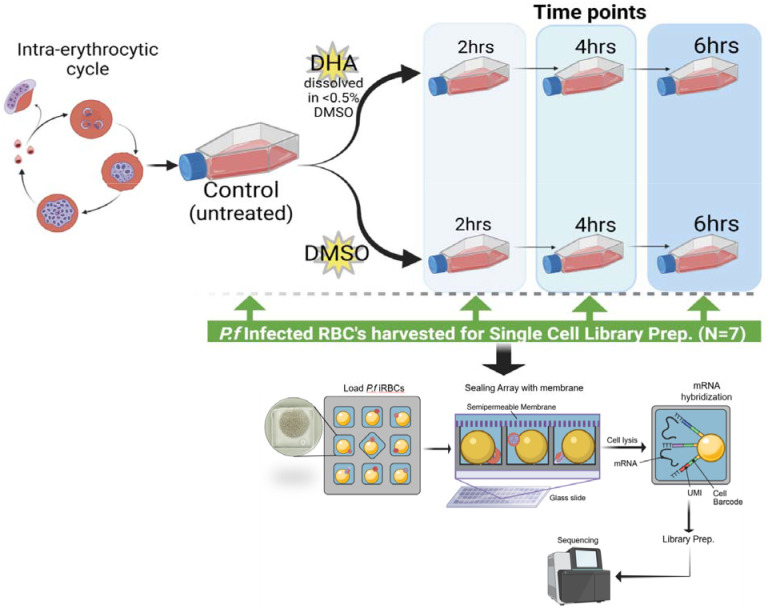
Figure 1: Schema of the experimental workflow applied in this study to explore transcriptome changes upon DHA exposure.
This schematic diagram illustrates the study design, points of sample collection and treatment, sample processing and single cell library preparation using the SeqWell platform. Both the wildtype (K13C580 strain) and mutant (K13580Y mutant strain) Pf parasite cell lines were cultured separately for 48hrs. The control (untreated) parasite was collected for library prep. The controls for each parasite strain were then divided into two groups, some treated with 700 nM DHA (lethal dose of DHA) and DMSO. After DHA and DMSO treatment or exposure, samples were collected after every 2 hrs for single cell library preparation using SeqWell.
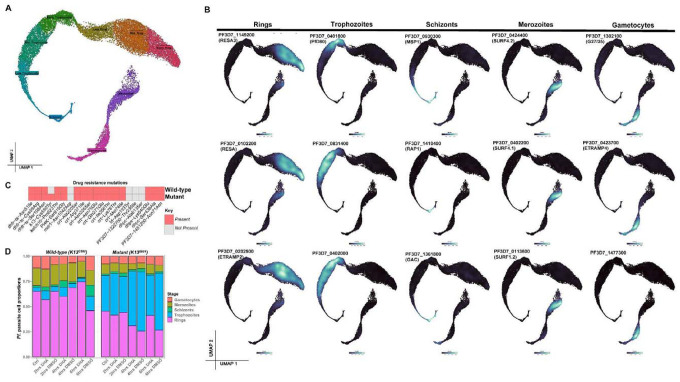
Figure 2: Single-cell gene expression identifies distinct life-cycle stages.
A. UMAP of single-cell transcriptomes sampled from all stages of the life cycle, with cells colored according to their stage. B. UMAP representations displaying the expression of parasite stage specific gene expression markers. With this single-cell gene expression profile of the Pf parasite, we can identify new unknown stage specific parasite marker genes. C. Presence and absence of known drug resistant mutations between the two Cambodian parasite strains (mutant and wildtype). We validated that the only variant difference between the two strains used in this study was the K13C580Y mutation is only present in the mutant strain. D. Distribution of parasite developmental stages across all sample groups (untreated controls, DHA and DMSO treated parasites).
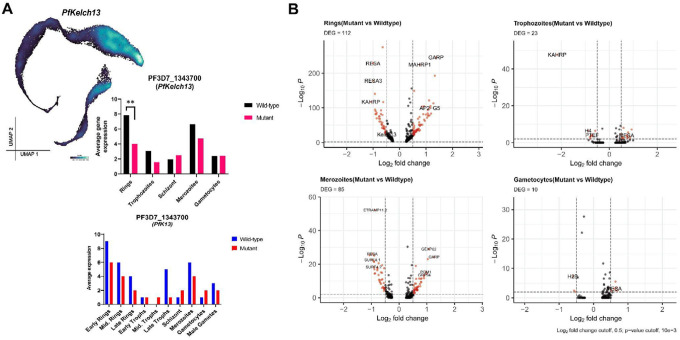
Figure 3: Gene expression differences between K13580Y and K13C580 parasites.
A. Kelch13 gene expression was observed to be downregulated in the K13580Y parasite population compared to the K13C580 (BH-adjusted p-value<0.01 and log2FC=1.5). PfKelch13 gene expression across the parasite stages in each line, shows PfKelch13 gene to be highly expressed in the K13C580 across all the stages compared to K13580Y. B. Volcano plot showing DEGs in the untreated K13580Y compared to untreated K13C580 on different parasite stages. Each red dot denotes genes differentially expressed between the K13C580 and the K13580Y (such as GARP and MAHRP1 which are upregulated in K13580Y rings compared to K13C580 rings).
Impact of DHA exposure on K13C580 parasites.
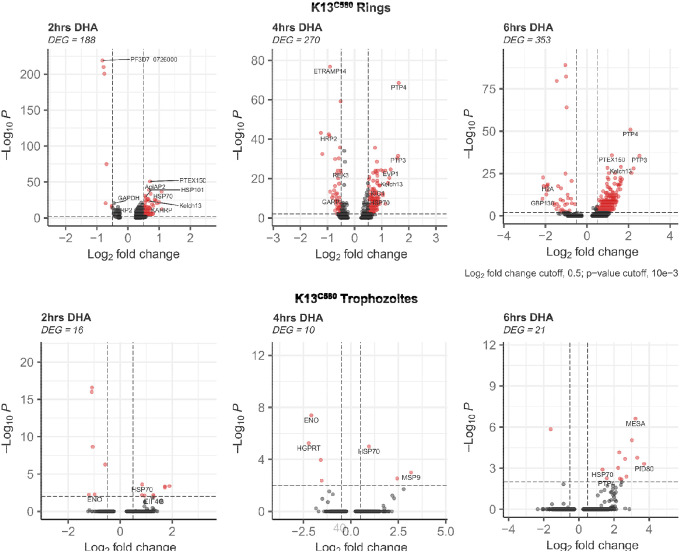
In K13C580 parasites, the transcriptional response to DHA dramatically differed based on parasite stages relative to DMSO (drug vehicle) treated controls. While we observed relative gene expression changes across all the erythrocytic stages of development (Supplementary Table S2), the ring stage had the greatest gene expression changes with 188, 270, and 353 genes identified as differentially expressed (DE) (p-value<0.05 and log2FC>0.4) at 2hrs, 4hrs and 6hrs after pulsed DHA exposure, respectively. The genes DE and downregulated across all the treatment timepoints in the rings were categorically associated with metabolic processes, such as glycolysis (GAPDH) and ATP generation (Supplemental Table S2), consistent with an arrest of parasite growth, as previously reported within 18h of DHA exposure (31). We observed the upregulation of stress-response genes such as HSP70, HSP90, HSP101, and NOT4 across all the treatment timepoints (Figure 4). We also observed the elevated expression of genes such as PTP3, PTP4, REX1, and MAHRP1, which are involved in protein trafficking and export within the parasite (Figure 4, Supplemental Table S2). The K13 gene and Kelch13 interaction candidate (KIC) genes (such as KIC1 and KIC5), known to code for proteins that associate with K13, were observed to be upregulated upon DHA treatment, impling that hemoglobin endocytosis is probably enhanced as a stress response and as a result enhancing drug potency within the parasite.
Figure 4: Gene expression differences associated with K13C580 (Rings and Trophozoites) DHA response.
Volcano plot showing DEGs in K13C580 DHA treated versus K13C580 DMSO treated (control) at 3 different timepoints. Each red dot denotes genes passing p-value and fold change difference (BH-adjusted p-value<0.01, average log2 fold-change>0.3) thresholds. Those on the right side of the plots are the upregulated genes induced due to DHA exposure. The red dots on the left side of the plots represent the downregulated genes passing the p-value and fold change difference between the DHA treated parasite and the DMSO treated parasite control (BH-adjusted p-value<0.05, average log2 fold-change<−0.3).
Since DHA is mainly known to impact phenotypic changes and parasite survival mainly in the ring and trophozoite stages (32), we also explored gene expression changes in the trophozoite stages of K13C580 at all timepoints, but we observed very few genes DE. We did observe some expression changes in the merozoite, schizont and gametocyte stages of K13C580 upon drug exposure (Supplemental Table S2 and Supplemental Figure S2).
Impact of DHA exposure on K13580Y parasites.
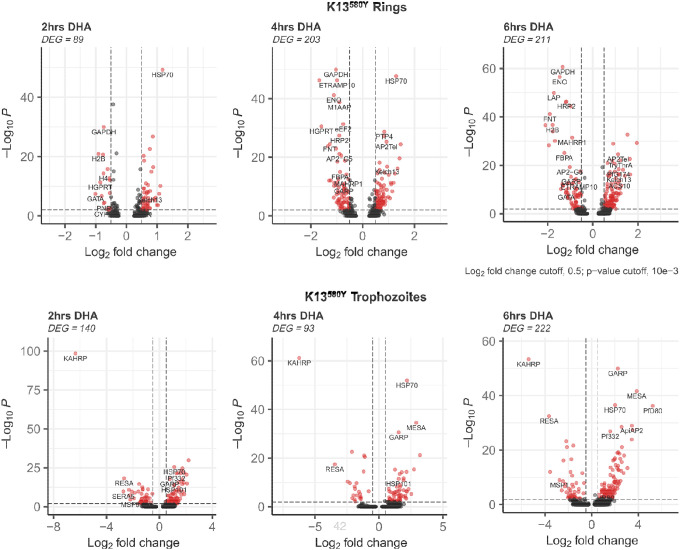
Examining K13580Y after DHA treatment, we observed greater gene expression changes in the ring and trophozoite stages compared to all the other erythrocytic stages. We identified 99, 203, and 211 genes to be DE in the ring stage in response to 2hrs, 4hrs and 6hrs DHA exposure, respectively. In the K13580Y ring stages, downregulated genes included those linked to glycolysis (GAPDH) and ATP generation, suggesting a developmental arrest similar to K13C580. Across all timepoints of drug exposure, we observed the downregulation of the GARP gene in the ring stage of the K13580Y strain (Supplemental Table S3 and Figure 5), but an upregulation of GARP in the trophozoite stage. We observed elevated expression of heat shock proteins (HSP70, HSP90 and HSP101), upregulation of genes associated with protein trafficking (PTP3, PTP4, REX1, and MAHRP1) and also upregulation of K13 and KIC genes (such as KIC1, KIC5) (Figure 5 and Supplemental Table S3). These findings imply that the gene expression response initiated by DHA on K13580Y involved the downregulation of metabolic process, upregulation of the unfolded protein response, elevated hemoglobin uptake and increased protein trafficking, which is a similar response initiated by the ring stage of K13C580, but a more enhanced cytoprotective response.
Figure 5: Gene expression differences associated with K13580Y (Rings and Trophozoites) response to DHA treatment in vitro.
Volcano plot showing DEGs in K13580Y mutant treated with DHA versus K13580Y treated with DMSO (control) at 3 different timepoints. Each red dot denotes genes passing p-value and fold change difference (BH-adjusted p-value<0.05, average log2 fold-change>0.3) thresholds. Those on the right are the upregulated genes induced due to DHA exposure. The red dots on the left represent the downregulated genes passing the p-value and fold change difference thresholds between the DHA treated parasite and the DMSO treated parasite control (BH-adjusted p-value<0.01, average log2 fold-change<−0.3).
The trophozoite stage of the K13580Y strain showed many gene expression changes upon drug exposure. We observed 140, 93, and 222 genes to be DE in the trophozoite stage of K13580Y in response to 2hrs, 4hrs and 6hrs DHA exposure, respectively. Within the trophozoite stage, we observed the upregulation of PTP3, PTP4, EXP1 (protein trafficking), HSP70, HSP90, HSP101 (unfolded protein response), ApiAP2 and, contrary to the rings stage, observed the upregulation of GARP across all timepoints of DHA exposure. The gene expression changes in the trophozoite also appear to be associated with the restriction of metabolic processes, initiation of the unfolded protein response, and elevated protein trafficking upon exposure to 700nM DHA over a 6hr period. We also did observe some expression changes in the K13580Y strain merozoite, schizont and gametocyte stages upon drug exposure primarily after 4 to 6hrs of DHA exposure (Supplemental Table S3).
K13580Y and K13C580 gene expression differences after treatment.
Upon DHA treatment, the transcriptional response appeared to be similar for both K13580Y and K13C580 parasite lines. Both K13580Y and K13C580 ring stages upregulated genes associated with unfolded protein response (UPR) and protein trafficking upon drug exposure, while genes associated with metabolic processes were downregulated (Supplemental Table S4). However, there were are some differences observed between these coisogenic lines upon DHA exposure, genes within the SERA gene family (PF3D7_0207600 (SERA5), PF3D7_0207500 (SERA6)) had significant decreased expression (log2FC=−1.9,s, adj-p-value = 4.54E-06) in K13580Y compared to K13C580 trophozoites (Figure 6 and Supplementary Table S4). Given SERA family genes are thought to be important in maintaining the erythrocytic cycle growth (33–35), downregulation of these genes in K13580Y compared to DHA treated K13C580 could contribute to the initiation of parasite dormancy through suppression of parasite development (Supplemental Table S3).
Figure 6: Gene expression differences between DHA treated K13580Y and K13C580 parasites.
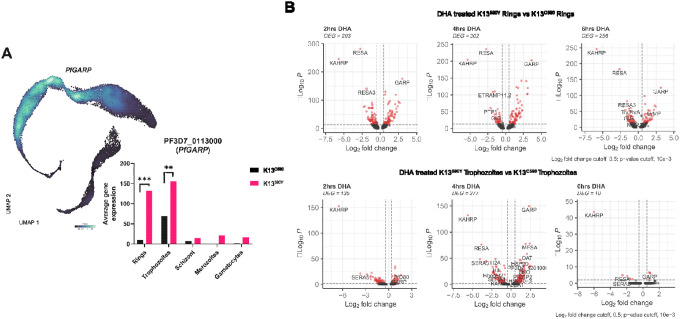
A. PfGARP was observed to be highly expressed in the K13580Y parasite population compared to the K13C580 parasite population (BH-adjusted p-value<0.01 and log2FC=3.8). Stratifying the PfGARP expression across the parasite stages in each strain shows PfGARP to be highly expressed in K13580Y across all the stages compared to K13C580. B. Volcano plot showing DEGs in K13580Y treated with DHA versus K13C580 treated with DHA at 3 different timepoints. Each red dot denotes genes passing p-value and fold change difference (BH-adjusted p-value<0.001, average log2 fold-change>0.3) thresholds. The dots on the right are the upregulated genes induced due to DHA exposure. The red dots on the left represent the downregulated genes passing the p-value and fold change difference thresholds between the DHA treated parasite and the DMSO treated parasite control (BH-adjusted p-value<0.001, average log2 fold-change<−0.3).
Elevated PfGARP expression in K13580Y.
GARP has recently been identified as a promising therapeutic target based on the impact of anti-GARP antibodies shown to trigger parasite death (36). The exact role and function of GARP still remains to be elucidated, however it has been reported to be restrictively expressed in the early trophozoite stage (37). In this study, we show that the PfGARP gene is expressed in more stages of the parasite’s life cycle, unlike previously reported (Figure 6). However, the expression level of this gene is variable across the stages, with higher expression between the late ring to the mid trophozoite stages (Figure 6A). We also observed variable GARP expression between the coisogenic lines used in the study. The K13580Y exhibited significantly higher GARP gene expression compared to K13C580 (log2FC=3.49, adj-p-value<0.00001) (Figure 6A and 6B). Upon DHA treatment of both K13C580 and K13580Y, we observed that GARP expression was significantly increased in the trophozoite stage (Figure 6B). These findings and observations imply that GARP may be involved in the parasite’s response to stress/drug exposure, and in this case the response seems to be more pronounced in the K13580Y parasite.
Fine grained analysis between K13580Y vs. K13C580 allows for quantification of treatment perturbation across the malaria asexual lifecycle
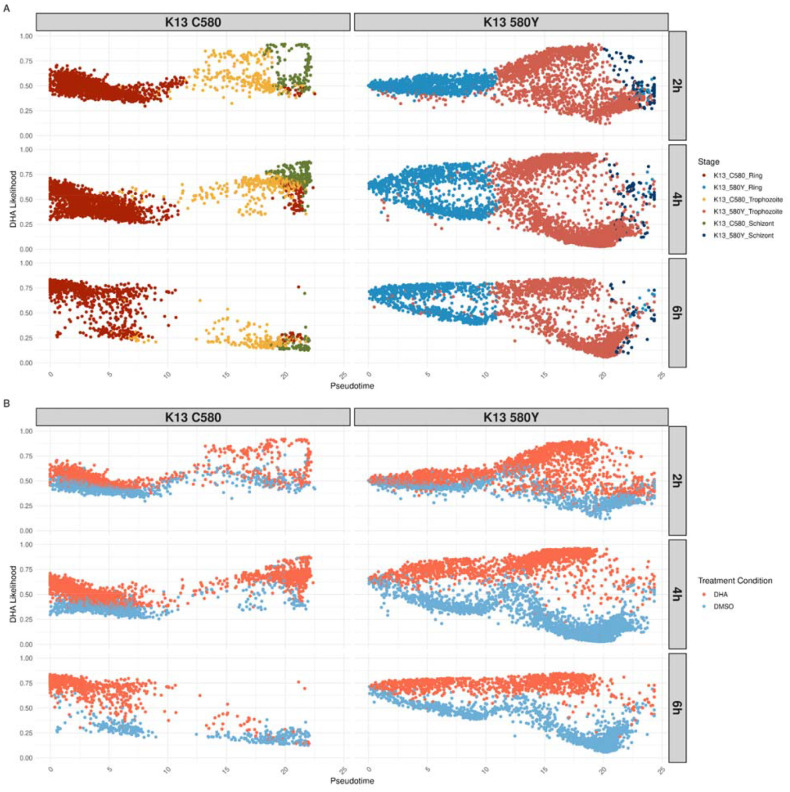
To quantify DHA perturbation across the malaria cell cycle stages, the MELD algorithm was utilized (see Supplemental Methods) (38). The output was a continuous measure, where a cell’s likelihood score that approaches 1 indicates a cell’s transcriptional profile is more similar to the DHA treatment condition, called the DHA likelihood. DHA likelihoods are scores ranging from 0 to 1, i.e. from less to more perturbed by DHA treatment. Pseudotime analysis was performed with slingshot (39), allowing visualization of the distribution of DHA likelihoods across the lifecycle, within each timepoint and strain (Supplemental Figure S3). Both lines show similar responses to DHA treatment, where there are separate transcriptional states based on treatment condition across the lifecycle (Figure 7A). As expected, DHA treated cells have a higher DHA likelihood than DMSO treated cells (Figure 7B). In K13C580, DHA likelihoods of the DHA treatment condition increased over the asexual lifecycle at the first two timepoints (2hrs and 4hrs) and remained elevated in the ring stages at 6hrs. In K13580Y, the same pattern was present, but with a greater representation of all the asexual stages across timepoints and higher levels of DHA likelihood values in the treatment condition.
Figure 7. Overall DHA transcriptional response trend is similar, but persistently elevated across the asexual cycle in the K13580Y strain.
Pseudotime, calculated per strain through slingshot (39), is plotted on the x-axis while the DHA likelihood output from MELD is plotted on the y-axis for each facet of the figure. Cells are colored by stage annotation (A) or treatment condition (B). The left column plots K13C580 cells while the right column plots K13580Y cells. Each row is a different timepoint, with the first row being the 2 hour timepoint, middle row being the 4 hour timepoint and the last row being the 6 hour timepoint.
When focusing on the ring and trophozoite stages, we isolated cell populations with the highest DHA likelihood within each timepoint using vertex frequency clustering (VFC) (38). We separated these highly enriched clusters of high DHA likelihood cells (Supplemental Figures S4 and S5) for fine grained differential expression analysis in rings and trophozoites.
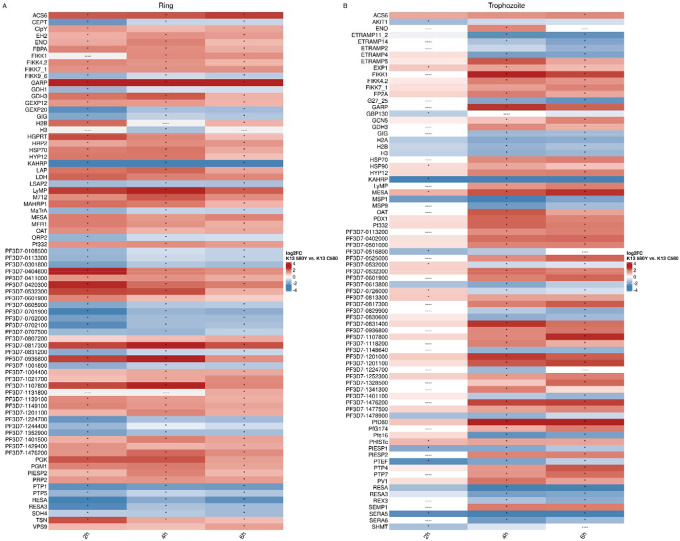
We compared the expression between the clusters with the highest DHA likelihood between K13580Y and K13C580 in the ring and trophozoite stages (Figure 8, Supplemental Figure S6). Rings and trophozoites in K13580Y showed consistent upregulation of GARP and downregulation of Knob Associated Histidine Rich Protein (KAHRP, PF3D7–0202000) and Ring Infected Erythrocyte Surface Antigen (RESA, PF3D7–0102200) compared to K13C580. To investigate differential gene pathway expression, gene ontology (GO) analysis was performed using the differentially expressed genes in K13580Y vs. K13C580 comparison (Supplemental Figure S7). In the ring stage, GO terms related to metabolic processes and translation were enriched in upregulated genes. In the trophozoite stage, GO terms related to nuclear regulation were enriched in downregulated genes in timepoints 4 hours and 6 hours.
Figure 8. Differential expression results show upregulation of GARP and stress response genes (HSP70, GCN5), perturbations in genes related to protein export (FIKK gene family, EXP1, ETRAMP genes, KAHRP, RESA), and downregulation of genes related to dormancy (SERA5, SERA6).
The union of the top50 differentially expressed genes (by absolute log2FC) per time point between the highest VFC subcluster in K13C580 compared with K13580Y in rings (A) and trophozoites (B). For each timepoint in (A) or (B), the top 50 genes by absolute log2FC were identified. The vectors of the top 50 genes were concatenated and filtered to unique genes. The average log2FC per timepoint is plotted, with “----” indicating genes that were not differentially expressed at that timepoint. An asterisk indicates genes that were significantly differentially expressed at an absolute log2FC > 0.3 and a BH-adjusted p-value of < 0.001.
Anti-GARP kills K13580Y equivalent to K13C580 in vitro.
GARP was elevated upon DHA treatment of K13580Y compared to K13C580. In order to evaluate if this elevated expression resulted in a functional or aberrant accumulation of the protein in the membrane, we performed growth-inhibition assays (GIAs) using anti-GARP-A antiserum that was prepared by immunizing mice with recombinant protein, as previously performed by Raj et al. 2020 (36), using the 3D7 strain as a positive control for anti-GARP-A inducted killing. We did not see a statistically significant difference in the rate of growth inhibition between K13580Y and K13C580 (Figure 9, Supplemental Figure S9) consistent with functionality of the pathway in both lines.
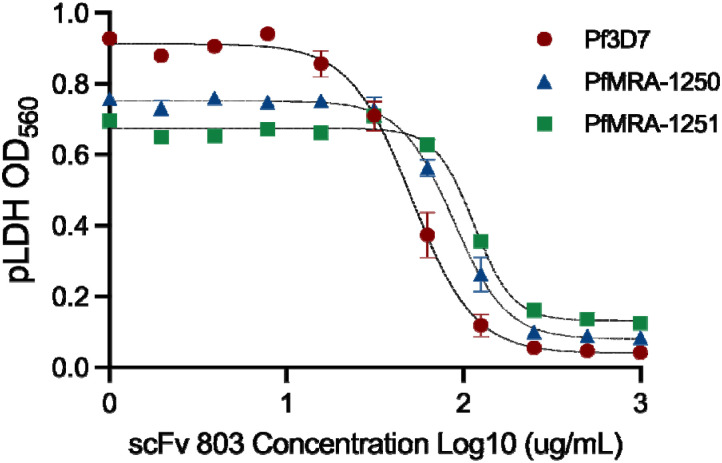
Figure 9: Anti-PfGARP mAb clone 803 kills both K13580Y and K13C580 parasites in vitro.
Ring-stage P. falciparum parasite strains Pf3D7, K13C580, and K13580Y were incubated for 72 hours with serially diluted anti-PfGARP scFv antibody clone 803 or media only. Parasitemia was evaluated by pLDH assay. Antibody treatment resulted in significant parasite growth inhibition. Data was fit to a four-parameter logistic regression model to obtain the following IC50 values: Pf3D7= 50μg/mL, K13C580= 87μg/mL, and K13580Y= 114μg/mL. Data was fit to a four-parameter logistic regression model to obtain the following IC50 values: Pf3D7= 50μg/mL, K13C580= 87μg/mL, and K13580Y= 114μg/mL. Data was fit to a four-parameter logistic regression model to obtain the following IC50 values: Pf3D7= 50μg/mL, K13C580= 87μg/mL, and K13580Y= 114μg/mL. Data points represent the mean of two (K13580Y) or three (Pf3D7 and K13C580) biologically independent replicates, with error bars indicating standard error of the mean (SEM).
DISCUSSION
To date, Plasmodium falciparum (Pf) gene expression changes triggered by drug treatment have focused on bulk gene expression data generated on synchronized parasites or from field isolates (13, 24, 31, 40). The utility of scRNAseq in P. falciparum to explore gene usage in K13580Y upon treatment, in the presence of the other stages in intra-erythrocytic stages development, has not been assessed previously. Here, we comprehensively surveyed the molecular behavior of P. falciparum K13C580 and K13580Y parasites upon artemisinin exposure, analyzing single cell transcriptomic changes across the erythrocyte life cycle stages of the coisogenic lines during a 6hr 700nM DHA treatment. Through treating asynchronized parasites to DHA exposure, specific subpopulations that are differentially affected by the perturbation can be isolated and studied. These populations were isolated through utilizing MELD and vertex frequency clustering, with similar results to traditional differential expression analysis (see Figures 6, 8 and Supplemental Figure S7). Our analyses corroborate findings from previous transcriptome studies on P. falciparum response to treatment (31) and further provides new insight into gene usage in response to drug exposure in both K13C580 and K13580Y.
When comparing K13580Y and K13C580 parasites, we found interesting differences in gene expression. GCN5 has recently been found to regulate stress response genes after artemisinin treatment (41) and this gene was upregulated in K13580Y trophozoites at all timepoints compared to K13C580 trophozoites (Figure 8). Additionally, FIKK1 and FIKK4.2 genes were upregulated in K13580Y rings and trophozoites compared to K13C580, genes that were both part of the “artemisinin resistance-associated transcriptional profile“ found when analyzing isolates from the Greater Mekong Subregion (GMS) (42). Downregulation of the SERA5 and 6 genes in K13580Y vs. K13C580 upon DHA exposure highlights some of the changes that could lead to parasite quiescence due to slowed development (Figure 8). We also revealed an increased expression of transport associated genes in K13580Y vs. K13C580 when exposed to DHA as well as a striking elevated expression of GARP, a potential P. falciparum therapeutic target (36), in the trophozoite stage of K13580Y upon DHA exposure, making it a viable therapeutic target candidate against the emergence of ART-R as a result of K13580Y mutation.
Our transcriptomic analysis revealed that untreated K13580Y had lower K13 gene expression compared to untreated K13C580. K13 is thought to aid hemoglobin endocytosis into the parasite, to be broken down as a source of amino acids to facilitate the assembly of parasite proteins and support the life of the parasite. The presence of the K13580Y mutation is associated with a slow rate of progression of the parasite from the ring to trophozoite to schizont stage compared to K13C580 (Figure 2C). This deceleration in development in the blood-stage is thought to be as a result of impaired hemoglobin uptake, which is necessary for parasite protein synthesis (23, 43). The K13 function of facilitating hemoglobin entry has been shown to be aided by a complex of other proteins, known as the K13 compartment proteins (KICs) (23). During a stress response, P. falciparum has been shown to elevate K13 gene and protein expression to facilitate hemoglobin entry, thereby availing an amino acid source which will be used to synthesize proteins necessary to control the stress response (31). Parasites exposed to DHA treatment did exhibit elevated expression of K13 and genes coding for the KIC’s mainly at the ring stages of both K13C580 and K13580Y. However, in K13580Y, K13 gene expression was lower in most of the parasite stages expressing the gene compared to K13C580 (Figure 6), despite some of the KIC genes being elevated upon DHA exposure (Supplementary Table 2 and 3). This would suggest that altered protein expression of K13 downstream of transcription likely alters parasite DHA sensitivity in K13580Y(23).
Artemisinin and its derivatives are known to exert their antimalarial activity by generating carbon-centered radicals that cause oxidative stress and subsequent protein alkylation (25, 44). Accumulation of the alkylated proteins increases proteotoxic stress within the parasites, causing a phenotype reminiscent of that induced by heat shock (45, 46), hence the observed upregulation of HSP70 and HSP90 in both K13580Y and K13C580 upon DHA treatment. This finding, together with the elevated expression of genes associated with protein synthesis (FIKK family of genes, HSP40) and transcriptional activation of genes (ApiAP2), implies the induction of the UPR in both parasite lines upon DHA exposure. We also observed a relatively high number of genes coding for export and trafficking proteins to be upregulated in the rings and trophozoites of both K13580Y and K13C580 upon DHA exposure, which could be as a result of ER stress response (47). Our findings show that both K13580Y and K13C580 exhibit a similar gene expression response to DHA exposure. However, K13580Y exhibits enhanced expression of UPR associated genes compared to K13C580, increasing the capacity of K13580Y to quickly repair or degrade proteins damaged by artemisinin. This capacity could minimize the damaging effects caused by the accumulation of DHA in K13580Y parasites. This elevated UPR in K13580Y, coupled with having lower levels of hemoglobin endocytosis and decreasing free heme-mediated artemisinin activation, contributes to the tolerance of K13580Y to artemisinin (48), leading to resistance/delayed killing of K13580Y.
GARP was recently identified as a therapeutic target (36) and previous studies have shown that the expression of GARP is highly restricted to the early trophozoite stage of the parasite’s life cycle; however, we show that the gene is also expressed in the late ring stages and trophozoites (Figure 6). GARP shares no homology with proteins of known function, it contains several complex repeat regions, and its function remains to be characterized. We observed elevated expression of GARP in K13580Y compared to K13C580 before treatment, which could imply it may be as a result of compensatory processes initiated by the parasite due to the K13 mutation. While not as elevated, other surface protein transcripts show upregulation suggesting a possible general effect. Given gene expression changes can correlate with function or represent dysregulation (e.g. increased turnover), we examined whether GARP still retained function as a therapeutic target, finding equivalent anti-GARP antibody killing of parasites between K13580Y and K13C580 (Figure 9). This suggests GARP and likely other surface proteins maintain normal function and suggests anti-GARP therapy can be a potential addition to our limited armamentarium as an effective therapy in the face of K13 artemisinin partial resistance mutations.
Conclusion:
The cellular response of K13580Y and K13C580 is similar. Both exhibit an arrest in metabolic functions, which is accompanied by elevated expression of genes associated with response to unfolded proteins and protein trafficking. However, K13580Y mediated consistent DHA perturbation throughout the asexual lifecycle and enhanced expression of UPR genes compared to K13C580. The elevated expression of GARP expression in both the K13580Y and K13C580 reveals a potential therapeutic intervention, regardless of K13 genotype. Future studies need to explore the gene expression changes in response to DHA treatment on other K13 mutant parasites coupled with protein expression of the differentially expressed gene. This observed ART response in different genetic backgrounds needs to be explored at the single cell level in field isolates of P. falciparum to find genetic dependencies of in vivo ART susceptibility and resistance.
METHODS
Parasites lines and culture.
The strains of parasite lines used in this study were MRA-1250 CamWT, an artemisinin susceptible fast-clearing Cambodian P. falciparum isolate, and MRA-1251 CamWT_C580Y, a Cambodian P. falciparum isolate which features a single nucleotide substitution leading to a C580Y amino acid change (MR4, BEI Resources, Manassas, VA). The P. falciparum parasites were maintained at 3% hematocrit in human O+ RBCs and P. falciparum culture media comprising RPMI 1640 (Thermo Fisher Scientific) supplemented with 0.5% (w/v) Albumax II, 50 mg/L hypoxanthine, 0.225% NaHCO3, 25mM HEPES, and 10 mg/L gentamycin. Parasites were cultured at 37 °C in 5% O2, 5% CO2, and 90% N2 (49). The parasite lines were cultured for 48hrs before we began exposing them to DHA and DMSO.
Drug susceptibility assays.
Drug stocks of dihydroartemisinin (DHA) were made in dimethyl sulfoxide (DMSO) and aliquots were stored at −20 °C. All drug assays were conducted such that the final DMSO concentration was <0.5%. Each parasite line was exposed to 700 nM DHA along with vehicle-treated 0.05% DMSO controls in independent experiments for 6hrs (Figure 1). Samples from each treated parasite line were collected at 2hr intervals for single cell library preparation and sequencing.
Enrichment of parasite infected RBCs.
To prepare the single cell RNAseq libraries, we needed to enrich the parasite infected RBCs thus increasing chances of loading and capturing mRNA transcripts from the parasites. Our unsynchronized parasite cultures were enriched using SLOPE as previously described (50, 51) with some modifications. Briefly, erythrocyte density was measured using a Cellometer Auto T4 (Nexcelom Biosciences, Lawrence, MA) and cell density adjusted to 2×109 erythrocytes/mL. The 22.5Units of the lytic agent streptolysin-O (SLO) per 50μl RBCs was added in a ratio of 2 parts SLO solution to 5 parts erythrocytes. Samples were mixed well by pipetting and incubated at room temperature for 6 min. 10 volumes of 1X PBS or media (RPMI 1640 HEPES) were added and cells were centrifuged at 2,500xg for 3 min. After removal of the supernatant, cells were washed twice more with 1X PBS. Following SLO lysis, cells suspended in 1X PBS were layered onto a 60% Percoll gradient (Sigma Aldrich, St Louis, MO) and centrifuged at 1,500xg for 10 min. The top layer of Percoll was discarded while the lower cell pellet, rich in infected erythrocytes, was collected and washed twice with 1xPBS and later suspended in 200μl RPMI to be loaded into the SeqWell arrays.
Single-Cell library preparation and sequencing.
To capture the transcriptome of Plasmodium single cells and generate single cell libraries, we utilized SeqWell, a massively parallel, low-input scRNA-seq platform (27, 28). Briefly, 10–15,000 Pf infected RBCs from each time point and treatment condition were loaded onto a functionalized polydimethylsiloxane (PDMS) array preloaded with uniquely barcoded mRNA capture beads (Chemgenes; Macosko-2011–10(V+), Wilmington, MA). After cells had settled into wells, the arrays were then sealed with a hydroxylated polycarbonate membrane with a pore size of 10 nm, facilitating buffer exchange while confining biological molecules (such as DNA and RNA) within each well. Following membrane-sealing, subsequent buffer exchange permitted cell lysis, mRNA transcript hybridization to beads, and bead removal before proceeding with reverse transcription. The obtained bead-bound cDNA product then underwent Exonuclease I treatment to remove excess primer before proceeding with second-strand synthesis, then PCR amplification of the cDNA product. The cDNA products were purified using the SPRIselect beads (Beckman Coulter, Brea, CA), and were run on a Fragment analyzer (Agilent Technologies, Santa Clara, CA) to verify the cDNA fragment sizes to be between 0.7–2kbps. The cDNA libraries were then fragmented and amplified for sequencing using the Illumina Nextera XT DNA Library Preparation kit (Illumina, FC-131–1096, San Diego, CA) using custom primers that enabled the specific amplification of only the 3’ ends. The libraries were purified, quantified, and then sequenced on a NextSeq550 system using High output 75 cycle kits (Illumina) at an average of 20,000 reads per cell. Raw read data and processed UMI count matrices from Pf MRA1250 and MRA1251 single cell experiments have been deposited and are publicly available through the SRA database (BioProject ID: PRJNA1049964).
Single-cell transcriptome analysis.
Raw reads were processed using version 2.3.0 of the Drop-seq pipeline (52), and according to the ‘Drop-seq Alignment Cookbook’, both found at http://mccarrolllab.com/dropseq/. Demultiplexed FASTQs were aligned to the P. falciparum 3D7 reference genome (PlasmoDB v.51) using STAR aligner. Individual reads were tagged with a 12bp barcode and 9bp unique molecular identifier (UMI) contained in Read 1 of each sequencing fragment. Following alignment, reads were grouped by the 12bp cell barcodes and subsequently collapsed by the 9bp UMI to generate a gene expression count matrix. Gene expression count matrices from Pf MRA1250 and Pf MRA1251 cell line single-cell experiments (n = 7 per Pf cell line) were analyzed and visualized in R (v.4.0.3) using the Seurat package (v.4.0.4) (53). Cells with less than 300 genes and 500 UMIs detected in less than ten cells were excluded from further analysis. We then proceeded to filter out 459 multigene family genes expressed such as vars (PfEMP1), rifins and stevors, retaining genes present across the different experimental conditions for each parasite line. Library size for each group in the study were internally normalized to 10,000 transcripts, log transformed and regressed on the number of UMIs per cell before dimensionality reduction and clustering. We integrated the datasets by utilizing the canonical correlation analysis (‘CCA’) in Seurat (54) to identify anchors/shared sources of variations between each treated and untreated groups to learn the parasite stage specific responses to DHA and DMSO treatment. Next, read data were scaled and analyzed via principal component analysis (PCA). The top PCs (n = 20) were dimensionally reduced via uniform manifold approximation (UMAP) (55) and unsupervised clustering was performed at several resolutions to identify parasite development stages and analyze differential gene expression.
A p-value cutoff of 0.05 was used to determine significance. For differential gene expression (DGE) between the treatment groups, a Bonferroni correction was applied to generate adjusted p-values. DGE analysis was done using the MAST package (56).
Details regarding fine-grained differential expression analysis utilizing MELD is in Supplemental Methods.
Growth inhibition assay on 3D7, K13C580 and K13580Y.
3D7, K13C580 and K13580Y cell lines were synchronized to the ring stage by treatment with 5% sorbitol for at least three successive replication cycles and cultured to the ring stage. Parasites were diluted with O+ RBCs to 0.5–0.75% parasitemia and 2% hematocrit in incomplete media (RPMI 1640 medium containing 25 mm HEPES and 50μg/mL hypoxanthine) supplemented with 10% AlbuMax II, 48mM sodium bicarbonate, and 20μg/mL gentamycin. Anti-PfGARP scFv clone 803 was adjusted to 2mg/mL in incomplete medium and serially diluted in 50μL total volume. 50μL of diluted parasites were added to each well, resulting in the following culture conditions: 0.5–0.75% parasitemia, 1% hematocrit in RPMI 1640 medium containing 25 mm HEPES, 50μg/mL hypoxanthine, 5% AlbuMax II, 24 mm sodium bicarbonate and 10 μg/mL gentamycin in 96-well round-bottom microtiter plates. After incubation for 72 hours, blood films and/or Plasmodium lactate dehydrogenase (pLDH) assays were performed to assess parasitemia levels. The half-maximum inhibitory concentration (IC50) for parasite growth was determined using a four-parameter logistic regression model.
Supplementary Material
IMPORTANCE.
Single cell RNA sequencing allows deconvolution of the cellular stages of malaria and investigation of their response to treatment conditions. Utilizing two different genetic backgrounds of a key resistance marker to artemisinin treatment, we compared the response to dihydroartemisinin between these genetic backgrounds. We found a distinct transcriptional profile post treatment in both genetic backgrounds, with downregulation of metabolic process genes and upregulation of stress response genes. Comparing these two genetic backgrounds post treatment using traditional differential expression, and a novel computational method called MELD, we found consistent increased expression of GARP and pathogenesis related genes, like PfEMP1. This study identifies possible gene dependencies of parasite survival post artemisinin treatment, providing targets for inhibiting the dormancy state.
Acknowledgments:
The following reagents were obtained through BEI Resources, NIAID, NIH: Plasmodium falciparum, strains CamWT, MRA-1250 and CamMT, MRA-1251, contributed by David A. Fidock; and Plasmodium falciparum, Strain 3D7, MRA-102, contributed by Daniel J. Carucci.
Funding:
This work was funded by the National Institute for Allergy and Infectious Diseases (K24AI134990 to JJJ; R01AI121558 to JJJ, and R01AI156267 to JAB and JJJ). Funds were received by the Falk Medical Research Trust Transformational Award (JK).
Funding Statement
This work was funded by the National Institute for Allergy and Infectious Diseases (K24AI134990 to JJJ; R01AI121558 to JJJ, and R01AI156267 to JAB and JJJ). Funds were received by the Falk Medical Research Trust Transformational Award (JK).
Data availability:
All sequence data has been submitted to SRA (PRJNA1049964). Code is available at https://github.com/sconnelly007/K13_mt_v_wt_sc.
REFERENCES
- 1.Programme GM. 2022. World malaria report 2022. World Health Organization. https://www.who.int/publications/i/item/9789240064898. Retrieved 14 November 2023. [Google Scholar]
- 2.Teuscher F, Gatton ML, Chen N, Peters J, Kyle DE, Cheng Q. 2010. Artemisinin induced dormancy in plasmodium falciparum: duration, recovery rates, and implications in treatment failure. J Infect Dis 202:1362–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rosenthal PJ. 2021. Has artemisinin resistance emerged in Africa? Lancet Infect Dis 21:1056–1057. [DOI] [PubMed] [Google Scholar]
- 4.Tacoli C, Gai PP, Bayingana C, Sifft K, Geus D, Ndoli J, Sendegeya A, Gahutu JB, Mockenhaupt FP. 2016. Artemisinin Resistance-Associated K13 Polymorphisms of Plasmodium falciparum in Southern Rwanda, 2010–2015. Am J Trop Med Hyg 95:1090–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nyunt MH, Soe MT, Myint HW, Oo HW, Aye MM, Han SS, Zaw NN, Cho C, Aung PZ, Kyaw KT, Aye TT, San NA, Ortega L, Thimasarn K, Bustos MDG, Galit S, Hoque MR, Ringwald P, Han E-T, Kyaw MP. 2017. Clinical and molecular surveillance of artemisinin resistant falciparum malaria in Myanmar (2009–2013). Malar J 16:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yasri S, Wiwanitkit V. 2021. Artemisinin resistance: an important emerging clinical problem in tropical medicine. Int J Physiol Pathophysiol Pharmacol 13:152–157. [PMC free article] [PubMed] [Google Scholar]
- 7.Ashley EA, Dhorda M, Fairhurst RM, Amaratunga C, Lim P, Suon S, Sreng S, Anderson JM, Mao S, Sam B, Sopha C, Chuor CM, Nguon C, Sovannaroth S, Pukrittayakamee S, Jittamala P, Chotivanich K, Chutasmit K, Suchatsoonthorn C, Runcharoen R, Hien TT, Thuy-Nhien NT, Thanh NV, Phu NH, Htut Y, Han K-T, Aye KH, Mokuolu OA, Olaosebikan RR, Folaranmi OO, Mayxay M, Khanthavong M, Hongvanthong B, Newton PN, Onyamboko MA, Fanello CI, Tshefu AK, Mishra N, Valecha N, Phyo AP, Nosten F, Yi P, Tripura R, Borrmann S, Bashraheil M, Peshu J, Faiz MA, Ghose A, Hossain MA, Samad R, Rahman MR, Hasan MM, Islam A, Miotto O, Amato R, MacInnis B, Stalker J, Kwiatkowski DP, Bozdech Z, Jeeyapant A, Cheah PY, Sakulthaew T, Chalk J, Intharabut B, Silamut K, Lee SJ, Vihokhern B, Kunasol C, Imwong M, Tarning J, Taylor WJ, Yeung S, Woodrow CJ, Flegg JA, Das D, Smith J, Venkatesan M, Plowe CV, Stepniewska K, Guerin PJ, Dondorp AM, Day NP, White NJ, Tracking Resistance to Artemisinin Collaboration (TRAC). 2014. Spread of artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 371:411–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fola AA, Feleke SM, Mohammed H, Brhane BG, Hennelly CM, Assefa A, Crudal RM, Reichert E, Juliano JJ, Cunningham J, Mamo H, Solomon H, Tasew G, Petros B, Parr JB, Bailey JA. 2023. Plasmodium falciparum resistant to artemisinin and diagnostics have emerged in Ethiopia. Nat Microbiol 8:1911–1919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Juliano JJ, Giesbrecht DJ, Simkin A, Fola AA, Lyimo BM, Pereus D, Bakari C, Madebe RA, Seth MD, Mandara CI, Popkin-Hall ZR, Moshi R, Mbwambo RB, Niaré K, MacInnis B, Francis F, Mbwambo D, Garimo I, Chacky F, Aaron S, Lusasi A, Molteni F, Njau R, Lazaro S, Mohamed A, Bailey JA, Ishengoma DS. 2023. Country wide surveillance reveals prevalent artemisinin partial resistance mutations with evidence for multiple origins and expansion of sulfadoxine-pyrimethamine resistance mutations in northwest Tanzania. bioRxiv. [Google Scholar]
- 10.Checchi F, Durand R, Balkan S, Vonhm BT, Kollie JZ, Biberson P, Baron E, Le Bras J, Guthmann JP. 2002. High Plasmodium falciparum resistance to chloroquine and sulfadoxine-pyrimethamine in Harper, Liberia: results in vivo and analysis of point mutations. Trans R Soc Trop Med Hyg 96:664–669. [DOI] [PubMed] [Google Scholar]
- 11.Roux AT, Maharaj L, Oyegoke O, Akoniyon OP, Adeleke MA, Maharaj R, Okpeku M. 2021. Chloroquine and Sulfadoxine-Pyrimethamine Resistance in Sub-Saharan Africa-A Review. Front Genet 12:668574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noedl H, Se Y, Schaecher K, Smith BL, Socheat D, Fukuda MM, Artemisinin Resistance in Cambodia 1 (ARC1) Study Consortium. 2008. Evidence of artemisinin-resistant malaria in western Cambodia. N Engl J Med 359:2619–2620. [DOI] [PubMed] [Google Scholar]
- 13.Dondorp AM, Nosten F, Yi P, Das D, Phyo AP, Tarning J, Lwin KM, Ariey F, Hanpithakpong W, Lee SJ, Ringwald P, Silamut K, Imwong M, Chotivanich K, Lim P, Herdman T, An SS, Yeung S, Singhasivanon P, Day NPJ, Lindegardh N, Socheat D, White NJ. 2009. Artemisinin resistance in Plasmodium falciparum malaria. N Engl J Med 361:455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ariey F, Witkowski B, Amaratunga C, Beghain J, Langlois A-C, Khim N, Kim S, Duru V, Bouchier C, Ma L, Lim P, Leang R, Duong S, Sreng S, Suon S, Chuor CM, Bout DM, Ménard S, Rogers WO, Genton B, Fandeur T, Miotto O, Ringwald P, Le Bras J, Berry A, Barale J-C, Fairhurst RM, Benoit-Vical F, Mercereau-Puijalon O, Ménard D. 2014. A molecular marker of artemisinin-resistant Plasmodium falciparum malaria. Nature 505:50–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Straimer J, Gnädig NF, Witkowski B, Amaratunga C, Duru V, Ramadani AP, Dacheux M, Khim N, Zhang L, Lam S, Gregory PD, Urnov FD, Mercereau-Puijalon O, Benoit-Vical F, Fairhurst RM, Ménard D, Fidock DA. 2015. Drug resistance. K13-propeller mutations confer artemisinin resistance in Plasmodium falciparum clinical isolates. Science 347:428–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Intharabut B, Kingston HW, Srinamon K, Ashley EA, Imwong M, Dhorda M, Woodrow C, Stepniewska K, Silamut K, Day NPJ, Dondorp AM, White NJ, Tracking Resistance to Artemisinin Collaboration. 2019. Artemisinin Resistance and Stage Dependency of Parasite Clearance in Falciparum Malaria. J Infect Dis 219:1483–1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ménard D, Khim N, Beghain J, Adegnika AA, Shafiul-Alam M, Amodu O, Rahim-Awab G, Barnadas C, Berry A, Boum Y, Bustos MD, Cao J, Chen J-H, Collet L, Cui L, Thakur G-D, Dieye A, Djallé D, Dorkenoo MA, Eboumbou-Moukoko CE, Espino F-E-CJ, Fandeur T, Ferreira-da-Cruz M-F, Fola AA, Fuehrer H-P, Hassan AM, Herrera S, Hongvanthong B, Houzé S, Ibrahim ML, Jahirul-Karim M, Jiang L, Kano S, Ali-Khan W, Khanthavong M, Kremsner PG, Lacerda M, Leang R, Leelawong M, Li M, Lin K, Mazarati J-B, Ménard S, Morlais I, Muhindo-Mavoko H, Musset L, Na-Bangchang K, Nambozi M, Niaré K, Noedl H, Ouédraogo J-B, Pillai DR, Pradines B, Quang-Phuc B, Ramharter M, Randrianarivelojosia M, Sattabongkot J, Sheikh-Omar A, Silué KD, Sirima SB, Sutherland C, Syafruddin D, Tahar R, Tang L-H, Touré OA, Tshibangu-wa-Tshibangu P, Vigan-Womas I, Warsame M, Wini L, Zakeri S, Kim S, Eam R, Berne L, Khean C, Chy S, Ken M, Loch K, Canier L, Duru V, Legrand E, Barale J-C, Stokes B, Straimer J, Witkowski B, Fidock DA, Rogier C, Ringwald P, Ariey F, Mercereau-Puijalon O, KARMA Consortium. 2016. A Worldwide Map of Plasmodium falciparum K13-Propeller Polymorphisms. N Engl J Med 374:2453–2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.World Health Organization. 2020. Report on antimalarial drug efficacy, resistance and response: 10 years of surveillance (2010–2019). World Health Organization, Genève, Switzerland. [Google Scholar]
- 19.Witkowski B, Amaratunga C, Khim N, Sreng S, Chim P, Kim S, Lim P, Mao S, Sopha C, Sam B, Anderson JM, Duong S, Chuor CM, Taylor WRJ, Suon S, Mercereau-Puijalon O, Fairhurst RM, Menard D. 2013. Novel phenotypic assays for the detection of artemisinin-resistant Plasmodium falciparum malaria in Cambodia: in-vitro and ex-vivo drug-response studies. Lancet Infect Dis 13:1043–1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Klonis N, Xie SC, McCaw JM, Crespo-Ortiz MP, Zaloumis SG, Simpson JA, Tilley L. 2013. Altered temporal response of malaria parasites determines differential sensitivity to artemisinin. Proc Natl Acad Sci U S A 110:5157–5162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saralamba S, Pan-Ngum W, Maude RJ, Lee SJ, Tarning J, Lindegårdh N, Chotivanich K, Nosten F, Day NPJ, Socheat D, White NJ, Dondorp AM, White LJ. 2011. Intrahost modeling of artemisinin resistance in Plasmodium falciparum. Proc Natl Acad Sci U S A 108:397–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ishengoma DS, Gosling R, Martinez-Vega R, Beshir KB, Bailey JA, Chimumbwa J, Sutherland C, Conrad MD, Tadesse FG, Juliano JJ, Kamya MR, Mbacham WF, Ménard D, Rosenthal PJ, Raman J, Tatarsky A, Tessema SK, Fidock DA, Djimde AA. 2024. Urgent action is needed to confront artemisinin partial resistance in African malaria parasites. Nat Med 30:1807–1808. [DOI] [PubMed] [Google Scholar]
- 23.Birnbaum J, Scharf S, Schmidt S, Jonscher E, Hoeijmakers WAM, Flemming S, Toenhake CG, Schmitt M, Sabitzki R, Bergmann B, Fröhlke U, Mesén-Ramírez P, Soares AB, Herrmann H, Bártfai R, Spielmann T. 2020. A Kelch13-defined endocytosis pathway mediates artemisinin resistance in malaria parasites. Science. [DOI] [PubMed] [Google Scholar]
- 24.Zhu L, Tripathi J, Rocamora FM, Miotto O, van der Pluijm R, Voss TS, Mok S, Kwiatkowski DP, Nosten F, Day NPJ, White NJ, Dondorp AM, Bozdech Z, Tracking Resistance to Artemisinin Collaboration I. 2018. The origins of malaria artemisinin resistance defined by a genetic and transcriptomic background. Nat Commun 9:5158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tilley L, Straimer J, Gnädig NF, Ralph SA, Fidock DA. 2016. Artemisinin Action and Resistance in Plasmodium falciparum. Trends Parasitol 32:682–696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Meshnick SR. 2002. Artemisinin: mechanisms of action, resistance and toxicity. Int J Parasitol 32:1655–1660. [DOI] [PubMed] [Google Scholar]
- 27.Gierahn TM, Wadsworth MH 2nd, Hughes TK, Bryson BD, Butler A, Satija R, Fortune S, Love JC, Shalek AK. 2017. Seq-Well: portable, low-cost RNA sequencing of single cells at high throughput. Nat Methods 14:395–398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aicher TP, Carroll S, Raddi G, Gierahn T, Wadsworth MH 2nd, Hughes TK, Love C, Shalek AK. 2019. Seq-Well: A Sample-Efficient, Portable Picowell Platform for Massively Parallel Single-Cell RNA Sequencing. Methods Mol Biol 1979:111–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Howick VM, Russell AJC, Andrews T, Heaton H, Reid AJ, Natarajan K, Butungi H, Metcalf T, Verzier LH, Rayner JC, Berriman M, Herren JK, Billker O, Hemberg M, Talman AM, Lawniczak MKN. 2019. The Malaria Cell Atlas: Single parasite transcriptomes across the complete Plasmodium life cycle. Science 365:eaaw2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mesén-Ramírez P, Bergmann B, Elhabiri M, Zhu L, von Thien H, Castro-Peña C, Gilberger T-W, Davioud-Charvet E, Bozdech Z, Bachmann A, Spielmann T. 2021. The parasitophorous vacuole nutrient channel is critical for drug access in malaria parasites and modulates the artemisinin resistance fitness cost. Cell Host Microbe 29:1774–1787.e9. [DOI] [PubMed] [Google Scholar]
- 31.Mok S, Stokes BH, Gnädig NF, Ross LS, Yeo T, Amaratunga C, Allman E, Solyakov L, Bottrill AR, Tripathi J, Fairhurst RM, Llinás M, Bozdech Z, Tobin AB, Fidock DA. 2021. Artemisinin-resistant K13 mutations rewire Plasmodium falciparum’s intra-erythrocytic metabolic program to enhance survival. Nat Commun 12:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xie SC, Dogovski C, Hanssen E, Chiu F, Yang T, Crespo MP, Stafford C, Batinovic S, Teguh S, Charman S, Klonis N, Tilley L. 2016. Haemoglobin degradation underpins the sensitivity of early ring stage Plasmodium falciparum to artemisinins. J Cell Sci 129:406–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller SK, Good RT, Drew DR, Delorenzi M, Sanders PR, Hodder AN, Speed TP, Cowman AF, de Koning-Ward TF, Crabb BS. 2002. A subset of Plasmodium falciparum SERA genes are expressed and appear to play an important role in the erythrocytic cycle. J Biol Chem 277:47524–47532. [DOI] [PubMed] [Google Scholar]
- 34.McCoubrie JE, Miller SK, Sargeant T, Good RT, Hodder AN, Speed TP, de Koning-Ward TF, Crabb BS. 2007. Evidence for a common role for the serine-type Plasmodium falciparum serine repeat antigen proteases: implications for vaccine and drug design. Infect Immun 75:5565–5574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas JA, Collins CR, Das S, Hackett F, Graindorge A, Bell D, Deu E, Blackman MJ. 2016. Development and Application of a Simple Plaque Assay for the Human Malaria Parasite Plasmodium falciparum. PLoS One 11:e0157873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Raj DK, Das Mohapatra A, Jnawali A, Zuromski J, Jha A, Cham-Kpu G, Sherman B, Rudlaff RM, Nixon CE, Hilton N, Oleinikov AV, Chesnokov O, Merritt J, Pond-Tor S, Burns L, Jolly G, Ben Mamoun C, Kabyemela E, Muehlenbachs A, Lambert L, Orr-Gonzalez S, Gnädig NF, Fidock DA, Park S, Dvorin JD, Pardi N, Weissman D, Mui BL, Tam YK, Friedman JF, Fried M, Duffy PE, Kurtis JD. 2020. Anti-PfGARP activates programmed cell death of parasites and reduces severe malaria. Nature 582:104–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.López-Barragán MJ, Lemieux J, Quiñones M, Williamson KC, Molina-Cruz A, Cui K, Barillas-Mury C, Zhao K, Su X-Z. 2011. Directional gene expression and antisense transcripts in sexual and asexual stages of Plasmodium falciparum. BMC Genomics 12:587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Burkhardt DB, Stanley JS, Tong A, Perdigoto AL, Gigante SA, Herold KC, Wolf G, Giraldez AJ, van Dijk D, Krishnaswamy S. 2021. Quantifying the effect of experimental perturbations at single-cell resolution. Nat Biotechnol 39:619–629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Street K, Risso D, Fletcher RB, Das D, Ngai J, Yosef N, Purdom E, Dudoit S. 2018. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19:477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mok S, Ashley EA, Ferreira PE, Zhu L, Lin Z, Yeo T, Chotivanich K, Imwong M, Pukrittayakamee S, Dhorda M, Nguon C, Lim P, Amaratunga C, Suon S, Hien TT, Htut Y, Faiz MA, Onyamboko MA, Mayxay M, Newton PN, Tripura R, Woodrow CJ, Miotto O, Kwiatkowski DP, Nosten F, Day NPJ, Preiser PR, White NJ, Dondorp AM, Fairhurst RM, Bozdech Z. 2015. Drug resistance. Population transcriptomics of human malaria parasites reveals the mechanism of artemisinin resistance. Science 347:431–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rawat M, Kanyal A, Sahasrabudhe A, Vembar SS, Lopez-Rubio J-J, Karmodiya K. 2021. Histone acetyltransferase PfGCN5 regulates stress responsive and artemisinin resistance related genes in Plasmodium falciparum. Sci Rep 11:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu L, van der Pluijm RW, Kucharski M, Nayak S, Tripathi J, White NJ, Day NPJ, Faiz A, Phyo AP, Amaratunga C, Lek D, Ashley EA, Nosten F, Smithuis F, Ginsburg H, von Seidlein L, Lin K, Imwong M, Chotivanich K, Mayxay M, Dhorda M, Nguyen HC, Nguyen TNT, Miotto O, Newton PN, Jittamala P, Tripura R, Pukrittayakamee S, Peto TJ, Hien TT, Dondorp AM, Bozdech Z. 2022. Artemisinin resistance in the malaria parasite, Plasmodium falciparum, originates from its initial transcriptional response. Commun Biol 5:274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yang T, Yeoh LM, Tutor MV, Dixon MW, McMillan PJ, Xie SC, Bridgford JL, Gillett DL, Duffy MF, Ralph SA, McConville MJ, Tilley L, Cobbold SA. 2019. Decreased K13 Abundance Reduces Hemoglobin Catabolism and Proteotoxic Stress, Underpinning Artemisinin Resistance. Cell Rep 29:2917–2928.e5. [DOI] [PubMed] [Google Scholar]
- 44.Paloque L, Ramadani AP, Mercereau-Puijalon O, Augereau J-M, Benoit-Vical F. 2016. Plasmodium falciparum: multifaceted resistance to artemisinins. Malar J 15:149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morano KA, Grant CM, Moye-Rowley WS. 2012. The response to heat shock and oxidative stress in Saccharomyces cerevisiae. Genetics 190:1157–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Roti Roti JL. 2008. Cellular responses to hyperthermia (40–46 degrees C): cell killing and molecular events. Int J Hyperthermia 24:3–15. [DOI] [PubMed] [Google Scholar]
- 47.Marti M, Spielmann T. 2013. Protein export in malaria parasites: many membranes to cross. Curr Opin Microbiol 16:445–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rosenthal MR, Ng CL. 2020. Plasmodium falciparum Artemisinin Resistance: The Effect of Heme, Protein Damage, and Parasite Cell Stress Response. ACS Infect Dis 6:1599–1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schuster FL. 2002. Cultivation of plasmodium spp. Clin Microbiol Rev 15:355–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brown AC, Guler JL. 2018. SLOPE: A two-part method for the enrichment of ring stage Plasmodium falciparum parasites. bioRxiv. [Google Scholar]
- 51.Brown AC, Moore CC, Guler JL. 2020. Cholesterol-dependent enrichment of understudied erythrocytic stages of human Plasmodium parasites. Sci Rep 10:1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, Trombetta JJ, Weitz DA, Sanes JR, Shalek AK, Regev A, McCarroll SA. 2015. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell 161:1202–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. 2018. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36:411–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM 3rd, Hao Y, Stoeckius M, Smibert P, Satija R. 2019. Comprehensive Integration of Single-Cell Data. Cell 177:1888–1902.e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Becht E, McInnes L, Healy J, Dutertre C-A, Kwok IWH, Ng LG, Ginhoux F, Newell EW. 2018. Dimensionality reduction for visualizing single-cell data using UMAP. Nat Biotechnol 10.1038/nbt.4314. [DOI] [PubMed] [Google Scholar]
- 56.Finak G, McDavid A, Yajima M, Deng J, Gersuk V, Shalek AK, Slichter CK, Miller HW, McElrath MJ, Prlic M, Linsley PS, Gottardo R. 2015. MAST: a flexible statistical framework for assessing transcriptional changes and characterizing heterogeneity in single-cell RNA sequencing data. Genome Biol 16:278. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequence data has been submitted to SRA (PRJNA1049964). Code is available at https://github.com/sconnelly007/K13_mt_v_wt_sc.