Abstract
A key reaction in the biosynthesis of menaquinone involves the conversion of the soluble bicyclic naphthalenoid compound 1,4-dihydroxy-2-naphthoic acid (DHNA) to the membrane-bound demethylmenaquinone. The enzyme catalyzing this reaction, DHNA-octaprenyltransferase, attaches a 40-carbon side chain to DHNA. The menA gene encoding this enzyme has been cloned and localized to a 2.0-kb region of the Escherichia coli genome between cytR and glpK. DNA sequence analysis of the cloned insert revealed a 308-codon open reading frame (ORF), which by deletion analyses was shown to restore anaerobic growth of a menA mutant. Reverse-phase high-performance liquid chromatography analysis of quinones extracted from the orf-complemented cells independently confirmed the restoration of menaquinone biosynthesis, and similarly, analyses of isolated cell membranes for DHNA octaprenyltransferase activity confirmed the introduction of the menA product into the orf-complemented menA mutant. The validity of an ORF-associated putative promoter sequence was confirmed by primer extension analyses.
Menaquinone (MK; vitamin K2) plays an essential role in several anaerobic electron transport systems. It is the major electron carrier during anaerobic growth with various electron acceptors (10). The genetics, biochemistry, and molecular biology of its biosynthetic pathway have been recently reviewed (12). Six genes, designated menA, -B, -C, -D, -E, and -F, have been implicated in the biosynthesis of MK from chorismate and 2-ketoglutarate. Five of these, menB, -C, -D, -E, and -F, all clustered at 51 min on the Escherichia coli chromosome, are responsible for the conversion of chorismate and 2-ketoglutarate to the soluble naphthalenoid aromatic compound 1,4-dihydroxy-2-naphthoic acid (DHNA) (12). In a subsequent reaction, the DHNA is attached to the membrane-bound octaprenylpyrophosphate and results in the formation of demethylmenaquinone (DMK). This latter step, prior to the methylation of DMK to MK, is under the control of menA, located at 88 min on the E. coli chromosome (12).
In 1975, Bentley (3) demonstrated that cell-free extracts of E. coli can convert DHNA in the presence of farnesyl pyrophosphate and Mg2+ to MK-3 and/or DMK-3. In a subsequent investigation, Shineberg and Young (21) showed that crude membranes of E. coli can carry out this reaction in the presence of either the synthetic compound solanesyl pyrophosphate or the natural octaprenyl pyrophosphate. Shineberg and Young named the enzyme 1,4-dihydroxy-2-naphthoate octaprenyltransferase and showed that the menA mutants lacked this enzyme (21).
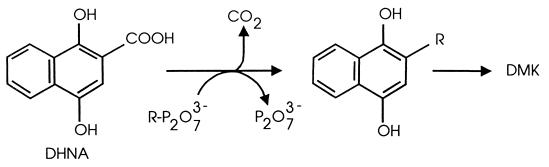
The conversion of DHNA to DMK proceeds in three stages as summarized in Fig. 1. These stages are the removal of the carboxyl group of DHNA as CO2, the attachment of the isoprenoid side chain, and a quinol-to-quinone oxidation, which is thought to be spontaneous (4).
FIG. 1.
Reactions involved in the prenylation of DHNA. R, prenyl.
In this study, we have cloned, sequenced, and identified the menA open reading frame (ORF) encoding the octaprenyltransferase.
(A preliminary communication of some of these findings has been published previously [22]).
Bacterial strains and growth conditions.
The strains of E. coli and the primary plasmids used are listed in Table 1. Other plasmids constructed in this study are shown in Fig. 2. Complementation of the menA mutant with plasmids was performed, as previously described (17, 20), by anaerobic growth on glycerol minimal medium with trimethylamine N-oxide (TMAO) as an electron acceptor (7). Cells for the isolation of quinones and membranes were grown anaerobically in 2-liter Erlenmeyer flasks with screw caps filled to the top with minimal medium (14) containing glucose as the carbon source. For anaerobic growth of the menA mutant, the glucose minimal medium was supplemented with 5 μg of uracil per ml (15).
TABLE 1.
Strains of E. coli K-12 and plasmids used in the study
| Strain or plasmid | Genotype | Reference or source |
|---|---|---|
| E. coli | ||
| PL2024 | gal trpA trpR iclR rpsL | 17, 20 |
| AN67 | Δ(gpt-proA)62 lacY1 tsx-29 supE44 galK2 λ− trpE81 pheA3::Mu tyrA4 xyl-5 mtl-1 menA401 thi-1 | 21 |
| HB101 | supE44 hsdS20 (rB− mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | Laboratory stock |
| JM83 | F−ara Δ(lac-proAB) rpsL (Strr) [φ80Δ(lacZ)M15] | Laboratory stock |
| Plasmid | ||
| pUC18 | Cloning vector | International Biotechnologies, Inc. |
| pUC19 | Cloning vector | International Biotechnologies, Inc. |
| pLC28-47 | Clarke-Carbon plasmid clone | 16 |
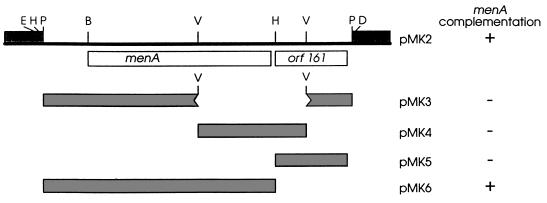
FIG. 2.
The menA plasmids. pMK2 was constructed by insertion into pUC18 of a 2.0-kb PstI fragment derived from pLC28-47; pMK3 has an EcoRV fragment deletion of pMK2; pMK4 contains only the EcoRV fragment of pMK2; and pMK5 and pMK6 contain the 0.5- and 1.5-kb HindIII/PstI fragments of pMK2, respectively. Complementation was assayed by anaerobic growth on glycerol medium with TMAO as the electron acceptor. B, BglII; D, HindIII; E, EcoRI; H, HincII; P, PstI; V, EcoRV.
DNA and RNA isolation and construction of plasmids.
Plasmid DNAs were isolated by the alkaline lysis procedure of Birnboim and Doly (5) and then purified in ethidium bromide-CsCl gradients. Total RNA from E. coli PL2024 cells was isolated as previously described (2). Cells for RNA isolation were grown either aerobically or anaerobically in Luria-Bertani medium supplemented with 10 mM glucose.
Plasmids used in this study contained inserts derived from plasmid pLC28-47 (16) and subcloned into pUC18. DNA fragments for cloning were electroeluted from 0.7% agarose gel slices, ligated, and transformed as previously described (17, 20). The host for initial transformation was either HB101 or JM83.
DNA sequencing and sequence analysis.
DNA sequence was determined by the dideoxynucleotide chain termination method (19) with the Sequenase 2.0 kit from Amersham Corp. (Arlington Heights, Ill.) and [α-32P]dATP (ICN, Costa Mesa, Calif.). Compressed regions were resolved with 7-deaza-dGTP. Sequencing reactions were primed with universal, reverse, and sequence-generated synthetic oligonucleotide primers synthesized on an Applied Biosystems (Foster City, Calif.) 391 DNA synthesizer. All sequences were confirmed by sequencing of the complementary strand. Nucleic acid and deduced protein sequences were analyzed with Pustell (IBI, New Haven, Conn.) and PC/Gene (Intelligenetics, Mountain View, Calif.) sequence analysis software.
Primer extensions.
Primer extensions were performed with the AMV reverse transcriptase primer extension system (Promega, Madison, Wis.). The manufacturer’s suggestions were followed, but the following modifications were made. After the completion of the primer extension reactions, the RNA-cDNA mixture was ethanol precipitated, and the pellet was washed twice with 70% ethanol and resuspended in 3 μl of diethyl pyrocarbonate-treated water. Following the addition of 1 μl of loading dye, the solution was heated to 90°C for 2 min and analyzed on 6% polyacrylamide sequencing gels. Equal quantities of anaerobically or aerobically produced total RNA (as determined spectrophotometrically) were used for the appropriate assays. Accompanying DNA sequence ladders were generated with the Amersham Corp. ΔTaq cycle sequencing kit according to the manufacturer’s directions.
Isolation and identification of MK and DMK by RPHPLC.
The isolation of quinones from cells was done with the chloroform-methanol technique of Bligh and Dyer (6). Approximately 2 g of wet cells was extracted with 25 ml of chloroform-methanol (2:1 [vol/vol]) for 3 h by constant stirring over a magnetic stirrer. The mixture was filtered through Whatman no. 1 filter paper to remove cell debris, and the filtrate was collected in a conical flask and dried under reduced pressure at 40°C in a rotary evaporator (13). The lipid extract was resuspended in a small volume of hexane and passed through a Sep-Pak silicon cartridge (Waters, Bedford, Mass.) to remove contaminating lipids. The cartridge was eluted with hexane-diethylether (96:4 [vol/vol]). The elutant was collected, dried under a stream of N2, and suspended in 20 μl of acetone. Five microliters was typically injected into the column for the identification of quinones. Extreme care was taken to minimize exposure to light at all stages. The separation and analysis of quinones were performed on a Waters 670 liquid chromatograph equipped with a Zorbax ODS prepacked (250 by 4.6 mm in diameter) reverse-phase high-performance liquid chromatography (RPHPLC) column (Chrompack Inc., Raritan, N.J.). Samples were eluted with methanol-isopropylether (3:1 [vol/vol]) at 0.8 ml/min. The quinones were detected with a UV detector at 245 nm. Menaquinones with 3 and 8 isoprene units (MK-3 and MK-8) were used as standards.
Isolation of membranes and assay of DHNA octaprenyltransferase.
Anaerobically grown cultures were harvested at late log phase by centrifugation at 3,000 × g for 10 min. The pellet was washed once by resuspension in 200 ml of 50 mM potassium phosphate buffer (pH 7.0) and recentrifuged. Each gram of washed pellet was resuspended in 1.5 ml of 50 mM potassium phosphate buffer (pH 7.0) containing 5 mM 2-mercaptoethanol and then passed twice through a French pressure cell at 15,000–20,000 lb/in2 at 4°C. A small amount of DNase was added to reduce viscosity. The extract was centrifuged for 30 min at 3,000 × g, and the supernatant was further centrifuged at 150,000 × g in a swinging bucket rotor (AH650 in a Sorval RC 70 ultracentrifuge) for 3 h at 4°C. The pellet was washed once by resuspension and centrifugation for a further 3 h. The washed pellet was resuspended in the buffer and used as the membrane fraction. The protein content of extracts was determined by the method of Lowry et al. (11) with bovine serum albumin as the standard.
For the assay of octaprenyltransferase, the procedure of Shineberg and Young (21) was used with modifications. The reaction mixture consisted of 10 μl of cell extract or membranes (0.2 to 0.25 mg of protein), 11.4 nmol of [3H]farnesyl pyrophosphate (specific activity, 44 μCi/mmol), 0.1 M Tris-HCl buffer (pH 8.0), 10 mM MgSO4, 10 mM dithiothreitol, and 0.4% Triton X-100 in a total volume of 0.5 ml. The reaction was started by the addition of 100 nmol of 1,4-dihydroxy-2-naphthoic acid in 10 μl of ethanol-diethyl ether (2:1 [vol/vol]). The tubes were incubated at 37°C for 30 min. The reaction was stopped by the addition of 2 ml of 0.1 M acetic acid in methanol and extracted twice with 3 ml of pentane. The combined pentane extracts were evaporated to dryness in a scintillation vial, 6 ml of Ultima Gold scintillation fluid (Packard Instrument Co., Inc.) was added, and the radioactivity was determined.
Cloning, sequencing, and identification of the menA gene.
The menA gene is located at 88 min on the E. coli linkage map (12) and has been shown to be cotransducible at frequencies of 60 and 90%, respectively, with the metB and glpK loci (12). Since the Clarke-Carbon plasmid pLC 28-47 (16) spans this region, it was used as a starting point for cloning experiments. A 2.0-kb, PstI fragment located between cytR and glpK (Fig. 2) was subcloned into the PstI site of pUC18 and designated pMK2. When transformed into the menA mutant, AN67 pMK2 was found to complement the mutant phenotype as assayed by the restoration of anaerobic growth on glycerol-TMAO medium.
The nucleotide sequence of the pMK2 insert was determined to identify a potential menA ORF. Two intact ORFs were located within the insert. One of these (orf308) encoded a 308-residue polypeptide, and the other (orf161) encoded a 161-residue protein. Appropriately positioned putative promoter sequences, with consensus −10 and −35 elements, and consensus ribosome binding site sequences were located 5′ to each ORF. To identify the ORF encoding the octaprenyltransferase, four pMK2 deletion plasmids (Fig. 2) were constructed and used to complement the menA mutant. Complementation occurred in the presence of pMK6, which retains an intact orf308 and a deleted orf161, but not in the presence of pMK5 (deleted orf308 and intact orf161) or of pMK3 or pMK4 (both reading frames disrupted). Thus, orf308 was designated the menA gene.
Analysis of menaquinones and octaprenyltransferase activity in various strains.
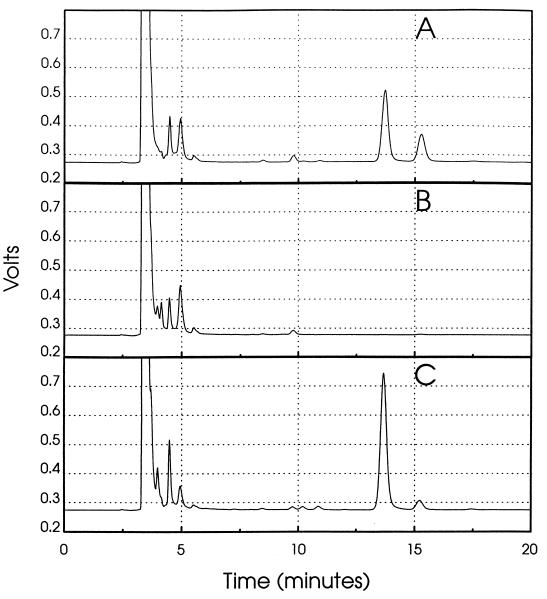
E. coli cells contain both MK-8 and DMK-8, both of which are known to be membrane bound (4). To confirm that the restoration of anaerobic growth on glycerol-TMAO by complementation was due to the restoration of the ability to synthesize menaquinones, the wild-type strain PL2024, the menA mutant AN67, and the two plasmid-containing strains AN67/pMK2 and AN67/pMK6 were grown for the analysis of quinone content. Quinones from these strains were extracted and analyzed by RPHPLC. The results shown in Fig. 3 clearly indicate the absence of both DMK and MK in the untransformed menA mutant. In contrast, the levels of both these napthoquinones in the orf308-complemented mutant (AN67/pMK6) indicate restoration of quinone synthesis. Similar results were obtained with AN67/pMK2 (data not shown).
FIG. 3.
HPLC tracing of menaquinones extracted from the wild type (A), the menA mutant (B), and the menA mutant complemented by plasmid pMK6 (C). Peaks were identified by authentic standards. DMK-8 and MK-8 eluted at 13.6 and 15.2 min, respectively.
Since the menA mutant showed restoration of MK-8 and DMK-8 when either pMK2 or pMK6 was carried, it was important to verify that the restoration correlated with the appearance of DHNA octaprenyltransferase in the plasmid-containing strains. Hence, the enzyme activity was assayed in membrane preparations from the wild-type strain PL2024, the menA mutant AN67, and the two plasmid-containing strains. As is evident from Table 2, enzyme activity is fully restored in both AN67/pMK2 and AN67/pMK6. However, the activity was notably higher in the presence of pMK6. This higher level agrees with an increased level of DMK in the complemented strain (Fig. 3).
TABLE 2.
DHNA-octaprenyltransferase activities in extracts of various strains
| Cell extract | Activitya
|
||
|---|---|---|---|
| 3,000 × g (supernatant) | 150,000 × g
|
||
| Pellet | Supernatant | ||
| AN67 (menA) | 20 | 30 | 8 |
| PL2024 (wild type) | 640 | 790 | 7 |
| AN67(pMK2) | 870 | 940 | 7 |
| AN67(pMK6) | 1,590 | 1,800 | 7 |
Activities are in picomoles per milligram of protein per hour and varied no more than 20%.
Similarities of octaprenyltransferases.
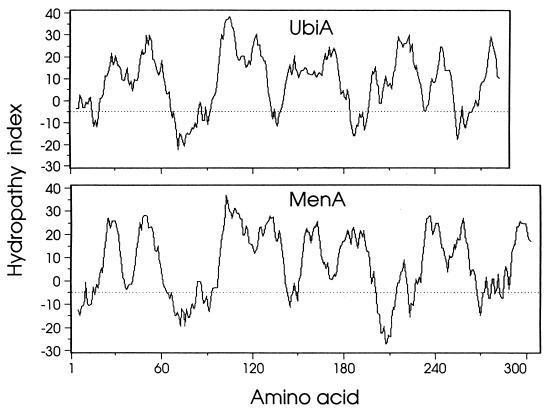
The menA region of the E. coli chromosome has also been sequenced as part of the genome sequencing project (18) and is identical to the sequence reported here. We now, however, identify the reported E. coli genome project orf308 reading frame as menA. A search of the GenBank database with the deduced DHNA octaprenyltransferase sequence identified two relevant categories of polypeptides. The first category included presumptive menA-encoded products in Haemophilus influenzae (accession no. U32732) and Synechocystis sp. (accession no. D90911), both of which are based upon our reporting of the E. coli menA sequence, and possibly the Bacillus subtilis 33.8-kDa gene product of ipa-6d (accession no. S39661 and X73124). All three of these proteins include the octaprenyl pyrophosphate binding motif noted below. A second category, however, was created by the ubiA gene product 4-hydroxybenzoate octaprenyltransferase from E. coli. Since menaquinone and ubiquinone have identical octaprenyl side chains, and since both have a common pool of octaprenyl pyrophosphate, we aligned the deduced polypeptide sequences of the menA and ubiA gene products. The results indicated 35% overall similarity (21% identity) (Fig. 4), which is even more apparent in a comparison of their hydrophobicity plots (Fig. 5). Highly conserved within the deduced DHNA octaprenyltransferase polypeptide is a sequence (LANDYGD) reminiscent of the putative polyprenyl pyrophosphate binding region (1) (Fig. 4).
FIG. 4.
Alignment of deduced menA and ubiA polypeptides. Amino acids are represented by the single-letter code. ∗, amino acid identity; ., amino acid similarity; −, gaps introduced to facilitate the alignment. The putative polyprenyl pyrophosphate binding site is indicated in bold-faced type.
FIG. 5.
Hydrophobicity comparison of the deduced menA- and ubiA-encoded polypeptides. Hydrophobic values are given on the ordinate. Positive hydrophobic values indicate hydrophobic regions, and negative values indicate hydrophilic regions. Amino acid residue numbers are shown on the abscissa. A moving 11-amino-acid window was used to generate the plots. E. coli ubiA polypeptide (25).
menA transcriptional analysis.
Following the confirmation of orf308 as the menA locus, the validity of the associated putative promoter was tested by primer extension analyses. Since DMK and MK are the primary quinones during anaerobic growth but are also synthesized under aerobic conditions, total RNA was prepared from both aerobically and anaerobically grown wild-type cells. Equal quantities of RNA from both sets of cells were independently assayed for transcriptional initiation sites with the two discrete primers indicated in Fig. 6. In all instances the +1 site was identified as 57 bp 5′ to the translational initiation codon. The levels of primed RNA were greater in aerobically than in anaerobically prepared cells (Fig. 6). To verify this result, additional RNA preparations from aerobically grown cells with minimal medium, plus either glucose or glycerol, were compared with anaerobically grown equivalents containing TMAO as the electron acceptor. While the levels of transcript were reduced in these sets of experiments compared to those in complex-media-grown cells, the greater level of transcript was still apparent in the aerobically grown samples.
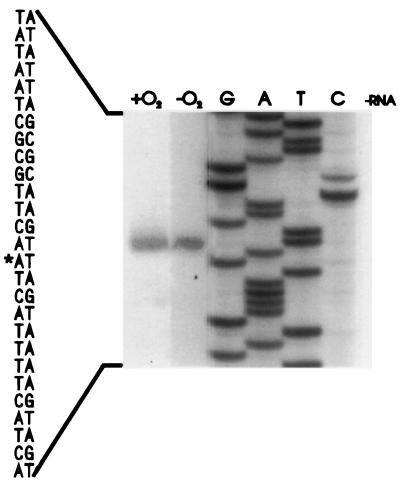
FIG. 6.
Primer extension identification of menA transcriptional initiation. Equivalent amounts of spectrophotometrically determined RNA from aerobically (+O2) and anaerobically (−O2) grown PL2024 cells were extended with the oligonucleotide GAGGGTTTTAGGTCGTAAACTTTCC derived from the menA sequence 3′ to the translational initiation codon. DNA sequence ladders (lanes G, A, T, and C) are extensions of pMK2 DNA with the same primer. ∗, transcriptional initiation origin. A second primer, located 25 bp upstream from the first and having the sequence GTTGTTCAGTCATAATACGCGCCAATAA, gave similar results.
The analysis of the 5′-flanking sequence of menA indicated the presence of presumptive ς70 promoter elements coupled with an appropriately positioned ribosomal binding site. Primer extension studies allowed us to confirm a transcriptional start site downstream from the putative perfect consensus (TATAAT) −10 promoter. However, 18 bp upstream, only a poorly matched (ATGAAG) −35 consensus sequence was evident, suggesting the possible need for additional regulatory factors to obtain higher levels of transcriptional initiation.
Transcriptional analyses show a greater abundance of menA transcript in aerobically grown than anaerobically grown cells. Previous studies have shown that aerobically grown cells contain about 10-fold-higher levels of DMK than of MK (23). Under anaerobic conditions, the level of MK increases, while the level of DMK decreases to a very low level (24). Recently, it has been shown that an enzyme (UbiE) involved in uniquinone (required for aerobic electron transport) biosynthesis can methylate DMK to MK (9). However, a more definitive assessment of the regulation of synthesis of DMK and MK must await the results of ongoing operon and protein lac fusion studies on the menA and ubiE genes.
Finally, we note a potential effect of the upstream hslU locus on menA transcripts. Plunkett et al. (18) described a potential stem-loop structure hypothesized as a transcriptional terminator for hslU. This structure must also be part of the menA transcript, as it lies directly between the menA transcriptional and translational initiation points. As such, it may play a role in menA mRNA stability in a manner similar to the more complex stem-loop arrangement found in ompA message (8). Such enhancement of mRNA stability would help explain the apparent paradox of aerobically elevated transcript levels for a menaquinone biosynthesis gene.
Nucleotide sequence accession number.
The sequence data reported in this study appear in the EMBL, GenBank, and DDBJ nucleotide sequence databases under accession no. U56082.
Acknowledgments
This research was supported by Public Health Service grant GM 50262 from the National Institutes of Health.
REFERENCES
- 1.Asby M N, Kutsunai S Y, Ackerman S, Tzagoloff A, Edwards P A. COQ2 is a candidate for the structural gene encoding para-hydroxybenzoate:octaprenyltransferase. J Biol Chem. 1992;267:4128–4136. [PubMed] [Google Scholar]
- 2.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley and Sons; 1989. pp. 4.4.1–4.9.8. [Google Scholar]
- 3.Bentley R. Biosynthesis of vitamin K and other natural naphthoquinones. Pure Appl Chem. 1975;41:47–68. [Google Scholar]
- 4.Bentley R, Meganathan R. Biosynthesis of vitamin K (menaquinone) in bacteria. Microbiol Rev. 1982;46:241–280. doi: 10.1128/mr.46.3.241-280.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birnboim B G, Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979;7:1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bligh E G, Dyer W J. A rapid method of total lipid extraction and purification. Can J Biochem Physiol. 1959;37:911–917. doi: 10.1139/o59-099. [DOI] [PubMed] [Google Scholar]
- 7.Daruwala R, Meganathan R. Dimethyl sulfoxide reductase is not required for trimethylamine N-oxide reduction in Escherichia coli. FEMS Microbiol Lett. 1991;83:255–260. doi: 10.1016/0378-1097(91)90485-s. [DOI] [PubMed] [Google Scholar]
- 8.Emory S A, Bouvet P, Belasco J G. A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992;6:135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- 9.Lee P T, Hsu A Y, Ha H T, Clarke C F. A C-methyltransferase involved in both ubiquinone and menaquinone biosynthesis: isolation and identification of the Escherichia coli ubiE gene. J Bacteriol. 1997;179:1748–1754. doi: 10.1128/jb.179.5.1748-1754.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lin E C C, Kuritzkes D. Pathways for anaerobic electron transport. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C: American Society for Microbiology; 1987. pp. 202–221. [Google Scholar]
- 11.Lowry O H, Rosebrough N J, Farr A L, Randall R J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–275. [PubMed] [Google Scholar]
- 12.Meganathan R. Biosynthesis of the isoprenoid quinones menaquinone (vitamin K2) and ubiquinone (coenzyme Q) In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Reznikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C: American Society for Microbiology; 1996. pp. 642–656. [Google Scholar]
- 13.Meganathan R, Coffell R. Identity of the quinone in Bacillus alcalophilus. J Bacteriol. 1985;164:911–913. doi: 10.1128/jb.164.2.911-913.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miguel L, Meganathan R. Electron donors and the quinone involved in dimethyl sulfoxide reduction in Escherichia coli. Curr Microbiol. 1991;22:109–115. [Google Scholar]
- 15.Newton N A, Cox G B, Gibson F. The function of menaquinone (vitamin K2) in Escherichia coli K12. Biochim Biophys Acta. 1971;244:155–166. doi: 10.1016/0304-4165(71)90132-2. [DOI] [PubMed] [Google Scholar]
- 16.Nishimura A, Akiyama K, Kohara Y, Horiuchi K. Correlation of a subset of the pLC plasmids to the physical map of Escherichia coli K-12. Microbiol Rev. 1992;56:137–151. doi: 10.1128/mr.56.1.137-151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palaniappan C, Sharma V, Hudspeth M E S, Meganathan R. Menaquinone (vitamin K2) biosynthesis: evidence that the Escherichia coli menD gene encodes both 2-succinyl-6-hydroxy-2,4-cyclohexadiene-1-carboxylic acid synthase and α-ketoglutarate decarboxylase activities. J Bacteriol. 1992;174:8111–8118. doi: 10.1128/jb.174.24.8111-8118.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Plunkett G, III, Burland V, Daniels D L, Blattner F R. Analysis of the Escherichia coli genome. III. DNA sequence of the region from 87.2 to 89.2 minutes. Nucleic Acids Res. 1993;21:3391–3398. doi: 10.1093/nar/21.15.3391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanger F, Nicklen S, Coulson A R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci USA. 1977;74:5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharma V, Suvarna K, Meganathan R, Hudspeth M E S. Menaquinone (vitamin K2) biosynthesis: nucleotide sequence and expression of the menB gene from Escherichia coli. J Bacteriol. 1992;174:5057–5062. doi: 10.1128/jb.174.15.5057-5062.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shineberg B, Young I G. Biosynthesis of bacterial menaquinones: the membrane associated 1,4-dihydroxy-2-naphthoate octaprenyltransferase of Escherichia coli. Biochemistry. 1976;15:2754–2758. doi: 10.1021/bi00658a007. [DOI] [PubMed] [Google Scholar]
- 22.Suvarna K, Meganathan R, Hudspeth M E S. Abstracts of the 94th General Meeting of the American Society for Microbiology 1994. Washington, D.C: American Society for Microbiology; 1994. Menaquinone (vitamin K2) biosynthesis: cloning and nucleotide sequence of the menA gene from Escherichia coli, abstr. K-159; p. 303. [Google Scholar]
- 23.Unden G. Differential roles of menaquinone and demethylmenaquinone in anaerobic electron transport of E. coli and their fnr-independent expression. Arch Microbiol. 1988;150:499–503. doi: 10.1007/BF00422294. [DOI] [PubMed] [Google Scholar]
- 24.Wallace B J, Young I G. Roles of quinone in electron transport to oxygen and nitrate in Escherichia coli studies with a ubiA− and menA− double mutant. Biochim Biophys Acta. 1977;461:84–100. doi: 10.1016/0005-2728(77)90071-8. [DOI] [PubMed] [Google Scholar]
- 25.Wu G, Williams H D, Gibson F, Poole R K. Mutants of Escherichia coli affected in respiration: the cloning and nucleotide sequence of ubiA, encoding the membrane-bound p-hydroxybenzoate:octaprenyltransferase. J Gen Microbiol. 1993;139:1795–1805. doi: 10.1099/00221287-139-8-1795. [DOI] [PubMed] [Google Scholar]