Abstract
Malignant brain tumors are aggressive and difficult to treat. Glioblastoma is the most common and lethal form of primary brain tumor, often found in patients with no genetic predisposition. The median life expectancy for individuals diagnosed with this condition is 6 months to 2 years and there is no known cure. New paradigms in cancer biology implicate a small subset of tumor cells in initiating and sustaining these incurable brain tumors. Here, we discuss the heterogenous nature of glioblastoma and theories behind its capacity for therapy resistance and recurrence. Within the cancer landscape, cancer stem cells are thought to be both tumor initiators and major contributors to tumor heterogeneity and therapy evasion and such cells have been identified in glioblastoma. At the cellular level, disruptions in the delicate balance between differentiation and self-renewal spur transformation and support tumor growth. While rapidly dividing cells are more sensitive to elimination by traditional treatments, glioblastoma stem cells evade these measures through slow division and reversible exit from the cell cycle. At the molecular level, glioblastoma tumor cells exploit several signaling pathways to evade conventional therapies through improved DNA repair mechanisms and a flexible state of senescence. We examine these common evasion techniques while discussing potential molecular approaches to better target these deadly tumors. Equally important, the presented information encourages the idea of augmenting conventional treatments with novel glioblastoma stem cell-directed therapies, as eliminating these harmful progenitors holds great potential to modulate tumor recurrence.
Keywords: Glioblastoma, Glioblastoma stem cells, heterogeneity, therapy resistance, tumor microenvironment, clinical treatments
1. Introduction
Glioblastoma (GBM) is the most common and deadly primary brain tumor diagnosis in Europe and North America1,2. It is an aggressive disease characterized by rapid tumor growth, potent invasive behavior, and high therapeutic resistance. Due to its severity, the prognosis for GBM is bleak with a median survival of ~15 months and a 5-year survival rate of only 5%3. While there is no cure for GBM, standard treatment entails surgical resection followed by dual therapy irradiation and the adjuvant chemotherapeutic temozolomide (TMZ)4. Years of clinical investigation indicate this regimen can improve patient survival rates5. Unfortunately, this combinatorial therapy does not comprehensively clear all tumor cells6. As a consequence, GBM is notorious for its inevitable recurrence and readministering these interventions provides only modest benefits7. Several features of GBM impair therapeutic efficacy, the first of which is the blood brain barrier (BBB). Drugs that improve tumor sensitivity to TMZ have difficulty penetrating the restrictive BBB, presenting a major challenge for chemotherapeutics8. Further, the vasculature network is leaky in GBM tumors, contributing to insufficient drug delivery across the tumor and decreasing their efficacy9. Also, due to their potent invasive nature, the boundary between tumor cells and healthy cells is poorly delineated, making complete surgical resection virtually impossible10.
Tumor heterogeneity is another key feature of GBM. A single GBM tumor is comprised of diverse cell types harboring a variety of genetic and transcriptomic phenotypes11,12. High levels of heterogeneity mean differing sensitivities to therapies, impairing their universal potency13-15. Based on these pleiomorphic features, there is growing support for the presence of cellular populations that exhibit stem cell-like properties termed “cancer stem cells” (CSCs). Co-existing symbiotically alongside bulk tumor cells, CSCs are thought to strongly contribute to tumor development, drug resistance, and cancer recurrence15-21. Distinct niches or microenvironments house CSC populations and amplify signals for tumor progression and maintain stemness. In addition to intrinsic resistance mechanisms, under therapeutic stress, CSCs and bulk tumor cells can reversibly exit the cell cycle, thereby evading therapies that rely on cell division before reinitiating tumorigenesis22.
As the recognition and importance of CSCs continue to grow, it is imperative to understand their molecular properties, invasive behavior, and interactions with surrounding cells. This review aims to better inform clinicians and scientists entering the complex field of GBM research. Here, we summarize issues related to cellular heterogeneity, tumor microenvironment, and therapeutic resistance while exploring potential therapeutic targets aimed at the eliminating CSCs.
2. Heterogeneity in glioblastoma
The variety of cell phenotypes present in GBM has been observed since it’s early diagnoses. Due to GBM’s hallmark complexity, it was the first cancer to be fully sequenced by The Cancer Genome Atlas Initiative23 and other –omics data quickly followed24-27. Molecular profiling now accompanies histology in identifying and classifying GBM28. Sequencing showed many differently expressed genes between GBM tumors but also several common alterations now used for diagnosis. These abnormalities include amplification of the endothelial growth factor receptor (EGFR), loss of chromosome 10, amplification of chromosome 7, and mutations of the telomerase (TERT) promoter23,27,29. One significant mutation used for GBM classification is in the isocitrate dehydrogenase (IDH1) gene25. Found in ~9% of patients, this mutation impairs tumor cell metabolism and influences overall survival rate – 1.1 years for wild type IDH1 (IDH-wt) compared to 3.8 years for mutant IDH1 (IDH-mut)30-32. So clinically divergent are IDH-wt and IDH-mut GBMs that the 2021 WHO nomenclature redefined ‘glioblastoma-mutant IDH’ as grade 4 ‘astrocytoma - mutant IDH’33.
Using molecular profiling criteria, GBM tumors have been clustered into three subtypes: proneural (PN), proliferative, and mesenchymal (MES)34. Transcriptome and single-cell analysis of multiple GBM tumors by Patel et. al identified that transcript signatures describing these bulk tumor subtypes also exist as geographically separate regions inside a single tumor26. Not only can a whole tumor be classified MES or PN, but regions within that tumor obeyed the same classification and expression signature, often with more than one such region in a single tumor26,35,36. In addition to spatial heterogeneity, tumors can also exhibit temporal heterogeneity, with ~45 - 63% of GBM tumors changing expression signatures during evolution and in response to environmental stimuli37-39. One of the most relevant examples of this change is the PN to MES transition observed in response to radiation therapy40. The MES signature is correlated with decreased therapy sensitivity, leading to an enrichment of MES cells in recurrent GBM which contributes to its intractability40,41.
2.1. The clonal evolution theory and the cancer stem cell theory
One hypothesis to explain tumor heterogeneity is the clonal evolution theory42,43. In this theory, a single transformed ancestor cell divides to establish and populate the tumor, using its selective growth advantage to outcompete normal cells and pass down cancerous mutations to each new generation. Each cancer cell then possesses the same tumor-initiating mutations that transformed its ancestor while accumulating new, potentially advantageous mutations through genetic instability. Heterogeneity then is the result of this Darwinian process, with naturally selected cells creating individual regions expressing a unique, advantageous genetic and epigenetic profile. This is, however, not the only explanation for the origins of heterogeneity.
In addition to clonal evolution, the cancer stem cell (CSC) theory can explain tumoral heterogeneity (Figure 1). By the clonal evolution theory, each cancer cell harbors the transformative mutations to make it tumorigenic and able to replicate indefinitely44. In contrast, in 1997, Bonnet et al. presented evidence in acute myeloid leukemia that only a small population of tumor cells are capable of recapitulating the tumor18. Specifically, isolated cancer cells expressing the stem cell marker CD34 formed tumors in xenograft transplants while cells lacking this marker did not18. Therefore, they concluded this small malignant population were “tumor-initiating cells (TICs)” and serve as progenitors for the remaining bulk of cancer cells. This theory contradicts clonal evolution in which all cells possess tumorigenic potential44. In a foundational review, Reya et al. expanded this idea to explain tumor growth mechanics and heterogeneity in all cancers, coining the term ‘cancer stem cell’ and laying the foundations for the CSC theory45.
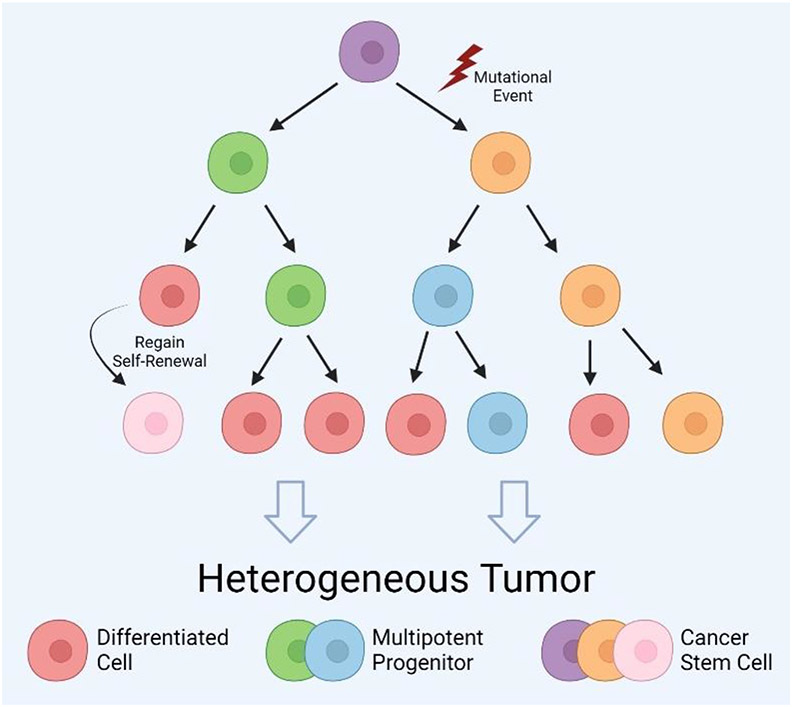
Figure 1. Tumor heterogeneity within glioblastoma lesions.
Tumor heterogeneity can be explained by the cancer stem cell (CSC) theory. In this hierarchical model, a single transformed stem cell (purple) can self-renew to create another CSC and a rapidly cycling multipotent progenitor (blue/green). These progenitors divide to supply the tumor bulk with differentiated cells (red). Heterogeneity arises from the degrees of differentiation through the tumor. The clonal evolution theory can also be incorporated into this framework through mutational events during CSC self-renewal to create a genetically novel CSC (orange) or by dedifferentiation of a differentiated cell back into a CSC (pink). These populations of CSCs and their descendants can then compete through natural selection of growth advantages. Created with BioRender.
According to this theory, CSCs are a rare and uniquely tumorigenic population of cells with the potential to self-renew and differentiate, similar to conventional stem cells46. Mimicking stem cell behavior in organogenesis, CSCs are slow cycling but generate lineage restricted progenitors47,48 that populate the bulk of the tumor with differentiated cells. In this model, the spectrum of differentiation is the source of tumoral heterogeneity and only CSCs can recapitulate it in a new tumor. Recently, the unidirectionality of this hierarchy has been reevaluated due to evidence that differentiated cells can regain stem-like characteristics via modulation of signaling pathways49,50 caused by intrinsic genetic instability or environmental factors. Numerous studies in varying cancer types have shown differentiated tumor cells adopting CSC characteristics through wingless/nuclearization factor kappa B (Wnt/NF-κB) signaling modulation, growth factor release, and during the epithelial to mesenchymal transition (EMT)51-54. This plasticity of differentiated cells has led some to merge the clonal evolution and CSC models55,56, proposing that CSCs can clonally evolve. The new model suggests CSCs can undergo random genetic mutation during cell division, thereby producing heterogeneous CSC populations. Most importantly, the hierarchy is not locked in one direction and differentiated cells can also accumulate novel mutations before regaining self-renewal and making a CSC distinct from its ancestor (Figure 1). These unique CSCs and their progeny can then follow Darwinian processing described by the clonal evolution model. Despite some controversy57-62, CSCs, and stemness in general, is recognized as an emerging hallmark of cancer63.
2.2. Cancer stem cells in glioblastoma
Cancer stem cells were identified in GBM by Singh et al. in 2003 and later termed glioblastoma stem cells (GSCs)64. While no single biomarker has been established to identify these cells, CD133, CD44, SSEA-1, L1CAM, Nestin and others have been used65,66. Satisfying the definition for CSCs, these cells possess self-renewal67,68, differentiate into neuronal, astroglial, oligodendroglial, and even endothelial lineages69-71, and initiate tumorigenesis in xenografts64. Originally, GSCs were believed to arise from transformed neural stem cells (NSCs) as many glioblastomas originate in the compartment housing healthy NSCs, the subventricular zone (SVZ), and they share similar gene expression profiles64,72,73. However, it is now understood NSC transformation is not the only origin for GSCs as differentiated brain cells have been shown to revert to a self-renewing, tumorigenic GSC state74-76. Astrocytes and mature neurons have been induced into GSCs by shRNA knockdown of Nf1 and p5377, expression of Oct4, Klf4, Sox2, c-Myc, and Nanog transcription factors76,78-81, and by environmental cues such as low oxygen availability (hypoxia). Alarmingly, chemotherapies and irradiation have also been shown to trigger dedifferentiation, underscoring the clinical relevance of these populations82-84. While GSCs do lose their stemness in response to differentiation cues like bone morphogenic protein (BMP), cells differentiated from GSCs display incomplete terminal differentiation and more easily reenter the cell cycle or dedifferentiate and form new GSCs, further amplifying heterogeneity85,86 (Figure 2).
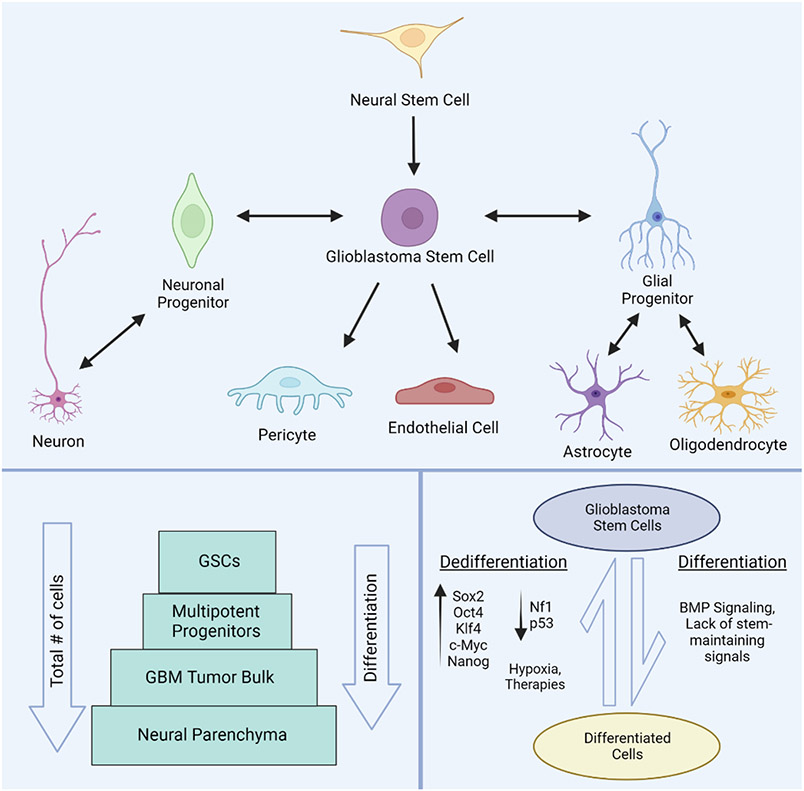
Figure 2. Organization and development of glioblastoma stem cells.
Glioblastoma stem cells (GSCs) are multipotent and capable of differentiation into multiple cell lineages including both neuronal and glial cells. Pericytes and endothelial cells can also be differentiated from GSCs to form the expanding vasculature in GBM tumors. Glioblastoma stem cells can originate from transformed neural stem cells or from dedifferentiation of differentiated brain cells (top panel). Representation of the sub-populations of GSCs relative to other tumor cell constituents and differentiation status (bottom left panel). The conversion between GSCs and differentiated cells within tumors is regulated by a variety of signaling molecules, oncogenes, and environmental conditions (bottom right panel). Created with BioRender.
Glioblastoma stem cells are crucial in tumor expansion, maintenance, and survival. Through an upregulation of genes involved in migration and extracellular matrix degradation, GSCs are more invasive than their differentiated peers87. In addition to expanding tumor boundaries, deep GSC infiltration makes complete surgical resection difficult10 and as few as 50 GSCs are capable of tumor recurrence88. Equally troublingly, GSCs exhibit strong chemo and radioresistance89,90 with high GSC numbers correlating to decreased therapy response and negative outcomes91,92. Recurrent GBMs are therefore enriched in GSCs90,93. Their DNA repair pathways are upregulated and along with their slow cell cycling, GSCs have ample time to repair therapy-induced DNA lesions, negating their cytotoxic effect90,94. Overall, GSCs are implicated in filling the tumor with bulk differentiated cells47,95 and driving tumor growth and survival. To facilitate this progression, these cells both influence and are influenced by their environment.
3. The tumor microenvironment and supporting niches
Tumor cells, non-tumor cells, and other biomolecules are constantly interacting within the bulk tumor and surrounding space defined by the tumor microenvironment (TME). Highly specialized zones within TMEs are defined as specific niches. Many cancers, including GBM, contain three major CSC niches: the perivascular region, the hypoxic zone, and the invasive niche96,97 (Figure 3).
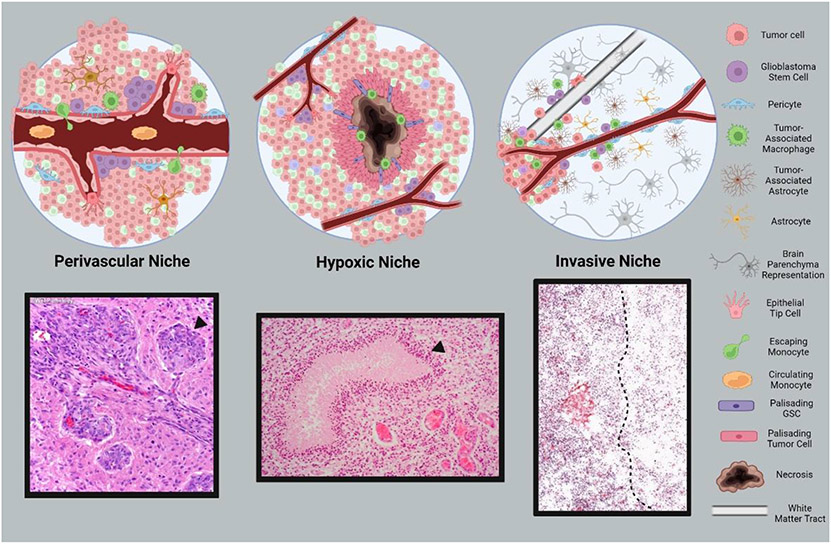
Figure 3. Niches that define the glioblastoma tumor microenvironment.
Defined regions within the tumor microenvironment are contained within specific niches. Glioblastoma tumors contain three major niches including the perivascular niche (left), the hypoxic niche (center), and the invasive niche (right). A variety of cell types and populations comprising each niche are depicted in the top panels. Corresponding histological features of niche components are displayed below each schematic. Denoted are: a glomeruloid microvascular proliferation (bottom left), pseudopalisading cells circumscribing necrosis (bottom center), leading tumor edge (bottom right). Histology images adapted from online resources98-99. Created with BioRender.
3.1. The perivascular niche
As a tumor grows, rapid cellular expansion eventually outpaces existing blood vessels’ supply capacity. New blood vessels are created through the process of angiogenesis to deliver the oxygen and other nutrients required to sustain the existing cell mass and fuel further development. Cells lacking adequate blood supply enter hypoxia, a state of oxygen deprivation, which triggers signaling responses that spur angiogenesis100,101. With a lack of available oxygen, the hypoxia inducible factor (HIF1/2) transcription factors are stabilized and upregulate vascular endothelial growth factor (VEGF), fibroblast growth factor (FGF), and platelet-derived growth factor (PDGF), all important angiogenesis agents102. Release of VEGF triggers matrix metalloproteinases (MMP) to remove the reinforcing pericytes coating blood vessels and induce remodeling103. Signaling molecules from the Notch pathway then cause leading endothelial cells to morph into tendrilled endothelial tip cells which extend in the direction of the VEGF signal, recruiting new endothelial cells and pericytes to construct blood vessels104.
Excessive VEGF-triggered angiogenesis can cause chronic vascular hyperplasia (CVH), a condition where excessive endothelial cell recruitment generates crowded, circular bundles of blood vessels termed glomeruloid microvascular proliferations (GMPs). This aberrant vasculature is a common histopathological feature of GBM often used for diagnosis,105,106. Importantly, because of its rapid growth, the new vascular structures are poorly assembled and prone to leakage and collapse107,108.
Small numbers of Nestin+/CD133+ GSCs are positioned in perivascular niche adjacent to existing vasculature and interact with endothelial cells to exacerbate angiogenesis while maintaining their stemness109. GSCs produce elevated levels of VEGF which accelerates and worsens angiogenesis110. Correspondingly, tumors generated in nude mice using CD133+ cells show increased vascular growth and a higher number of branching points than CD133− tumors110. The perivascular niche may also be a refuge for GSCs during therapies as DNA repair capacity, and therefore resistance, is elevated in this region111.
3.2. The hypoxic niche
While hypoxic stress and necrosis may be expected to inhibit tumor growth, the opposite reaction occurs in GBM which develops a hypoxic niche with pseudopalisading tumor cells surrounding and escaping from a central site of hypoxia-induced necrosis112. The extent of necrosis is positively associated with a more severe prognosis113. Hypoxic conditions greatly support GSC stemness through transcription upregulation of stem-promoting genes such as Sox2, Oct4, NANOG, Klf4 and c-Myc while decreasing differentiation signals like BMPs114-116. Mesenchymal GSCs are especially sensitive to HIF2α signaling through CD44 which may explain the severity of the MES signature117. Hypoxia also pushes GSCs to be more resistant to therapeutics118-121. Following cessation of chemotherapeutics, the surviving GSCs repopulate the tumor and can adopt a therapy-resistant state to evade immune attack and preserve GSC stemness93.
In addition to contributing to hypoxic conditions, porous blood vessels produced during cancer-related angiogenesis enable circulating immune cells, predominantly bone marrow-derived monocytes, to enter the brain112,122. After squeezing through leaky vessels, the tumor environment converts these monocytes into tumor-associated macrophages (TAMs)123,124 which promote oncogenesis, tumor proliferation, and contribute to tumor survival125-128. In addition, necrotic cell death causes proinflammatory signals such as interleukin 6 (IL-6), VEGF, and stromal cell derived factor 1 (SDF-1) to be released in the tumor129,130. These factors promote monocyte polarization into M2 macrophages, imparting immunosuppressive effects and disabling the monocyte’s ability to clear the necrotic debris127,131. Hypoxia itself, through HIF1α activity, improves the immune suppression activity of TAMs and accelerates their polarization132. Release of interleukin 1 (IL-1) by TAMs disrupts the BBB and allows for more immune cell invasion, fueling a vicious cycle of immune cell recruitment, immunosuppression, and tumor development133.
3.3. The invasive niche
At the edge of the growing tumor is the invasive niche, where the tumor contacts normal brain tissue containing astrocytes, neurons, and extracellular matrix (ECM), among other brain features. Astrocytes comprise 50% of brain tissue134,135, and when the CNS is injured, astrocytes convert from their normal quiescent state into a reactive state through astrogliosis136. Reactive astrocytes upregulate the production of growth factors, including VEGF, cytokines (especially IL-6), and MMPs, which help the brain recover from injury by activating numerous pathways including phosphoinositide-3 kinase/protein kinase B (PI3K/AKT), Sonic hedgehog (Shh), p53, and NF-κB137,138. Cell sorting has shown converted astrocyte populations surrounding and inside GBM regions with a unique astrogliosis-associated transcriptome termed tumor-associated astrocytes (TAA)135,139,140. Within TAAs, the same signaling pathway proteins normally responsible for repair post-injury promote GBM invasion and/or proliferation134,141. In-vitro studies have shown an increase in proliferation and invasion when cancer cells are cultured in astrocyte-conditioned media142-144. TAAs also show high levels of connexin-43 (CX43) and Janus kinase/signal transducer and activator of transcription (JAK/STAT) signaling to enhance cell survival by inhibiting apoptosis and promoting immunosuppression145.
The resident GSC population is an active participant in the invasive niche with Nestin+ cells often found on the leading edge146,147. White matter tracts, which are utilized for migration, express Jagged1, activating Notch signaling and upregulating the transcription factor Sox2148 which creates a stemness-favoring environment that encourages GSC migration along the tracks. Many of the products secreted from TAAs also act to enforce cell stemness. For example, Shh signaling activates the Gli transcription factor which promotes stemness and self-renewal149. Upregulation of IL-6 and STAT3 signaling by TAAs is critical for stem maintenance, while STAT3 knockdown eliminates GSC multipotency and proliferation150,151. Macroscopically, the larger the area GBM invades, the more GSCs escape resection. As GBM can be revived by small colonies of GSCs even after treatment, the expansion of GSCs outside the tumor bulk will allow relapse.
GBM niches are highly complex and have been the focus of much research. Significantly, niches are dynamic in their temporal and spatial properties. As the invasive edge expands outward, new perivascular niches are assembled in its wake and can eventually expand the necrotic region. As niches change during GBM progression, GSCs accumulate and fluctuate, especially when exposed to therapies. The robust nature of GSCs within the different niches are thought to improve tumor cell survival and assist with therapeutic resistance.
4. Therapy evasion and resistance
4.1. Ionizing radiation resistance
As a complement to surgical resection for GBM treatment, radiation and chemotherapeutics are employed to preferentially inflict DNA damage to cancer cells. Cancer cells are vulnerable to DNA damage due to their inherent genomic instability, DNA stress from unrestrained proliferation, and mutated damage response proteins152. Ionizing radiation (IR) therapy bombards the tumor area with high energy particles that directly damage the DNA backbone153. Additionally, the high energy particles instantaneously create reactive oxygen species (ROS),such as hydroxyl radicals, inside cells and trigger mitochondria to increase their own ROS production154. These radicals oxidize DNA bases to form double stranded breaks (DSBs), a form of damage monitored by the ataxia telangiectasia mutated (ATM) kinase. Upon encountering DSBs, ATM kinase is phosphorylated and helps facilitate cell cycle arrest at the G1/S phase, utilizing cell cycle checkpoint 2 (CHK2) and p53155. Additionally, ATM acts as a scaffold to recruit repair elements to the DSB with an increase in phosphorylated ATM associated with better DNA repair155. A parallel system using ATM and Rad3-related protein (ATR) and CHK1 acts on single strand breaks (SSBs) in DNA and regulates entrance from G2 into M phase. Both axes pause the cell cycle for DNA damage repair and to prevent apoptosis. While elevated DNA damage fuels genomic instability, once the cancer phenotype is achieved, tumor cells rely more strongly on repair mechanisms for survival156,157. The diverse genetic makeup of GBM means that radioresistance in cells will also be variable in nature.
It’s no surprise that GSCs are a main culprit of radioresistance as they possess a greater ability to survive than their more differentiated counterparts. Following IR dosing, GSCs, identified as CD133+ cells, exhibited less apoptosis, higher ATM kinase activity, and an equal tumor-forming capacity as non-irradiated CD133+ cells90,158. Other upregulated repair genes in GSCs includes ATR, CHK1, and Poly ADP ribose polymerase (PARP1)158,159. As a result, it is common following IR therapy to see an enrichment in CD133+ populations in the tumor environment160. Since these cells maintain their differentiation capabilities, they can proceed to repopulate the tumor91,161. This new population often retains radioresistance from their surviving GSC ancestors. Finally, autophagy is suspected to play a role in radioresistance. As a defense mechanism, autophagy is triggered by cell stress such as hypoxia or elevated ROS and markers for autophagy such as LP3 and ATG5 and ATG12 increase in CD133+ cells following IR therapy162.
4.2. Chemotherapeutic resistance
In addition to IR therapy, GBM cells are often resistant to chemotherapies, including temozolomide (TMZ), the foundational adjuvant chemotherapeutic prescribed for GBM. Capable of penetrating the BBB, TMZ is an alkylating agent that interacts with DNA to preferentially methylate the N3 position of adenine and the N7 and O6 positions of guanine163. Creation of O6-methylguanine, although only 5-10% of the modifications made by TMZ, is the most cytotoxic of TMZ’s effects163. O6-methylguanine leads to a nucleotide mismatch during DNA replication where thymine is incorrectly inserted instead of cytosine. During repeated cycles of mismatch repair, DNA breaks are generated, causing cell cycle arrest at the G2/M transition and eventual apoptosis164.
The most well characterized and arguably most important mechanism of alkylating agent resistance occurs through O6-methylguanine-DNA methyltransferase (MGMT), a mismatch repair protein that repairs the damage caused by TMZ. Unfortunately, MGMT is one of the most common differentially expressed proteins in GBM tumors, meaning that many patients possess some immunity against TMZ treatment. In fact, because of the prevalence of MGMT upregulation, TMZ effect is reduced in ~50% of patients163 and GSCs possess greater MGMT and DDR mechanisms, making them more tolerant to TMZ treatment94,165. Tumors with diminished MGMT show better upfront reaction to TMZ and exhibit longer progression-free survival. Decreased levels of MGMT have been shown to be related to methylation status of the MGMT promoter, with a methylated promoter transcribing less MGMT and an unmethylated promoter more166. This epigenetic marker can be easily identified using methylation-specific PCR and is now considered a useful and accurate predictor of TMZ efficacy167,168.
4.3. Resistance via senescence
Some cancer cells do not undergo apoptosis in response to therapy-induced DNA damage and instead adopt therapy induced senescence (TIS)169. Senescent cells exit the cell cycle at the G2/M phase and upregulate DNA repair and apoptosis survival signals170. While conventional understanding suggests senescence is permanent, some studies indicate senescent tumor cells can reenter the cell cycle to become more aggressive171. During senescence, tumor cells secrete cytokines, chemokines, matrix metalloproteinases, and growth factors collectively known as the senescence-associated secretory phenotype (SASP)172 (Figure 4). In addition to fueling proliferation and invasion, the SASP also assists with GSC maintenance by activating Wnt signaling to maintain self-renewal22,173. The SASP can reprogram non-senescent cells to possess stem cell like qualities by releasing factors that modulate the transcription factors Oct4, Sox2, Klf4, and c-Myc, as seen in other cancers22,173-175.
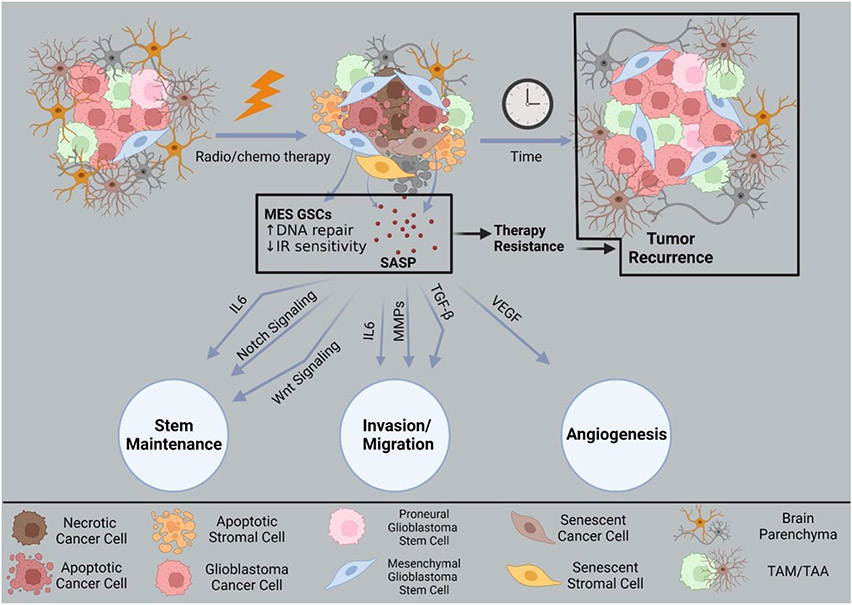
Figure 4. Therapy evasion of glioblastoma cells through senescence.
Glioblastoma cells (top left) that resist radio/chemotherapy are enriched for therapy-resistant mesenchymal (MES) signatures. Some assume therapy induced senescence, bypassing normal apoptosis processes. Senescent tumor cells (top middle) over time can recur to give rise to new masses (top right). During senescence, cells secrete cytokines, chemokines, matrix metalloproteinases, and other growth factors known as the senescence-associated secretory phenotype (SASP). These collective signals support stem maintenance by activating the Wnt pathway. Factors from the SASP can also contribute to invasion, migration, and angiogenesis supporting tumor recurrence. Created with BioRender.
5. Therapeutic avenues and clinical trials
5.1. Receptor tyrosine kinase signaling as a therapeutic target
Targeting the molecular pathways implicated in GBM pathology is a promising avenue for therapy development (summarized in Table 1). Proteins of interest include receptor tyrosine kinases, stem maintenance factors, and angiogenic markers. Crosstalk among these pathways is also an important consideration since the combination and balance/imbalance of signaling effects drives cell growth, proliferation, stemness, immune evasion, and invasion. Among the most appealing targets are the receptor tyrosine kinases (RTKs) such as EGFR and its downstream effector PI3K/AKT. Malignant amplification of these proteins is common in GBM cells with EGFR mutations occurring in 57.4% of GBM27. Enhanced EGFR signaling in GBM supports proliferation, angiogenesis, invasion, and survival. Small molecule RTK inhibitors that target receptors like EGFR have shown promise in other cancer types along with GBM176,177. Erlotinib is one such FDA approved RTK inhibitor for non-small cell lung cancer where it improved progression-free survival178. In combination with TMZ and IR therapy, Erlotinib showed some improvement in overall survival for newly diagnosed GBM patients, although its use as a monotherapy falls short179,180. Another promising EGFR inhibitor, Gefitinib, showed a decrease in EGFR phosphorylation, but this reduction did not translate to a reduction in tumor severity181. A combination of both Erlotinib and Gefitinib, however, was shown to be ineffective in GBM tissue samples182. Similarly, second-generation EGFR inhibitors Dacomitinib and Afatinib failed to show meaningful improvement as single-agent treatments183,184. A third-generation EGFR inhibitor, AZD9291 (osimertinib), was approved by the FDA for non-small cell lung cancer and is being evaluated for GBM with positive preclinical results185-187.
Table 1.
Summary of molecular drug therapies in glioblastoma treatment.
| Pathway | Drug therapy | GBM status and treatment |
Efficacy | Ref | Clinical Trial ID |
|---|---|---|---|---|---|
| EGFR (drug) | Erlotinib | Newly diagnosed and recurrent | Minimally effective as a mono therapy | Raizer180 | NCT00045110 |
| Erlotinib + IR + TMZ | Newly diagnosed | Improved PFS and OS | Prados179 | NCT00187486 | |
| Gefitinib | Recurrent | Ineffective in reducing EGFR pathway | Hegi181 | NCT00250887 | |
| Dacomitinib | Recurrent | Ineffective as a monotherapy | Sepúlveda-Sánchez183 | NCT01520870 | |
| Afatinib | Recurrent | Moderate improvement for EGFRvIII+ patients | Reardon184 | NCT00727506 | |
| AZD9291 (osimertinib) | Advanced NSCLC with EGFR inhibitor resistance | Improved PFS, highly active | Jänne185 | NCT01802632 | |
| EGFR (antibody) | Cetuximab | Recurrent | Ineffective as a monotherapy, minority saw modest OS improvement | Belda-Iniesta, Neyns188,189 | [N/A] |
| Nimotuzumab + IR + TMZ | Newly diagnosed | Improved PFS and OS | She, Du190,191 | [N/A] | |
| PI3K | Buparlisib | Recurrent | Minimally effective as a monotherapy | Wen199 | NCT01339052 |
| Buparlisib + Bevacuzimab | Recurrent | No additional effect compared to Bevacuzimab alone | Hainsworth200 | NCT01349660 | |
| PX-866 | First recurrence | No reduction in PI3K signaling | Pitz203 | NCT01259869 | |
| AKT | Perifosine | Recurrent | Ineffective as a monotherapy | Kaley208 | NCT00590954 |
| Perifosine + Temsirolimus | Recurrent | Ongoing | Lassman182 | NCT02238496 | |
| mTOR | RMC-5552 | Recurrent | Ongoing | Burnett207 | NCT05557292 |
| Temsirolimus + Erlotinib | Recurrent | High toxicity, no reduction in AKT pathway | Wen209 | NCT00112736 | |
| Everolimus + IR + TMZ | Newly diagnosed | Moderate toxicity, no appreciable survival benefit | Ma210 | NCT00553150 | |
| Notch | MK-0752 | Advanced solid tumors | Variable induction of stable disease | Krop217 | [N/A] |
| RO4929097 + IR + TMZ | Newly diagnosed | Tolerable toxicity, CD133+ cells diminished | Xu219 | NCT01119599 | |
| RO4929097 | Recurrent | Ineffective as a monotherapy | Peereboom262 | NCT01122901 | |
| Wnt/COX2 | Celecoxib + low dose TMZ | Recurrent | Improved benefit compared to IR + TMZ alone | Stockhammer231 | [N/A] |
| Shh | Glasdegib + IRT + TMZ | Newly diagnosed | Promising preliminary effects | Vaz263 | NCT03466450 |
| Vismodegib + IR + TMZ | Newly diagnosed w/nonmethylated MGMT | Ongoing | Wick239 | NCT03158389 | |
| Vismodegib | Recurrent GBM | Ineffective as monotherapy | Sloan264 | NCT00980343 | |
| VEGF | Bevacuzimab + IR | Recurrent | Improved PFS, no improved OS | Tsien265 | NCT01730950 |
| Proteosome | Bortezimab + IR + TMZ | Newly diagnosed | Effective for MGMT methylated GBM | Kong261 | NCT00998010 |
| PARPi | Olaparib + Cediranib maleate | Recurrent | No improved benefit to Bevacuzimab alone | Arrillaga-Romany266 | NCT02974621 |
| Olaparib + IR + TMZ | Newly diagnosed | Ongoing | Leseur254 | NCT03212742 |
In addition to small molecule inhibitors, monoclonal antibodies against EGFR have also been explored, namely Cetuximab. Although most patients experienced minimal benefit, a small population showed some improvement, though the molecular underpinnings of the improvements are unclear188,189. Combined with standard therapy, the monoclonal antibody Nimotuzumab increased survival and tolerability in Phase II trials of MGMT-methylated GBM190,191. Due to their size, antibody-based therapies have difficulty breaching the BBB with only 0.1-0.2% of circulating antibodies showing BBB penetration192. New engineering approaches to overcome this limitation are themselves a novel field for therapeutic development193-195.
The PI3K/AKT/mTOR pathway is another popular frontier for drug design. Buparlisib (BKM-120) and PX-866 are two pan-PI3K inhibitors that affect multiple PI3K isoforms. Buparlisib induces apoptosis through mitotic disfunction and reduced tumor growth while increasing survival in mice with xenografted GBM tumors196,197. In recent clinical trials, the drug was deemed to be less effective198,199. Buparlisib is still being explored as a combination therapy alongside the VEGF inhibitor, Bevacuzimab200 and the PARP inhibitor Rucaparib201. Similarly, PX-866 demonstrated powerful effects in GBM cells but came up short in clinical trials with a similarly vexing absence of explanation202,203. Another avenue being explored is targeting individual PI3K isoforms, as it has been suggested that each isoform plays stronger roles in certain cancer characteristics204. Individually targeting one isoform may be specific enough to avoid redundancies while maintaining therapeutic efficacy205,206. Downstream from PI3K targets, new mTOR inhibitors are being developed for clinical trials207. They may have value in a combination therapy if monotherapies are unsuccessful. The well-tolerated drug, Perifosine, targets AKT and is under investigation in combination with PI3K and mTOR inhibitors208. Drugs like Temsirolimus and Everolimus that target mTOR suffer from the same theme plaguing most inhibitors of this pathway: monotherapies fail due to insufficient intertumoral drug levels or compensatory signaling209,210.
5.3. Notch, Wnt and Sonic hedgehog targets
Other potential targets in GSC maintenance include Notch, Wnt, and Shh. Notch signaling is part of neural stem cell compartmentalization, preventing apoptosis and ensuring a stock of stem cells during brain development211. The Notch pathway also has a protective role in GSC self-renewal211. Gamma-secretase inhibitors (GSIs) are used to inhibit activation of the Notch signaling pathway and early reports indicate their effectiveness at depleting GSCs or sensitizing them to radiotherapy212-214. Clinical trials using the GSI MK-0752 for refractory CNS malignancies (including GBM) showed good tolerance and favorable Notch inhibition215-217. Another Notch inhibitor, RO4929097, successfully induced differentiation and decreased self-renewal in GSCs in orthotropic mouse models218. In a Phase 0/1 combinatorial trial of RO4929097 with TMZ and radiotherapy, Notch signaling was diminished and explanted tumors showed decreased CD133+ markers. Gene expression analysis of recurrent tumors in the treatment group revealed adaptive Notch downregulation but the upregulation of mesenchymal genes and angiogenic factors as compensatory mechanisms219. The modulation of Notch with MK-0752 affected PI3K/AKT while RO4929097 altered VEGF levels220,221, a good indication that targeting several pathways simultaneously may be a productive path forward. Notably, GSIs have the potential to impair Notch regulation in healthy cells as well as in tumor cells222,223. As such, new generations of GSIs are being developed and tested in GBM cell lines224 with the hope of reducing impacts on healthy tissue.
Another upregulated pathway in GBM is the Wnt network, having roles in stem maintenance, therapy evasion, proliferation, and invasion225-227. Due to crosstalk between Wnt and cyclooxygenase-2 (COX-2) pathways, non-steroidal anti-inflammatory drugs (NSAIDs) that inhibit COX-2 also suppress Wnt signaling228,229. Small molecule inhibition with the NSAID celecoxib sensitized GBM tumors to TMZ by regulating the expression of MGMT225. Celecoxib was also effective at sensitizing tumor-derived GSCs to radiotherapy and increased the mean survival rates of mice with xenografted tumors230. In one clinical trial of 28 patients with recurrent GBM, treating with celecoxib and continuous low dose TMZ showed improvements compared to alternating TMZ dosage, with median survival time rising from 11.1 months to 16.8 months231. An additional clinical trial incorporating celecoxib in a multi-drug cocktail accompanied by TMZ showed anti-neoplastic effect, though the effect of celecoxib in this result is unreported232. Moreover, Wnt inhibitors continue to be explored in other solid tumors233.
Sonic hedgehog signaling mediates stem cell differentiation, and its overexpression is implicated in sustaining GSCs, providing therapy resistance, and promoting GBM invasion234-236. The transcriptional effector Gli1 in Shh cross-talks with the PI3K/AKT and VEGF pathways237. Clement et al. sensitized GSCs to TMZ through blockade of Shh using cyclopamine in GBM cell lines149. A dual approach with a Shh pathway inhibitor, NVP-LDE-225, and a PI3K/mTOR inhibitor, NVP-BEZ-235, diminished proliferation, growth, and stemness of isolated GSCs238, demonstrating the benefits of Shh inhibition. Furthermore, another Shh inhibitor, Vismodegib, is among five drugs under investigation in the NCT Master Match, an ongoing clinical trial for newly diagnosed GBM patients for whom TMZ is excluded239. Vismodegib has also showed promising anti-cancer activity in medulloblastoma clinical trials240.
5.4. Targeting angiogenesis and DNA repair
Angiogenesis targets such as VEGF are highly appealing to minimize nutrient delivery to GBM tumors. The humanized monoclonal antibody, Bevacuzimab, binds the VEGF ligand to prevent it from interacting with cell-surface receptors241. In a Phase II clinical trial, Bevacuzimab elicited a decrease in tumor size, lessened cerebral edema, and offered longer benefits to patients with recurrent GBM242. In light of this success, the FDA granted Bevacuzimab accelerated approval as both a monotherapy and in combination with the cytotoxic agent Irinotecan243. Full authorization was granted in 2017. Some GBM subtypes may decrease Bevacizumab efficacy in GBM as GSCs persist to enable therapy resistance and invasion244-246. Anti-angiogenesis treatments also enhance delivery of other therapeutics by repairing the leaky vessels needed to transport drugs to the tumor247. For example, the addition of an anti-VEGF agent was shown to improve delivery of immunotherapeutic CAR-T cells to tumors in mice248. This is especially valuable to reach the outer, invasive edges of the tumor249. Importantly, appropriate dosing is needed to balance angiogenesis prevention as over-inhibition will create necrosis through lack of nutrient supply and trigger increased tumor invasion and worse prognosis250,251. Despite potential risks, normalization of the vasculature through anti-angiogenic means continues to be a promising avenue to improve chemotherapy252.
Finally, DNA repair mechanisms are being explored as an approach for most cancer treatments including GBM. By disabling the machinery needed to overcome DNA damage, GBM tumors are sensitized to the cytotoxic effects of alkylating agents such as TMZ and the nitrosureas, lomustine and carmustine. PARP inhibitors have also become a popular sensitizing agent under considerable investigation. These agents disable the base excision repair mechanism that GSCs rely on for single strand break repair and improve radiotherapy effectiveness253,254. Efficacy in xenografts depends on the ability of PARP inhibitors to penetrate the BBB255,256. In clinical trials, the PARP inhibitor ABT-888 failed to overcome TMZ resistant in GBM, but modifications to PARP treatment remains an active area of research257,258. Likewise, MGMT is a main culprit chemoresistance and is a popular DNA repair target being explored for combinatorial treatments. Drug-induced knockdown of MGMT may improve outcomes for patients with unmethylated MGMT. In addition, proteosome inhibitors, such as Bortezomib, can deplete MGMT levels leading to prolonged survival in mouse models with unmethylated MGMT promoters259,260. An initial clinical trial showed adequate tolerance and efficacy in newly diagnosed GBM, though MGMT-methylated patients show a greater effect261.
6. Conclusions
Many challenges remain as we seek to reduce the burden of GBM in newly diagnosed patients and those experiencing recurrence. Historically recognized hurdles such as heterogeneity and therapy resistance have been reexamined with the discovery of cancer stem cells. The development of the cancer stem cell theory offers putative explanation for tumor initiation and the origin and prevalence of the highly damaging heterogeneity observed in GBM. Through this heterogeneity, therapies are less universally effective. Inclusion of GSCs into the GBM model also offers further explanations for GBM’s radiation and chemotherapeutic evasion as GSCs possess improved DNA repair mechanisms, a slower cell cycle, and reversible senescence phenotype, all of which allow them to overcome therapies' cytotoxic effects.
As modern paradigms in cancer biology implicate GSCs in sustaining incurable GBMs, research aimed at eliminating this sub-population is imperative. New therapeutic approaches targeting GSCs alongside bulk tumor cells may enhance treatment outcomes. Many clinical trials have been performed and more are currently underway that target pathways critical to GSC function such as RTKs, angiogenesis, DNA repair and the stem-maintenance pathways Notch, Wnt, and Shh. Compared to other cancer types, brain tumors have to navigate the restrictive BBB which limits the delivery of novel drugs, but new drug delivery may facilitate penetrance across the BBB. As a result, therapies that prove effective in eliminating GSCs can likely be easily applied to CSCs in other cancer types.
Finally, new pharmacological discovery tools are needed to close the gap between cellular success stories and those of clinical trials. Better structure-activity-relationship predictive tools are imperative to elevate rational drug design based on native protein structures. The combined use of automated drug screening tools and cryo-Electron Microscopy (EM) structure determination creates a new era in therapeutic development. As many protein structures are now amenable to three-dimensional analysis at high-resolution, the next wave of drug design may include pinpointing protein binding sites to improve inhibitors of signaling pathways or DNA repair. Overall, improved outcomes for GBM patients are expected through the use of new platforms to target GSC-specific properties at the nanoscale. Future research efforts aimed at eliminating GSC self-renewal is a necessary consideration for scientists and clinicians working in the GBM field.
Acknowledgements.
The authors thank Maria J. Solares and Liam Kaylor for their insights and help in the initial stages of the manuscript development.
Funding statement.
The work was funded by the Center for Structural Oncology supported through the Huck Institutes of the Life Sciences at Pennsylvania State University.
Footnotes
Conflict of interest statement.
The authors have no conflicts of interest to declare.
References:
- 1.Rock K, Mcardle O, Forde P, et al. A clinical review of treatment outcomes in glioblastoma multiforme—the validation in a non-trial population of the results of a randomised Phase III clinical trial: has a more radical approach improved survival? Br J Radiol. 2012;85(1017):e729–e733. doi: 10.1259/bjr/83796755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hanif F, Muzaffar K, Perveen K, Malhi SM, Simjee SU. Glioblastoma Multiforme: A Review of its Epidemiology and Pathogenesis through Clinical Presentation and Treatment. Asian Pac J Cancer Prev APJCP. 2017;18(1):3–9. doi: 10.22034/APJCP.2017.18.1.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ostrom QT, Gittleman H, Liao P, et al. CBTRUS Statistical Report: Primary Brain and Central Nervous System Tumors Diagnosed in the United States in 2007–2011. Neuro-Oncol. 2014;16(suppl_4):iv1–iv63. doi: 10.1093/neuonc/nou223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330 [DOI] [PubMed] [Google Scholar]
- 5.Schapira AHV. Neurology and Clinical Neuroscience E-Book. Elsevier Health Sciences; 2006 [Google Scholar]
- 6.Kim J, Lee IH, Cho HJ, et al. Spatiotemporal Evolution of the Primary Glioblastoma Genome. Cancer Cell. 2015;28(3):318–328. doi: 10.1016/j.ccell.2015.07.013 [DOI] [PubMed] [Google Scholar]
- 7.Weller M, van den Bent M, Hopkins K, et al. EANO guideline for the diagnosis and treatment of anaplastic gliomas and glioblastoma. Lancet Oncol. 2014;15(9):e395–e403. doi: 10.1016/S1470-2045(14)70011-7 [DOI] [PubMed] [Google Scholar]
- 8.Agarwal S, Mittapalli RK, Zellmer DM, et al. Active Efflux of Dasatinib from the Brain Limits Efficacy against Murine Glioblastoma: Broad Implications for the Clinical Use of Molecularly Targeted Agents. Mol Cancer Ther. 2012;11(10):2183–2192. doi: 10.1158/1535-7163.MCT-12-0552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Overcoming the blood–brain tumor barrier for effective glioblastoma treatment - ClinicalKey. Accessed April 15, 2023. https://www.clinicalkey.com/#!/content/playContent/1-s2.0-S1368764615000126?returnurl=null&referrer=null [DOI] [PubMed]
- 10.Shergalis A, Bankhead A, Luesakul U, Muangsin N, Neamati N. Current Challenges and Opportunities in Treating Glioblastoma. Pharmacol Rev. 2018;70(3):412–445. doi: 10.1124/pr.117.014944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Della Monica R, Cuomo M, Buonaiuto M, et al. MGMT and Whole-Genome DNA Methylation Impacts on Diagnosis, Prognosis and Therapy of Glioblastoma Multiforme. Int J Mol Sci. 2022;23(13):7148. doi: 10.3390/ijms23137148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Etcheverry A, Aubry M, de Tayrac M, et al. DNA methylation in glioblastoma: impact on gene expression and clinical outcome. BMC Genomics. 2010;11(1):701. doi: 10.1186/1471-2164-11-701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bonavia R, Inda M del M, Cavenee WK, Furnari FB. Heterogeneity Maintenance in Glioblastoma: A Social Network. Cancer Res. 2011;71(12):4055–4060. doi: 10.1158/0008-5472.CAN-11-0153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parker NR, Khong P, Parkinson JF, Howell VM, Wheeler HR. Molecular Heterogeneity in Glioblastoma: Potential Clinical Implications. Front Oncol. 2015;5. Accessed April 27, 2023. https://www.frontiersin.org/articles/10.3389/fonc.2015.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Heppner GH, Miller BE. Tumor heterogeneity: biological implications and therapeutic consequences. Cancer Metastasis Rev. 1983;2(1):5–23. doi: 10.1007/BF00046903 [DOI] [PubMed] [Google Scholar]
- 16.Dick JE. Breast cancer stem cells revealed. Proc Natl Acad Sci. 2003;100(7):3547–3549. doi: 10.1073/pnas.0830967100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O’Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007;445(7123):106–110. doi: 10.1038/nature05372 [DOI] [PubMed] [Google Scholar]
- 18.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730–737. doi: 10.1038/nm0797-730 [DOI] [PubMed] [Google Scholar]
- 19.Batlle E, Clevers H. Cancer stem cells revisited. Nat Med. 2017;23(10):1124–1134. doi: 10.1038/nm.4409 [DOI] [PubMed] [Google Scholar]
- 20.Ayob AZ, Ramasamy TS. Cancer stem cells as key drivers of tumour progression. J Biomed Sci. 2018;25(1):20. doi: 10.1186/s12929-018-0426-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Najafi M, Farhood B, Mortezaee K. Cancer stem cells (CSCs) in cancer progression and therapy. J Cell Physiol. 2019;234(6):8381–8395. doi: 10.1002/jcp.27740 [DOI] [PubMed] [Google Scholar]
- 22.Milanovic M, Fan DNY, Belenki D, et al. Senescence-associated reprogramming promotes cancer stemness. Nature. 2018;553(7686):96–100. doi: 10.1038/nature25167 [DOI] [PubMed] [Google Scholar]
- 23.McLendon R, Friedman A, Bigner D, et al. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Verhaak RGW, Hoadley KA, Purdom E, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parsons DW, Jones S, Zhang X, et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science. 2008;321(5897):1807. doi: 10.1126/science.1164382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Patel AP, Tirosh I, Trombetta JJ, et al. Single-cell RNA-seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396–1401. doi: 10.1126/science.1254257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brennan CW, Verhaak RGW, McKenna A, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155(2):462–477. doi: 10.1016/j.cell.2013.09.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol (Berl). 2016;131(6):803–820. doi: 10.1007/s00401-016-1545-1 [DOI] [PubMed] [Google Scholar]
- 29.Ohgaki H, Kleihues P. Genetic Pathways to Primary and Secondary Glioblastoma. Am J Pathol. 2007;170(5):1445–1453. doi: 10.2353/ajpath.2007.070011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han S, Liu Y, Cai SJ, et al. IDH mutation in glioma: molecular mechanisms and potential therapeutic targets. Br J Cancer. 2020;122(11):1580–1589. doi: 10.1038/s41416-020-0814-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nobusawa S, Watanabe T, Kleihues P, Ohgaki H. IDH1 Mutations as Molecular Signature and Predictive Factor of Secondary Glioblastomas. Clin Cancer Res. 2009;15(19):6002–6007. doi: 10.1158/1078-0432.CCR-09-0715 [DOI] [PubMed] [Google Scholar]
- 32.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 Mutations in Gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Louis DN, Perry A, Wesseling P, et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro-Oncol. 2021;23(8):1231–1251. doi: 10.1093/neuonc/noab106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019 [DOI] [PubMed] [Google Scholar]
- 35.Sottoriva A, Spiteri I, Piccirillo SGM, et al. Intratumor heterogeneity in human glioblastoma reflects cancer evolutionary dynamics. Proc Natl Acad Sci. 2013;110(10):4009–4014. doi: 10.1073/pnas.1219747110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bergmann N, Delbridge C, Gempt J, et al. The Intratumoral Heterogeneity Reflects the Intertumoral Subtypes of Glioblastoma Multiforme: A Regional Immunohistochemistry Analysis. Front Oncol. 2020;10. Accessed February 20, 2023. https://www.frontiersin.org/articles/10.3389/fonc.2020.00494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Swanton C. Intratumor Heterogeneity: Evolution through Space and Time. Cancer Res. 2012;72(19):4875–4882. doi: 10.1158/0008-5472.CAN-12-2217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Becker AP, Sells BE, Haque SJ, Chakravarti A. Tumor Heterogeneity in Glioblastomas: From Light Microscopy to Molecular Pathology. Cancers. 2021;13(4):761. doi: 10.3390/cancers13040761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang J, Cazzato E, Ladewig E, et al. Clonal evolution of glioblastoma under therapy. Nat Genet. 2016;48(7):768–776. doi: 10.1038/ng.3590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fedele M, Cerchia L, Pegoraro S, Sgarra R, Manfioletti G. Proneural-Mesenchymal Transition: Phenotypic Plasticity to Acquire Multitherapy Resistance in Glioblastoma. Int J Mol Sci. 2019;20(11):2746. doi: 10.3390/ijms20112746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segerman A, Niklasson M, Haglund C, et al. Clonal Variation in Drug and Radiation Response among Glioma-Initiating Cells Is Linked to Proneural-Mesenchymal Transition. Cell Rep. 2016;17(11):2994–3009. doi: 10.1016/j.celrep.2016.11.056 [DOI] [PubMed] [Google Scholar]
- 42.Nowell PC. The Clonal Evolution of Tumor Cell Populations. Science. 1976;194(4260):23–28. doi: 10.1126/science.959840 [DOI] [PubMed] [Google Scholar]
- 43.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481(7381):306–313. doi: 10.1038/nature10762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shackleton M, Quintana E, Fearon ER, Morrison SJ. Heterogeneity in Cancer: Cancer Stem Cells versus Clonal Evolution. Cell. 2009;138(5):822–829. doi: 10.1016/j.cell.2009.08.017 [DOI] [PubMed] [Google Scholar]
- 45.Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414(6859):105–111. doi: 10.1038/35102167 [DOI] [PubMed] [Google Scholar]
- 46.Clarke MF, Dick JE, Dirks PB, et al. Cancer Stem Cells—Perspectives on Current Status and Future Directions: AACR Workshop on Cancer Stem Cells. Cancer Res. 2006;66(19):9339–9344. doi: 10.1158/0008-5472.CAN-06-3126 [DOI] [PubMed] [Google Scholar]
- 47.Lan X, Jörg DJ, Cavalli FMG, et al. Fate mapping of human glioblastoma reveals an invariant stem cell hierarchy. Nature. 2017;549(7671):227–232. doi: 10.1038/nature23666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jackson M, Hassiotou F, Nowak A. Glioblastoma stem-like cells: at the root of tumor recurrence and a therapeutic target. Carcinogenesis. 2015;36(2):177–185. doi: 10.1093/carcin/bgu243 [DOI] [PubMed] [Google Scholar]
- 49.Takahashi K, Yamanaka S. Induction of Pluripotent Stem Cells from Mouse Embryonic and Adult Fibroblast Cultures by Defined Factors. Cell. 2006;126(4):663–676. doi: 10.1016/j.cell.2006.07.024 [DOI] [PubMed] [Google Scholar]
- 50.Visvader JE, Lindeman GJ. Cancer Stem Cells: Current Status and Evolving Complexities. Cell Stem Cell. 2012;10(6):717–728. doi: 10.1016/j.stem.2012.05.007 [DOI] [PubMed] [Google Scholar]
- 51.Schwitalla S, Fingerle AA, Cammareri P, et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell. 2013;152(1-2):25–38. doi: 10.1016/j.cell.2012.12.012 [DOI] [PubMed] [Google Scholar]
- 52.Vermeulen L, De Sousa E Melo F, van der Heijden M, et al. Wnt activity defines colon cancer stem cells and is regulated by the microenvironment. Nat Cell Biol. 2010;12(5):468–476. doi: 10.1038/ncb2048 [DOI] [PubMed] [Google Scholar]
- 53.Pradella D, Naro C, Sette C, Ghigna C. EMT and stemness: flexible processes tuned by alternative splicing in development and cancer progression. Mol Cancer. 2017;16(1):8. doi: 10.1186/s12943-016-0579-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang S sha, Jiang J, Liang X hua, Tang Y ling. Links between cancer stem cells and epithelial ndash; mesenchymal transition. OncoTargets Ther. 2015;8:2973–2980. doi: 10.2147/OTT.S91863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kreso A, Dick JE. Evolution of the Cancer Stem Cell Model. Cell Stem Cell. 2014;14(3):275–291. doi: 10.1016/j.stem.2014.02.006 [DOI] [PubMed] [Google Scholar]
- 56.Wang W, Quan Y, Fu Q, et al. Dynamics between Cancer Cell Subpopulations Reveals a Model Coordinating with Both Hierarchical and Stochastic Concepts. PLOS ONE. 2014;9(1):e84654. doi: 10.1371/journal.pone.0084654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dirkse A, Golebiewska A, Buder T, et al. Stem cell-associated heterogeneity in Glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat Commun. 2019;10(1):1787. doi: 10.1038/s41467-019-09853-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.van Niekerk G, Davids LM, Hattingh SM, Engelbrecht AM. Cancer stem cells: A product of clonal evolution? Int J Cancer. 2017;140(5):993–999. doi: 10.1002/ijc.30448 [DOI] [PubMed] [Google Scholar]
- 59.Meacham CE, Morrison SJ. Tumour heterogeneity and cancer cell plasticity. Nature. 2013;501(7467):328–337. doi: 10.1038/nature12624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hung KF, Yang T, Kao SY. Cancer stem cell theory: Are we moving past the mist? J Chin Med Assoc. 2019;82(11):814. doi: 10.1097/JCMA.0000000000000186 [DOI] [PubMed] [Google Scholar]
- 61.Yoo MH, Hatfield DL. The cancer stem cell theory: Is it correct? Mol Cells. 2008;26(5):514–516. [PMC free article] [PubMed] [Google Scholar]
- 62.Wang J, Sakariassen PØ, Tsinkalovsky O, et al. CD133 negative glioma cells form tumors in nude rats and give rise to CD133 positive cells. Int J Cancer. 2008;122(4):761–768. doi: 10.1002/ijc.23130 [DOI] [PubMed] [Google Scholar]
- 63.Hanahan D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022;12(1):31–46. doi: 10.1158/2159-8290.CD-21-1059 [DOI] [PubMed] [Google Scholar]
- 64.Singh SK, Clarke ID, Terasaki M, et al. Identification of a Cancer Stem Cell in Human Brain Tumors. Cancer Res. 2003;63(18):5821–5828. [PubMed] [Google Scholar]
- 65.Brescia P, Richichi C, Pelicci G. Current Strategies for Identification of Glioma Stem Cells: Adequate or Unsatisfactory? J Oncol. 2012;2012:e376894. doi: 10.1155/2012/376894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tang X, Zuo C, Fang P, et al. Targeting Glioblastoma Stem Cells: A Review on Biomarkers, Signal Pathways and Targeted Therapy. Front Oncol. 2021;11. Accessed February 20, 2023. https://www.frontiersin.org/articles/10.3389/fonc.2021.701291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gimple RC, Bhargava S, Dixit D, Rich JN. Glioblastoma stem cells: lessons from the tumor hierarchy in a lethal cancer. Genes Dev. 2019;33(11-12):591–609. doi: 10.1101/gad.324301.119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lathia JD, Mack SC, Mulkearns-Hubert EE, Valentim CLL, Rich JN. Cancer stem cells in glioblastoma. Genes Dev. 2015;29(12):1203–1217. doi: 10.1101/gad.261982.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Galli R, Binda E, Orfanelli U, et al. Isolation and Characterization of Tumorigenic, Stem-like Neural Precursors from Human Glioblastoma. Cancer Res. 2004;64(19):7011–7021. doi: 10.1158/0008-5472.CAN-04-1364 [DOI] [PubMed] [Google Scholar]
- 70.Soda Y, Marumoto T, Friedmann-Morvinski D, et al. Transdifferentiation of glioblastoma cells into vascular endothelial cells. Proc Natl Acad Sci. 2011;108(11):4274–4280. doi: 10.1073/pnas.1016030108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Scully S, Francescone R, Faibish M, et al. Transdifferentiation of Glioblastoma Stem-Like Cells into Mural Cells Drives Vasculogenic Mimicry in Glioblastomas. J Neurosci. 2012;32(37):12950–12960. doi: 10.1523/JNEUROSCI.2017-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lee JH, Lee JE, Kahng JY, et al. Human glioblastoma arises from subventricular zone cells with low-level Cell driver mutations. Nature. 2018;560(7717):243–247. doi: 10.1038/s41586-018-0389-3 [DOI] [PubMed] [Google Scholar]
- 73.Jiang Y, Uhrbom L. On the origin of glioma. Ups J Med Sci. 2012;117(2). doi: 10.3109/03009734.2012.658976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Safa AR, Saadatzadeh MR, Cohen-Gadol AA, Pollok KE, Bijangi-Vishehsaraei K. Glioblastoma stem cells (GSCs) epigenetic plasticity and interconversion between differentiated non-GSCs and GSCs. Genes Dis. 2015;2(2):152–163. doi: 10.1016/j.gendis.2015.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Olmez I, Shen W, McDonald H, Ozpolat B. Dedifferentiation of patient-derived glioblastoma multiforme cell lines results in a cancer stem cell-like state with mitogen-independent growth. J Cell Mol Med. 2015;19(6):1262–1272. doi: 10.1111/jcmm.12479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moon JH, Kwon S, Jun EK, et al. Nanog-induced dedifferentiation of p53-deficient mouse astrocytes into brain cancer stem-like cells. Biochem Biophys Res Commun. 2011;412(1):175–181. doi: 10.1016/j.bbrc.2011.07.070 [DOI] [PubMed] [Google Scholar]
- 77.Friedmann-Morvinski D, Bushong EA, Ke E, et al. Dedifferentiation of Neurons and Astrocytes by Oncogenes Can Induce Gliomas in Mice. Science. 2012;338(6110):1080–1084. doi: 10.1126/science.1226929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Rios ÁFL, Tirapelli DP da C, Cirino ML de A, Rodrigues AR, Ramos ES, Carlotti CG Jr. Expression of pluripotency-related genes in human glioblastoma. Neuro-Oncol Adv. 2022;4(1):vdab163. doi: 10.1093/noajnl/vdab163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fang X, Yoon JG, Li L, et al. The SOX2 response program in glioblastoma multiforme: an integrated ChIP-seq, expression microarray, and microRNA analysis. BMC Genomics. 2011;12(1):11. doi: 10.1186/1471-2164-12-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kim JB, Sebastiano V, Wu G, et al. Oct4-Induced Pluripotency in Adult Neural Stem Cells. Cell. 2009;136(3):411–419. doi: 10.1016/j.cell.2009.01.023 [DOI] [PubMed] [Google Scholar]
- 81.Wang ML, Chiou SH, Wu CW. Targeting cancer stem cells: emerging role of Nanog transcription factor. OncoTargets Ther. 2013;6:1207–1220. doi: 10.2147/OTT.S38114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Lee G, Auffinger B, Guo D, et al. Dedifferentiation of Glioma Cells to Glioma Stem-like Cells By Therapeutic Stress-induced HIF Signaling in the Recurrent GBM Model. Mol Cancer Ther. 2016;15(12):3064–3076. doi: 10.1158/1535-7163.MCT-15-0675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Dahan P, Martinez Gala J, Delmas C, et al. Ionizing radiations sustain glioblastoma cell dedifferentiation to a stem-like phenotype through survivin: possible involvement in radioresistance. Death Dis. 2014;5(11):e1543–e1543. doi: 10.1038/cddis.2014.509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Auffinger B, Tobias AL, Han Y, et al. Conversion of differentiated cancer cells into cancer stem-like cells in a glioblastoma model after primary chemotherapy. Cell Death Differ. 2014;21(7):1119–1131. doi: 10.1038/cdd.2014.31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Piccirillo SGM, Reynolds BA, Zanetti N, et al. Bone morphogenetic proteins inhibit the tumorigenic potential of human brain tumour-initiating cells. Nature. 2006;444(7120):761–765. doi: 10.1038/nature05349 [DOI] [PubMed] [Google Scholar]
- 86.Carén H, Stricker SH, Bulstrode H, et al. Glioblastoma Stem Cells Respond to Differentiation Cues but Fail to Undergo Commitment and Terminal Cell-Cycle Arrest. Stem Cell Rep. 2015;5(5):829–842. doi: 10.1016/j.stemcr.2015.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Inoue A, Takahashi H, Harada H, et al. Cancer stem-like cells of glioblastoma characteristically express MMP-13 and display highly invasive activity. Int J Oncol. 2010;37(5):1121–1131. doi: 10.3892/ijo_00000764 [DOI] [PubMed] [Google Scholar]
- 88.Suvà ML, Rheinbay E, Gillespie SM, et al. Reconstructing and Reprogramming the Tumor-Propagating Potential of Glioblastoma Stem-like Cells. Cell. 2014;157(3):580–594. doi: 10.1016/j.cell.2014.02.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Eramo A, Ricci-Vitiani L, Zeuner A, et al. Chemotherapy resistance of glioblastoma stem cells. Cell Death Differ. 2006;13(7):1238–1241. doi: 10.1038/sj.cdd.4401872 [DOI] [PubMed] [Google Scholar]
- 90.Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756–760. doi: 10.1038/nature05236 [DOI] [PubMed] [Google Scholar]
- 91.Baumann M, Krause M, Hill R. Exploring the role of cancer stem cells in radioresistance. Nat Rev Cancer. 2008;8(7):545–554. doi: 10.1038/nrc2419 [DOI] [PubMed] [Google Scholar]
- 92.Zeppernick F, Ahmadi R, Campos B, et al. Stem Cell Marker CD133 Affects Clinical Outcome in Glioma Patients. Clin Cancer Res. 2008;14(1):123–129. doi: 10.1158/1078-0432.CCR-07-0932 [DOI] [PubMed] [Google Scholar]
- 93.Chen J, Li Y, Yu TS, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522–526. doi: 10.1038/nature11287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Liu G, Yuan X, Zeng Z, et al. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5(1):67. doi: 10.1186/1476-4598-5-67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Couturier CP, Ayyadhury S, Le PU, et al. Single-cell RNA-seq reveals that glioblastoma recapitulates a normal neurodevelopmental hierarchy. Nat Commun. 2020;11(1):3406. doi: 10.1038/s41467-020-17186-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hambardzumyan D, Bergers G. Glioblastoma: Defining Tumor Niches. Trends Cancer. 2015;1(4):252–265. doi: 10.1016/j.trecan.2015.10.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Plaks V, Kong N, Werb Z. The Cancer Stem Cell Niche: How Essential Is the Niche in Regulating Stemness of Tumor Cells? Cell Stem Cell. 2015;16(3):225–238. doi: 10.1016/j.stem.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.WebPathology. Accessed April 29, 2023. http://webpathology.com
- 99.Specimen Detail :: Ivy Glioblastoma Atlas Project. Accessed April 29, 2023. https://glioblastoma.alleninstitute.org/ish/specimen/show/268000655
- 100.Krock BL, Skuli N, Simon MC. Hypoxia-Induced Angiogenesis: Good and Evil. Genes Cancer. 2011;2(12):1117–1133. doi: 10.1177/1947601911423654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Pugh CW, Ratcliffe PJ. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat Med. 2003;9(6):677–684. doi: 10.1038/nm0603-677 [DOI] [PubMed] [Google Scholar]
- 102.Hardee ME, Zagzag D. Mechanisms of glioma-associated neovascularization. Am J Pathol. 2012;181(4):1126–1141. doi: 10.1016/j.ajpath.2012.06.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dvorak HF. Tumors: Wounds That Do Not Heal—Redux. Cancer Immunol Res. 2015;3(1):1–11. doi: 10.1158/2326-6066.CIR-14-0209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Rosińska S, Gavard J. Tumor Vessels Fuel the Fire in Glioblastoma. Int J Mol Sci. 2021;22(12):6514. doi: 10.3390/ijms22126514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Brat DJ, Van Meir EG. Glomeruloid Microvascular Proliferation Orchestrated by VPF/VEGF: A New World of Angiogenesis Research. Am J Pathol. 2001;158(3):789–796. doi: 10.1016/S0002-9440(10)64025-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Birner P, Piribauer M, Fischer I, et al. Vascular Patterns in Glioblastoma Influence Clinical Outcome and Associate with Variable Expression of Angiogenic Proteins: Evidence for Distinct Angiogenic Subtypes. Brain Pathol. 2003;13(2):133–143. Doi: 10.1111/j.1750-3639.2003.tb00013.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Bergers G, Song S. The role of pericytes in blood-vessel formation and maintenance. Neuro-Oncol. 2005;7(4):452–464. doi: 10.1215/S1152851705000232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nagy JA, Dvorak AM, Dvorak HF. VEGF-A and the induction of pathological angiogenesis. Annu Rev Pathol. 2007;2:251–275. doi: 10.1146/annurev.pathol.2.010506.134925 [DOI] [PubMed] [Google Scholar]
- 109.Calabrese C, Poppleton H, Kocak M, et al. A perivascular niche for brain tumor stem cells. Cancer Cell. 2007;11(1):69–82. doi: 10.1016/j.ccr.2006.11.020 [DOI] [PubMed] [Google Scholar]
- 110.Bao S, Wu Q, Sathornsumetee S, et al. Stem Cell–like Glioma Cells Promote Tumor Angiogenesis through Vascular Endothelial Growth Factor. Cancer Res. 2006;66(16):7843–7848. doi: 10.1158/0008-5472.CAN-06-1010 [DOI] [PubMed] [Google Scholar]
- 111.Hardee ME, Marciscano AE, Medina-Ramirez CM, et al. Resistance of Glioblastoma-Initiating Cells to Radiation Mediated by the Tumor Microenvironment Can Be Abolished by Inhibiting Transforming Growth Factor-β. Cancer Res. 2012;72(16):4119–4129. doi 10.1158/0008-5472.CAN-12-0546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Rong Y, Durden DL, Van Meir EG, Brat DJ. “Pseudopalisading” necrosis in glioblastoma: a familiar morphologic feature that links vascular pathology, hypoxia, and angiogenesis. J Neuropathol Exp Neurol. 2006;65(6):529–539. doi: 10.1097/00005072-200606000-00001 [DOI] [PubMed] [Google Scholar]
- 113.Necrosis as a prognostic factor in glioblastoma multiforme - Barker - 1996. - Cancer - Wiley Online Library. Accessed February 20, 2023. https://acsjournals.onlinelibrary.wiley.com/doi/abs/10.1002/%28SICI%291097-0142%2819960315%2977%3A6%3C1161%3A%3AAID-CNCR24%3E3.0.CO%3B2-Z [DOI] [PubMed] [Google Scholar]
- 114.Heddleston JM, Li Z, Hjelmeland AB, Rich JN. The Hypoxic Microenvironment Maintains Glioblastoma Stem Cells and Promotes Reprogramming towards a Cancer Stem Cell Phenotype. Cell Cycle Georget Tex. 2009;8(20):3274–3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.McCord AM, Jamal M, Shankavarum UT, Lang FF, Camphausen K, Tofilon PJ. Physiologic Oxygen Concentration Enhances the Stem-Like Properties of CD133+ Human Glioblastoma Cells In vitro. Mol Cancer Res. 2009;7(4):489–497. doi: 10.1158/1541-7786.MCR-08-0360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Pistollato F, Chen HL, Rood BR, et al. Hypoxia and HIF1α Repress the Differentiative Effects of BMPs in High-Grade Glioma. Stem Cells. 2009;27(1):7–17. doi: 10.1634/stemcells.2008-0402 [DOI] [PubMed] [Google Scholar]
- 117.Johansson E, Grassi ES, Pantazopoulou V, et al. CD44 Interacts with HIF-2α to Modulate the Hypoxic Phenotype of Perinecrotic and Perivascular Glioma Cells. Cell Rep. 2017;20(7):1641–1653. doi: 10.1016/j.celrep.2017.07.049 [DOI] [PubMed] [Google Scholar]
- 118.Tejero R, Huang Y, Katsyv I, et al. Gene signatures of quiescent glioblastoma cells reveal mesenchymal shift and interactions with niche microenvironment. EBioMedicine. 2019;42:252–269. doi: 10.1016/j.ebiom.2019.03.064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Ishii A, Kimura T, Sadahiro H, et al. Histological Characterization of the Tumorigenic “Peri-Necrotic Niche” Harboring Quiescent Stem-Like Tumor Cells in Glioblastoma. PLOS ONE. 2016;11(1):e0147366. doi: 10.1371/journal.pone.0147366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Kahlert UD, Maciaczyk D, Dai F, et al. Resistance to Hypoxia-Induced, BNIP3-Mediated Cell Death Contributes to an Increase in a CD133-Positive Cell Population in Human Glioblastomas In Vitro. J Neuropathol Exp Neurol. 2012;71(12):1086–1099. doi: 10.1097/NEN.0b013e3182772d83 [DOI] [PubMed] [Google Scholar]
- 121.Wang P, Wan W, Xiong S, et al. HIF1α regulates glioma chemosensitivity through the transformation between differentiation and dedifferentiation in various oxygen levels. Sci Rep. 2017;7(1):7965. doi: 10.1038/s41598-017-06086-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Parney IF, Waldron JS, Parsa AT. Flow cytometry and in vitro analysis of human glioma-associated macrophages. Laboratory investigation. J Neurosurg. 2009;110(3):572–582. doi: 10.3171/2008.7.JNS08475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.D’Alessio A, Proietti G, Sica G, Scicchitano BM. Pathological and Molecular Features of Glioblastoma and Its Peritumoral Tissue. Cancers. 2019;11(4):469. doi: 10.3390/cancers11040469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hambardzumyan D, Gutmann DH, Kettenmann H. The role of microglia and macrophages in glioma maintenance and progression. Nat Neurosci. 2016;19(1):20–27. doi: 10.1038/nn.4185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Morisse MC, Jouannet S, Dominguez-Villar M, Sanson M, Idbaih A. Interactions between tumor-associated macrophages and tumor cells in glioblastoma: unraveling promising targeted therapies. Expert Rev Neurother. 2018;18(9):729–737. doi: 10.1080/14737175.2018.1510321 [DOI] [PubMed] [Google Scholar]
- 126.Grégoire H, Roncali L, Rousseau A, et al. Targeting Tumor Associated Macrophages to Overcome Conventional Treatment Resistance in Glioblastoma. Front Pharmacol. 2020;11:368. doi: 10.3389/fphar.2020.00368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Qian BZ, Pollard JW. Macrophage Diversity Enhances Tumor Progression and Metastasis. Cell. 2010;141(1):39–51. doi: 10.1016/j.cell.2010.03.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Ruffell B, Coussens LM. Macrophages and Therapeutic Resistance in Cancer. Cancer Cell. 2015;27(4):462–472. doi: 10.1016/j.ccell.2015.02.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Yeung Y, McDonald K, Grewal T, Munoz L. Interleukins in glioblastoma pathophysiology: implications for therapy. Br J Pharmacol. 2013;168(3):591–606. doi: 10.1111/bph.12008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rempel SA, Dudas S, Ge S, Gutierrez JA. Identification and Localization of the Cytokine SDF1 and Its Receptor, CXC Chemokine Receptor 4, to Regions of Necrosis and Angiogenesis in Human Glioblastoma. Clin Cancer Res. 2000;6(1):102–111. [PubMed] [Google Scholar]
- 131.Wu A, Wei J, Kong LY, et al. Glioma cancer stem cells induce immunosuppressive macrophages/microglia. Neuro-Oncol. 2010;12(11):1113–1125. doi: 10.1093/neuonc/noq082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kumar V, Gabrilovich DI. Hypoxia-inducible factors in regulation of immune responses in tumour microenvironment. Immunology. 2014;143(4):512–519. doi: 10.1111/imm.12380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Herting CJ, Chen Z, Maximov V, et al. Tumour-associated macrophage-derived interleukin-1 mediates glioblastoma-associated cerebral oedema. Brain J Neurol. 2019;142(12):3834–3851. doi: 10.1093/brain/awz331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Mega A, Hartmark Nilsen M, Leiss LW, et al. Astrocytes enhance glioblastoma growth. Glia. 2020;68(2):316–327. doi: 10.1002/glia.23718 [DOI] [PubMed] [Google Scholar]
- 135.Brandao M, Simon T, Critchley G, Giamas G. Astrocytes, the rising stars of the glioblastoma microenvironment. Glia. 2019;67(5):779–790. doi: 10.1002/glia.23520 [DOI] [PubMed] [Google Scholar]
- 136.Pekny M, Pekna M. Astrocyte Reactivity and Reactive Astrogliosis: Costs and Benefits. Physiol Rev. 2014;94(4):1077–1098. doi: 10.1152/physrev.00041.2013 [DOI] [PubMed] [Google Scholar]
- 137.Placone AL, Quiñones-Hinojosa A, Searson PC. The role of astrocytes in the progression of brain cancer: complicating the picture of the tumor microenvironment. Tumour Biol J Int Soc Oncodevelopmental Biol Med. 2016;37(1):61–69. doi: 10.1007/s13277-015-4242-0 [DOI] [PubMed] [Google Scholar]
- 138.Oushy S, Hellwinkel JE, Wang M, et al. Glioblastoma multiforme-derived extracellular vesicles drive normal astrocytes towards a tumour-enhancing phenotype. Philos Trans R Soc Lond B Biol Sci. 2018;373(1737):20160477. doi: 10.1098/rstb.2016.0477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.John Lin CC, Yu K, Hatcher A, et al. Identification of diverse astrocyte populations and their malignant analogs. Nat Neurosci. 2017;20(3):396–405. doi: 10.1038/nn.4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Schiffer D, Annovazzi L, Casalone C, Corona C, Mellai M. Glioblastoma: Microenvironment and Niche Concept. Cancers. 2019;11(1):5. doi: 10.3390/cancers11010005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Westphal M, Lamszus K. The neurobiology of gliomas: from cell biology to the development of therapeutic approaches. Nat Rev Neurosci. 2011;12(9):495–508. doi: 10.1038/nrn3060 [DOI] [PubMed] [Google Scholar]
- 142.Rath BH, Fair JM, Jamal M, Camphausen K, Tofilon PJ. Astrocytes Enhance the Invasion Potential of Glioblastoma Stem-Like Cells. PLOS ONE. 2013;8(1):e54752. doi: 10.1371/journal.pone.0054752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Seike T, Fujita K, Yamakawa Y, et al. Interaction between lung cancer cells and astrocytes via specific inflammatory cytokines in the microenvironment of brain metastasis. Clin Exp Metastasis. 2011;28(1):13–25. doi: 10.1007/s10585-010-9354-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Li R, Li G, Deng L, et al. IL-6 augments the invasiveness of U87MG human glioblastoma multiforme cells via up-regulation of MMP-2 and fascin-1. Oncol Rep. 2010;23(6):1553–1559. doi: 10.3892/or_00000795 [DOI] [PubMed] [Google Scholar]
- 145.Zhang H, Zhou Y, Cui B, Liu Z, Shen H. Novel insights into astrocyte-mediated signaling of proliferation, invasion and tumor immune microenvironment in glioblastoma. Biomed Pharmacother. 2020;126:110086. doi: 10.1016/j.biopha.2020.110086 [DOI] [PubMed] [Google Scholar]
- 146.Kitai R, Horita R, Sato K, et al. Nestin expression in astrocytic tumors delineates tumor infiltration. Brain Tumor Pathol. 2010;27(1):17–21. doi: 10.1007/s10014-009-0261-0 [DOI] [PubMed] [Google Scholar]
- 147.Ishiwata T, Teduka K, Yamamoto T, Kawahara K, Matsuda Y, Naito Z. Neuroepithelial stem cell marker nestin regulates the migration, invasion and growth of human gliomas. Oncol Rep. 2011;26(1):91–99. doi: 10.3892/or.2011.1267 [DOI] [PubMed] [Google Scholar]
- 148.Wang J, Xu SL, Duan JJ, et al. Invasion of white matter tracts by glioma stem cells is regulated by a NOTCH1–SOX2 positive-feedback loop. Nat Neurosci. 2019;22(1):91–105. doi: 10.1038/s41593-018-0285-z [DOI] [PubMed] [Google Scholar]
- 149.Clement V, Sanchez P, de Tribolet N, Radovanovic I, Ruiz i Altaba A. HEDGEHOG-GLI1 signaling regulates human glioma growth, cancer stem cell self-renewal, and tumorigenicity. Curr Biol CB. 2007;17(2):165–172. doi: 10.1016/j.cub.2006.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Sherry MM, Reeves A, Wu JK, Cochran BH. STAT3 is required for proliferation and maintenance of multipotency in glioblastoma stem cells. Stem Cells Dayt Ohio. 2009;27(10):2383–2392. doi: 10.1002/stem.185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Hossain A, Gumin J, Gao F, et al. Mesenchymal Stem Cells Isolated From Human Gliomas Increase Proliferation and Maintain Stemness of Glioma Stem Cells Through the IL-6/gp130/STAT3 Pathway. Stem Cells Dayt Ohio. 2015;33(8):2400–2415. doi: 10.1002/stem.2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Trenner A, Sartori AA. Harnessing DNA Double-Strand Break Repair for Cancer Treatment. Front Oncol. 2019;9. Accessed April 28, 2023. https://www.frontiersin.org/articles/10.3389/fonc.2019.01388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Srinivas US, Tan BWQ, Vellayappan BA, Jeyasekharan AD. ROS and the DNA damage response in cancer. Redox Biol. 2019;25:101084. doi: 10.1016/j.redox.2018.101084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Chen Q, Chai YC, Mazumder S, et al. The late increase in intracellular free radical oxygen species during apoptosis is associated with cytochrome c release, caspase activation, and mitochondrial dysfunction. Cell Death Differ. 2003;10(3):323–334. doi: 10.1038/sj.cdd.4401148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Smith HL, Southgate H, Tweddle DA, Curtin NJ. DNA damage checkpoint kinases in cancer. Expert Rev Mol Med. 2020;22:e2. doi: 10.1017/erm.2020.3 [DOI] [PubMed] [Google Scholar]
- 156.Squatrito M, Brennan CW, Helmy K, Huse JT, Petrini JH, Holland EC. Loss of ATM/Chk2/p53 Pathway Components Accelerates Tumor Development and Contributes to Radiation Resistance in Gliomas. Cancer Cell. 2010;18(6):619–629. doi: 10.1016/j.ccr.2010.10.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Mladenov E, Magin S, Soni A, Iliakis G. DNA Double-Strand Break Repair as Determinant of Cellular Radiosensitivity to Killing and Target in Radiation Therapy. Front Oncol. 2013;3. Accessed April 15, 2023. https://www.frontiersin.org/articles/10.3389/fonc.2013.00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Zhou W, Sun M, Li GH, et al. Activation of the phosphorylation of ATM contributes to radioresistance of glioma stem cells. Oncol Rep. 2013;30(4):1793–1801. doi: 10.3892/or.2013.2614 [DOI] [PubMed] [Google Scholar]
- 159.Ahmed SU, Carruthers R, Gilmour L, Yildirim S, Watts C, Chalmers AJ. Selective Inhibition of Parallel DNA Damage Response Pathways Optimizes Radiosensitization of Glioblastoma Stem-like Cells. Cancer Res. 2015;75(20):4416–4428. doi: 10.1158/0008-5472.CAN-14-3790 [DOI] [PubMed] [Google Scholar]
- 160.Tamura K, Aoyagi M, Wakimoto H, et al. Accumulation of CD133-positive glioma cells after high-dose irradiation by Gamma Knife surgery plus external beam radiation: Clinical article. J Neurosurg. 2010;113(2):310–318. doi: 10.3171/2010.2.JNS091607 [DOI] [PubMed] [Google Scholar]
- 161.Tamura K, Aoyagi M, Ando N, et al. Expansion of CD133-positive glioma cells in recurrent de novo glioblastomas after radiotherapy and chemotherapy: Laboratory investigation. J Neurosurg. 2013;119(5):1145–1155. doi: 10.3171/2013.7.JNS122417 [DOI] [PubMed] [Google Scholar]
- 162.Lomonaco SL, Finniss S, Xiang C, et al. The induction of autophagy by γ-radiation contributes to the radioresistance of glioma stem cells. Int J Cancer. 2009;125(3):717–722. doi: 10.1002/ijc.24402 [DOI] [PubMed] [Google Scholar]
- 163.Lee SY. Temozolomide resistance in glioblastoma multiforme. Genes Dis. 2016;3(3):198–210. doi: 10.1016/j.gendis.2016.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Ohba S, Yamashiro K, Hirose Y. Inhibition of DNA Repair in Combination with Temozolomide or Dianhydrogalactiol Overcomes Temozolomide-Resistant Glioma Cells. Cancers. 2021;13(11):2570. doi: 10.3390/cancers13112570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Pistollato F, Abbadi S, Rampazzo E, et al. Intratumoral Hypoxic Gradient Drives Stem Cells Distribution and MGMT Expression in Glioblastoma. Stem Cells. 2010;28(5):851–862. doi: 10.1002/stem.415 [DOI] [PubMed] [Google Scholar]
- 166.Hegi ME, Diserens AC, Gorlia T, et al. MGMT Gene Silencing and Benefit from Temozolomide in Glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331 [DOI] [PubMed] [Google Scholar]
- 167.van Nifterik KA, van den Berg J, van der Meide WF, et al. Absence of the MGMT protein as well as methylation of the MGMT promoter predict the sensitivity for temozolomide. Br J Cancer. 2010;103(1):29–35. doi: 10.1038/sj.bjc.6605712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Brandes AA, Franceschi E, Tosoni A, et al. O6-methylguanine DNA-methyltransferase methylation status can change between first surgery for newly diagnosed glioblastoma and second surgery for recurrence: clinical implications. Neuro-Oncol. 2010;12(3):283–288. doi: 10.1093/neuonc/nop050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Ewald JA, Desotelle JA, Wilding G, Jarrard DF. Therapy-Induced Senescence in Cancer. JNCI J Natl Cancer Inst. 2010;102(20):1536–1546. doi: 10.1093/jnci/djq364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Prasanna PG, Citrin DE, Hildesheim J, et al. Therapy-Induced Senescence: Opportunities to Improve Anticancer Therapy. JNCI J Natl Cancer Inst. 2021;113(10):1285–1298. doi: 10.1093/jnci/djab064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Saleh T, Tyutyunyk-Massey L, Gewirtz DA. Tumor Cell Escape from Therapy-Induced Senescence as a Model of Disease Recurrence after Dormancy. Cancer Res. 2019;79(6):1044–1046. doi: 10.1158/0008-5472.CAN-18-3437 [DOI] [PubMed] [Google Scholar]
- 172.Sharpless NE, Sherr CJ. Forging a signature of in vivo senescence. Nat Rev Cancer. 2015;15(7):397–408. doi: 10.1038/nrc3960 [DOI] [PubMed] [Google Scholar]
- 173.Cahu J, Bustany S, Sola B. Senescence-associated secretory phenotype favors the emergence of cancer stem-like cells. Cell Death Dis. 2012;3(12):e446–e446. doi: 10.1038/cddis.2012.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Mosteiro L, Pantoja C, de Martino A, Serrano M. Senescence promotes in vivo reprogramming through p16INK4a and IL-6. Aging Cell. 2018;17(2):e12711. doi: 10.1111/acel.12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Nacarelli T, Fukumoto T, Zundell JA, et al. NAMPT Inhibition Suppresses Cancer Stem-like Cells Associated with Therapy-Induced Senescence in Ovarian Cancer. Cancer Res. 2020;80(4):890–900. doi: 10.1158/0008-5472.CAN-19-2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ciardiello F, Tortora G. EGFR Antagonists in Cancer Treatment. N Engl J Med. 2008;358(11):1160–1174. doi: 10.1056/NEJMra0707704 [DOI] [PubMed] [Google Scholar]
- 177.Bhullar KS, Lagarón NO, McGowan EM, et al. Kinase-targeted cancer therapies: progress, challenges and future directions. Mol Cancer. 2018;17(1):48. doi: 10.1186/s12943-018-0804-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Cohen MH, Johnson JR, Chen YF, Sridhara R, Pazdur R. FDA Drug Approval Summary: Erlotinib (Tarceva®) Tablets. The Oncologist. 2005;10(7):461–466. doi: 10.1634/theoncologist.10-7-461 [DOI] [PubMed] [Google Scholar]
- 179.Prados MD, Chang SM, Butowski N, et al. Phase II Study of Erlotinib Plus Temozolomide During and After Radiation Therapy in Patients With Newly Diagnosed Glioblastoma Multiforme or Gliosarcoma. J Clin Oncol. 2009;27(4):579–584. doi: 10.1200/JCO.2008.18.9639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Raizer JJ, Abrey LE, Lassman AB, et al. A phase II trial of erlotinib in patients with recurrent malignant gliomas and nonprogressive glioblastoma multiforme postradiation therapy†. Neuro-Oncol. 2010;12(1):95–103. doi: 10.1093/neuonc/nop015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hegi ME, Diserens AC, Bady P, et al. Pathway Analysis of Glioblastoma Tissue after Preoperative Treatment with the EGFR Tyrosine Kinase Inhibitor Gefitinib—A Phase II Trial. Mol Cancer Ther. 2011;10(6):1102–1112. doi: 10.1158/1535-7163.MCT-11-0048 [DOI] [PubMed] [Google Scholar]
- 182.Lassman AB, Rossi MR, Razier JR, et al. Molecular Study of Malignant Gliomas Treated with Epidermal Growth Factor Receptor Inhibitors: Tissue Analysis from North American Brain Tumor Consortium Trials 01-03 and 00-01. Clin Cancer Res. 2005;11(21):7841–7850. doi: 10.1158/1078-0432.CCR-05-0421 [DOI] [PubMed] [Google Scholar]
- 183.Sepúlveda-Sánchez JM, Vaz MÁ, Balañá C, et al. Phase II trial of dacomitinib, a pan–human EGFR tyrosine kinase inhibitor, in recurrent glioblastoma patients with EGFR amplification. Neuro-Oncol. 2017;19(11):1522–1531. doi: 10.1093/neuonc/nox105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Reardon DA, Nabors LB, Mason WP, et al. Phase I/randomized phase II study of afatinib, an irreversible ErbB family blocker, with or without protracted temozolomide in adults with recurrent glioblastoma. Neuro-Oncol. 2015;17(3):430–439. doi: 10.1093/neuonc/nou160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Jänne PA, Yang JCH, Kim DW, et al. AZD9291 in EGFR Inhibitor–Resistant Non–Small-Cell Lung Cancer. N Engl J Med. 2015;372(18):1689–1699. doi: 10.1056/NEJMoa1411817 [DOI] [PubMed] [Google Scholar]
- 186.Liu X, Chen X, Shi L, et al. The third-generation EGFR inhibitor AZD9291 overcomes primary resistance by continuously blocking ERK signaling in glioblastoma. J Exp Clin Cancer Res. 2019;38(1):219. doi: 10.1186/s13046-019-1235-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Cardona AF, Jaramillo-Velásquez D, Ruiz-Patiño A, et al. Efficacy of osimertinib plus bevacizumab in glioblastoma patients with simultaneous EGFR amplification and EGFRvIII mutation. J Neurooncol. 2021;154(3):353–364. doi: 10.1007/s11060-021-03834-3 [DOI] [PubMed] [Google Scholar]
- 188.Belda-Iniesta C, de Castro Carpeño J, Saenz EC, Gutiérrez M, Perona R, González-Barón M. Long term responses with cetuximab therapy in glioblastoma multiforme. Cancer Biol Ther. 2006;5(8):912–914. doi: 10.4161/cbt.5.8.3118 [DOI] [PubMed] [Google Scholar]
- 189.Neyns B, Sadones J, Joosens E, et al. Stratified phase II trial of cetuximab in patients with recurrent high-grade glioma. Ann Oncol. 2009;20(9):1596–1603. doi: 10.1093/annonc/mdp032 [DOI] [PubMed] [Google Scholar]
- 190.She L, Gong X, Su L, Liu C. Radiotherapy Plus Temozolomide With or Without Nimotuzumab Against the Newly Diagnosed EGFR-Positive Glioblastoma: A Retrospective Cohort Study. The Oncologist. 2023;28(1):e45–e53. doi: 10.1093/oncolo/oyac202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Du XJ, Li XM, Cai LB, et al. Efficacy and safety of nimotuzumab in addition to radiotherapy and temozolomide for cerebral glioblastoma: a phase II multicenter clinical trial. J Cancer. 2019;10(14):3214–3223. doi: 10.7150/jca.30123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yu YJ, Watts RJ. Developing Therapeutic Antibodies for Neurodegenerative Disease. Neurotherapeutics. 2013;10(3):459–472. doi: 10.1007/s13311-013-0187-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Niewoehner J, Bohrmann B, Collin L, et al. Increased Brain Penetration and Potency of a Therapeutic Antibody Using a Monovalent Molecular Shuttle. Neuron. 2014;81(1):49–60. doi: 10.1016/j.neuron.2013.10.061 [DOI] [PubMed] [Google Scholar]
- 194.Bajracharya R, Caruso AC, Vella LJ, Nisbet RM. Current and Emerging Strategies for Enhancing Antibody Delivery to the Brain. Pharmaceutics. 2021;13(12):2014. doi: 10.3390/pharmaceutics13122014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Zeiadeh I, Najjar A, Karaman R. Strategies for Enhancing the Permeation of CNS-Active Drugs through the Blood-Brain Barrier: A Review. Molecules. 2018;23(6):1289. doi: 10.3390/molecules23061289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 196.Burger MT, Pecchi S, Wagman A, et al. Identification of NVP-BKM120 as a Potent, Selective, Orally Bioavailable Class I PI3 Kinase Inhibitor for Treating Cancer. ACS Med Chem Lett. 2011;2(10):774–779. doi: 10.1021/ml200156t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Netland IA, Førde HE, Sleire L, et al. Treatment with the PI3K inhibitor buparlisib (NVP-BKM120) suppresses the growth of established patient-derived GBM xenografts and prolongs survival in nude rats. J Neurooncol. 2016;129(1):57–66. doi: 10.1007/s11060-016-2158-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Wen PY, Touat M, Alexander BM, et al. Buparlisib in Patients With Recurrent Glioblastoma Harboring Phosphatidylinositol 3-Kinase Pathway Activation: An Open-Label, Multicenter, Multi-Arm, Phase II Trial. J Clin Oncol. 2019;37(9):741–750. doi: 10.1200/JCO.18.01207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Wen PY, Rodon JA, Mason W, et al. Phase I, open-label, multicentre study of buparlisib in combination with temozolomide or with concomitant radiation therapy and temozolomide in patients with newly diagnosed glioblastoma. ESMO Open. 2020;5(4). doi: 10.1136/esmoopen-2020-000673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Hainsworth JD, Becker KP, Mekhail T, et al. Phase I/II study of bevacizumab with BKM120, an oral PI3K inhibitor, in patients with refractory solid tumors (phase I) and relapsed/refractory glioblastoma (phase II). J Neurooncol. 2019;144(2):303–311. doi: 10.1007/s11060-019-03227-7 [DOI] [PubMed] [Google Scholar]
- 201.Zhang S, Peng X, Li X, et al. BKM120 sensitizes glioblastoma to the PARP inhibitor rucaparib by suppressing homologous recombination repair. Cell Death Dis. 2021;12(6):1–14. doi: 10.1038/s41419-021-03805-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Harder BG, Peng S, Sereduk CP, et al. Inhibition of phosphatidylinositol 3-kinase by PX-866 suppresses temozolomide-induced autophagy and promotes apoptosis in glioblastoma cells. Mol Med. 2019;25(1):49. doi: 10.1186/s10020-019-0116-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Pitz MW, Eisenhauer EA, MacNeil MV, et al. Phase II study of PX-866 in recurrent glioblastoma. Neuro-Oncol. 2015;17(9):1270–1274. doi: 10.1093/neuonc/nou365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Xie S, Ni J, McFaline-Figueroa JR, et al. Divergent Roles of PI3K Isoforms in PTEN-Deficient Glioblastomas. Cell Rep. 2020;32(13):108196. doi: 10.1016/j.celrep.2020.108196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Hutchings K, Pridham K, Liu M, Sheng Z. DDDR-19. NEXT-GENERATION PI3K INHIBITOR FOR GLIOBLASTOMA TREATMENT. Neuro-Oncol. 2022;24(Supplement_7):vii102. doi: 10.1093/neuonc/noac209.384 [DOI] [Google Scholar]
- 206.Pridham KJ, Le L, Guo S, et al. PIK3CB/p110β is a selective survival factor for glioblastoma. Neuro-Oncol. 2018;20(4):494–505. doi: 10.1093/neuonc/nox181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Burnett GL, Yang YC, Aggen JB, et al. Discovery of RMC-5552, a Selective Bi-Steric Inhibitor of mTORC1, for the Treatment of mTORC1 - Activated Tumors. J Med Chem. 2023;66(1):149–169. doi: 10.1021/acs.jmedchem.2c01658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Kaley TJ, Panageas KS, Mellinghoff IK, et al. Phase II trial of an AKT inhibitor (perifosine) for recurrent glioblastoma. J Neurooncol. 2019;144(2):403–407. doi: 10.1007/s11060-019-03243-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Wen PY, Chang SM, Lamborn KR, et al. Phase I/II study of erlotinib and temsirolimus for patients with recurrent malignant gliomas: North American Brain Tumor Consortium trial 04-02. Neuro-Oncol. 2014;16(4):567–578. doi: 10.1093/neuonc/not247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210.Ma DJ, Galanis E, Anderson SK, et al. A phase II trial of everolimus, temozolomide, and radiotherapy in patients with newly diagnosed glioblastoma: NCCTG N057K. Neuro-Oncol. 2015;17(9):1261–1269. doi: 10.1093/neuonc/nou328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Jin J, Grigore F, Chen CC, Li M. Self-renewal signaling pathways and differentiation therapies of glioblastoma stem cells (Review). Int J Oncol. 2021;59(1):1–11. doi: 10.3892/ijo.2021.5225 [DOI] [PubMed] [Google Scholar]
- 212.Wang J, Wakeman TP, Lathia JD, et al. Notch Promotes Radioresistance of Glioma Stem Cells. Stem Cells. 2010;28(1):17–28. doi: 10.1002/stem.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Fan X, Khaki L, Zhu TS, et al. NOTCH Pathway Blockade Depletes CD133-Positive Glioblastoma Cells and Inhibits Growth of Tumor Neurospheres and Xenografts. Stem Cells. 2010;28(1):5–16. doi: 10.1002/stem.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Hu YY, Zheng MH, Cheng G, et al. Notch signaling contributes to the maintenance of both normal neural stem cells and patient-derived glioma stem cells. BMC Cancer. 2011;11(1):82. doi: 10.1186/1471-2407-11-82 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Hoffman LM, Fouladi M, Olson J, et al. Phase I trial of weekly MK-0752 in children with refractory central nervous system malignancies: a pediatric brain tumor consortium study. Childs Nerv Syst. 2015;31(8):1283–1289. doi: 10.1007/s00381-015-2725-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Fouladi M, Stewart CF, Olson J, et al. Phase I Trial of MK-0752 in Children With Refractory CNS Malignancies: A Pediatric Brain Tumor Consortium Study. J Clin Oncol. 2011;29(26):3529–3534. doi: 10.1200/JCO.2011.35.7806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Krop I, Demuth T, Guthrie T, et al. Phase I Pharmacologic and Pharmacodynamic Study of the Gamma Secretase (Notch) Inhibitor MK-0752 in Adult Patients With Advanced Solid Tumors. J Clin Oncol. 2012;30(19):2307–2313. doi: 10.1200/JCO.2011.39.1540 [DOI] [PubMed] [Google Scholar]
- 218.Saito N, Fu J, Zheng S, et al. A High Notch Pathway Activation Predicts Response to γ Secretase Inhibitors in Proneural Subtype of Glioma Tumor-Initiating Cells. Stem Cells. 2014;32(1):301–312. doi: 10.1002/stem.1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 219.Xu R, Shimizu F, Hovinga K, et al. Molecular and Clinical Effects of Notch Inhibition in Glioma Patients: A Phase 0/I Trial. Clin Cancer Res. 2016;22(19):4786–4796. doi: 10.1158/1078-0432.CCR-16-0048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Yi L, Zhou X, Li T, et al. Notch1 signaling pathway promotes invasion, self-renewal and growth of glioma initiating cells via modulating chemokine system CXCL12/CXCR4. J Exp Clin Cancer Res. 2019;38(1):339. doi: 10.1186/s13046-019-1319-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Saito N, Aoki K, Hirai N, et al. Effect of Notch expression in glioma stem cells on therapeutic response to chemo-radiotherapy in recurrent glioblastoma. Brain Tumor Pathol. 2015;32(3):176–183. doi: 10.1007/s10014-015-0215-7 [DOI] [PubMed] [Google Scholar]
- 222.Wolfe MS. Aspartic Proteases of Alzheimer’s Disease: β and γ-Secretases. In: Bradshaw RA, Stahl PD, eds. Encyclopedia of Cell Biology. Academic Press; 2016:661–669. doi: 10.1016/B978-0-12-394447-4.10077-X [DOI] [Google Scholar]
- 223.Doody RS, Raman R, Farlow M, et al. A Phase 3 Trial of Semagacestat for Treatment of Alzheimer’s Disease. N Engl J Med. 2013;369(4):341–350. doi: 10.1056/NEJMoa1210951 [DOI] [PubMed] [Google Scholar]
- 224.Ding X, Ding C, Wang F, et al. Effects of NOTCH1 signaling inhibitor γ-secretase inhibitor II on growth of cancer stem cells. Oncol Lett. 2018;16(5):6095–6099. doi: 10.3892/ol.2018.9377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Wickström M, Dyberg C, Milosevic J, et al. Wnt/β-catenin pathway regulates MGMT gene expression in cancer and inhibition of Wnt signalling prevents chemoresistance. Nat Commun. 2015;6(1):8904. doi: 10.1038/ncomms9904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Lee Y, Kim KH, Kim DG, et al. FoxM1 Promotes Stemness and Radio-Resistance of Glioblastoma by Regulating the Master Stem Cell Regulator Sox2. PLOS ONE. 2015;10(10):e0137703. doi: 10.1371/journal.pone.0137703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Binda E, Visioli A, Giani F, et al. Wnt5a Drives an Invasive Phenotype in Human Glioblastoma Stem-like Cells. Cancer Res. 2017;77(4):996–1007. doi: 10.1158/0008-5472.CAN-16-1693 [DOI] [PubMed] [Google Scholar]
- 228.Giles RH, van Es JH, Clevers H. Caught up in a Wnt storm: Wnt signaling in cancer. Biochim Biophys Acta BBA - Rev Cancer. 2003;1653(1):1–24. doi: 10.1016/S0304-419X(03)00005-2 [DOI] [PubMed] [Google Scholar]
- 229.Qiu J, Shi Z, Jiang J. Cyclooxygenase-2 in glioblastoma multiforme. Drug Discov Today. 2017;22(1):148–156. doi: 10.1016/j.drudis.2016.09.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Ma HI, Chiou SH, Hueng DY, et al. Celecoxib and radioresistant glioblastoma-derived CD133+ cells: improvement in radiotherapeutic effects: Laboratory investigation. J Neurosurg. 2011;114(3):651–662. doi: 10.3171/2009.11.JNS091396 [DOI] [PubMed] [Google Scholar]
- 231.Stockhammer F, Misch M, Koch A, et al. Continuous low-dose temozolomide and celecoxib in recurrent glioblastoma. J Neurooncol. 2010;100(3):407–415. doi: 10.1007/s11060-010-0192-y [DOI] [PubMed] [Google Scholar]
- 232.Halatsch ME, Dwucet A, Schmidt CJ, et al. In Vitro and Clinical Compassionate Use Experiences with the Drug-Repurposing Approach CUSP9v3 in Glioblastoma. Pharmaceuticals. 2021;14(12):1241. doi: 10.3390/ph14121241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Latour M, Her NG, Kesari S, Nurmemmedov E. WNT Signaling as a Therapeutic Target for Glioblastoma. Int J Mol Sci. 2021;22(16):8428. doi: 10.3390/ijms22168428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Carballo GB, Honorato JR, de Lopes GPF, Spohr TCL de S e. A highlight on Sonic hedgehog pathway. Cell Commun Signal. 2018;16(1):11. doi: 10.1186/s12964-018-0220-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Cochrane CR, Szczepny A, Watkins DN, Cain JE. Hedgehog Signaling in the Maintenance of Cancer Stem Cells. Cancers. 2015;7(3):1554–1585. doi: 10.3390/cancers7030851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Uchida H, Arita K, Yunoue S, et al. Role of sonic hedgehog signaling in migration of cell lines established from CD133-positive malignant glioma cells. J Neurooncol. 2011;104(3):697–704. doi: 10.1007/s11060-011-0552-2 [DOI] [PubMed] [Google Scholar]
- 237.Santoni M, Burattini L, Nabissi M, et al. Essential Role of Gli Proteins in Glioblastoma Multiforme. Curr Protein Pept Sci. 14(2):133–140. [DOI] [PubMed] [Google Scholar]
- 238.Nanta R, Shrivastava A, Sharma J, Shankar S, Srivastava RK. Inhibition of sonic hedgehog and PI3K/Akt/mTOR pathways cooperate in suppressing survival, self-renewal and tumorigenic potential of glioblastoma-initiating cells. Mol Cell Biochem. 2019;454(1):11–23. doi: 10.1007/s11010-018-3448-z [DOI] [PubMed] [Google Scholar]
- 239.Wick W, Dettmer S, Berberich A, et al. N2M2 (NOA-20) phase I/II trial of molecularly matched targeted therapies plus radiotherapy in patients with newly diagnosed non-MGMT hypermethylated glioblastoma. Neuro-Oncol. 2019;21(1):95–105. doi: 10.1093/neuonc/noy161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Li Y, Song Q, Day BW. Phase I and phase II sonidegib and vismodegib clinical trials for the treatment of paediatric and adult MB patients: a systemic review and meta-analysis. Acta Neuropathol Commun. 2019;7(1):123. doi: 10.1186/s40478-019-0773-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Ferrara N, Hillan KJ, Novotny W. Bevacizumab (Avastin), a humanized anti-VEGF monoclonal antibody for cancer therapy. Biochem Biophys Res Commun. 2005;333(2):328–335. doi: 10.1016/j.bbrc.2005.05.132 [DOI] [PubMed] [Google Scholar]
- 242.Vredenburgh JJ, Desjardins A, Herndon JE II, et al. Phase II Trial of Bevacizumab and Irinotecan in Recurrent Malignant Glioma. Clin Cancer Res. 2007;13(4):1253–1259. doi: 10.1158/1078-0432.CCR-06-2309 [DOI] [PubMed] [Google Scholar]
- 243.Cohen MH, Shen YL, Keegan P, Pazdur R. FDA Drug Approval Summary: Bevacizumab (Avastin®) as Treatment of Recurrent Glioblastoma Multiforme. The Oncologist. 2009;14(11):1131–1138. doi: 10.1634/theoncologist.2009-0121 [DOI] [PubMed] [Google Scholar]
- 244.Sandmann T, Bourgon R, Garcia J, et al. Patients With Proneural Glioblastoma May Derive Overall Survival Benefit From the Addition of Bevacizumab to First-Line Radiotherapy and Temozolomide: Retrospective Analysis of the AVAglio Trial. J Clin Oncol. 2015;33(25):2735–2744. doi: 10.1200/JCO.2015.61.5005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245.Piao Y, Liang J, Holmes L, et al. Glioblastoma resistance to anti-VEGF therapy is associated with myeloid cell infiltration, stem cell accumulation, and a mesenchymal phenotype. Neuro-Oncol. 2012;14(11):1379–1392. doi: 10.1093/neuonc/nos158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Piao Y, Liang J, Holmes L, Henry V, Sulman E, de Groot JF. Acquired Resistance to Anti-VEGF Therapy in Glioblastoma Is Associated with a Mesenchymal Transition. Clin Cancer Res. 2013;19(16):4392–4403. doi: 10.1158/1078-0432.CCR-12-1557 [DOI] [PubMed] [Google Scholar]
- 247.Jain RK. Normalizing Tumor Microenvironment to Treat Cancer: Bench to Bedside to Biomarkers. J Clin Oncol. 2013;31(17):2205–2218. doi: 10.1200/JCO.2012.46.3653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Dong X, Ren J, Amoozgar Z, et al. Anti-VEGF therapy improves EGFR-vIII-CAR-T cell delivery and efficacy in syngeneic glioblastoma models in mice. J Immunother Cancer. 2023;11(3):e005583. doi: 10.1136/jitc-2022-005583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.de Vries NA, Beijnen JH, Boogerd W, van Tellingen O. Blood–brain barrier and chemotherapeutic treatment of brain tumors. Expert Rev Neurother. 2006;6(8):1199–1209. doi: 10.1586/14737175.6.8.1199 [DOI] [PubMed] [Google Scholar]
- 250.Rivera LB, Bergers G. Intertwined regulation of angiogenesis and immunity by myeloid cells. Trends Immunol. 2015;36(4):240–249. doi: 10.1016/j.it.2015.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Lu-Emerson C, Duda DG, Emblem KE, et al. Lessons From Anti–Vascular Endothelial Growth Factor and Anti–Vascular Endothelial Growth Factor Receptor Trials in Patients With Glioblastoma. J Clin Oncol. 2015;33(10):1197–1213. doi: 10.1200/JCO.2014.55.9575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Martin JD, Seano G, Jain RK. Normalizing Function of Tumor Vessels: Progress, Opportunities, and Challenges. Annu Rev Physiol. 2019;81(1):505–534. doi: 10.1146/annurev-physiol-020518-114700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Venere M, Hamerlik P, Wu Q, et al. Therapeutic targeting of constitutive PARP activation compromises stem cell phenotype and survival of glioblastoma-initiating cells. Cell Death Differ. 2014;21(2):258–269. doi: 10.1038/cdd.2013.136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Lesueur P, Lequesne J, Grellard JM, et al. Phase I/IIa study of concomitant radiotherapy with olaparib and temozolomide in unresectable or partially resectable glioblastoma: OLA-TMZ-RTE-01 trial protocol. BMC Cancer. 2019;19(1):198. doi: 10.1186/s12885-019-5413-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Donawho CK, Luo Y, Luo Y, et al. ABT-888, an Orally Active Poly(ADP-Ribose) Polymerase Inhibitor that Potentiates DNA-Damaging Agents in Preclinical Tumor Models. Clin Cancer Res. 2007;13(9):2728–2737. doi: 10.1158/1078-0432.CCR-06-3039 [DOI] [PubMed] [Google Scholar]
- 256.Parrish KE, Cen L, Murray J, et al. Efficacy of PARP Inhibitor Rucaparib in Orthotopic Glioblastoma Xenografts Is Limited by Ineffective Drug Penetration into the Central Nervous System. Mol Cancer Ther. 2015;14(12):2735–2743. doi: 10.1158/1535-7163.MCT-15-0553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Robins HI, Zhang P, Gilbert MR, et al. A randomized phase I/II study of ABT-888 in combination with temozolomide in recurrent temozolomide resistant glioblastoma: an NRG oncology RTOG group study. J Neurooncol. 2016;126(2):309–316. doi: 10.1007/s11060-015-1966-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Sim HW, Galanis E, Khasraw M. PARP Inhibitors in Glioma: A Review of Therapeutic Opportunities. Cancers. 2022;14(4):1003. doi: 10.3390/cancers14041003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 259.Rahman MA, Gras Navarro A, Brekke J, et al. Bortezomib administered prior to temozolomide depletes MGMT, chemosensitizes glioblastoma with unmethylated MGMT promoter and prolongs animal survival. Br J Cancer. 2019;121(7):545–555. doi: 10.1038/s41416-019-0551-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Tang JH, Yang L, Chen JX, et al. Bortezomib inhibits growth and sensitizes glioma to temozolomide (TMZ) via down-regulating the FOXM1–Survivin axis. Cancer Commun. 2019;39(1):81. doi: 10.1186/s40880-019-0424-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Kong XT, Nguyen NT, Choi YJ, et al. Phase 2 Study of Bortezomib Combined With Temozolomide and Regional Radiation Therapy for Upfront Treatment of Patients With Newly Diagnosed Glioblastoma Multiforme: Safety and Efficacy Assessment. Int J Radiat Oncol. 2018;100(5):1195–1203. doi: 10.1016/j.ijrobp.2018.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Peereboom DM, Ye X, Mikkelsen T, et al. A Phase II and Pharmacodynamic Trial of RO4929097 for Patients with Recurrent/ Progressive Glioblastoma. Neurosurgery. 2021;88(2):246. doi: 10.1093/neuros/nyaa412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Vaz MÁ, Gironés R, Del Barco S, et al. Safety and efficacy of glasdegib in combination with temozolomide and radiotherapy in patients with newly diagnosed glioblastoma: Phase Ib/II GEINO 1602 trial. J Clin Oncol. 2022;40(16_suppl):2060–2060. doi: 10.1200/JCO.2022.40.16_suppl.2060 [DOI] [Google Scholar]
- 264.Sloan AE, Nock CJ, Ye X, et al. ABTC-0904: targeting glioma stem cells in GBM: a phase 0/II study of hedgehog pathway inhibitor GDC-0449. J Neurooncol. 2023;161(1):33–43. doi: 10.1007/s11060-022-04193-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.CI Tsien, Pugh SL, Dicker AP, et al. NRG Oncology/RTOG1205: A Randomized Phase II Trial of Concurrent Bevacizumab and Reirradiation Versus Bevacizumab Alone as Treatment for Recurrent Glioblastoma. J Clin Oncol. 2023;41(6):1285–1295. doi: 10.1200/JCO.22.00164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Arrillaga-Romany I, Sahebjam S, Picconi D, et al. ACTR-61. A RANDOMIZED PHASE 2 TRIAL OF CEDIRANIB IN COMBINATION WITH OLAPARIB VERSUS BEVACIZUMAB IN PATIENTS WITH RECURRENT GLIOBLASTOMA. Neuro-Oncol. 2019;21(Supplement_6):vi27. doi: 10.1093/neuonc/noz175.103 [DOI] [Google Scholar]