Abstract
The localization of FtsI (PBP3), a penicillin-binding protein specifically required for cell division in Escherichia coli, was investigated by immunofluorescence microscopy and found to localize to the septum. The localization of FtsI was not observed in ftsZ or ftsA mutants, indicating that it was dependent on the prior localization of these proteins. Addition of furazlocillin, a specific inhibitor of FtsI, prevented localization of FtsI even though FtsZ and FtsA localization occurred. Interestingly, the localization of FtsN was also prevented by furazlocillin. FtsZ displayed limited localization in furazlocillin-treated cells, whereas it was efficiently localized in FtsI-depleted cells. FtsW, another essential cell division protein, was also localized to the septum.
Many genes that are required for cell division have been identified in Escherichia coli. These include the fts genes ftsA, ftsZ, ftsQ, ftsN, ftsL, ftsK, ftsW, and ftsI and the recently identified zipA gene (21, 31). These genes, and probably others that are not yet identified, are responsible for the formation of the septum at midcell. The septum is formed through invagination of the cytoplasmic membrane and the accompanying synthesis of peptidoglycan at the leading edge of the invagination. This septal specific peptidoglycan biosynthesis occurs in two stages (36, 46, 47), an early stage that is penicillin insensitive and a later stage that is sensitive to penicillin and requires PBP3, the product of the ftsI gene (9, 40).
Genetic and inhibitor studies suggest that there are two modes of peptidoglycan synthesis involved in cell growth: elongation and septal. The elongation mode of synthesis requires PBP2 and RodA (41), whereas the septal mode requires FtsI (PBP3) and possibly FtsW (23, 40). PBP2 and FtsI are both high-molecular-weight penicillin-binding proteins that have transpeptidase activity (24, 25). Inhibition of PBP2 leads to loss of rod shape, whereas inhibition of FtsI results in a block to cell division (9, 26, 38). RodA is also required for rod morphology (41) and appears to augment the synthetic activity of PBP2, raising the possibility that they function in concert (25). Since FtsW is homologous to RodA but is specifically required for cell division, it has been suggested that FtsW, by analogy to RodA and PBP2, may function in concert with FtsI (23).
The synthesis of septal peptidoglycan involves a switch from the elongation mode of synthesis, which involves diffuse insertion of new peptidoglycan along the cylinder of the cell, to a septal mode, which involves insertion at the leading edge of the invaginating septum (46). These two distinct modes of growth of the peptidoglycan were inferred from genetic and inhibitor studies and demonstrated by autoradiography of sacculi following the incorporation of a radioactive precursor into peptidoglycan. More recently, the growth of the sacculus has been examined by prelabeling peptidoglycan and then watching the segregation of the label (16). The results agree with the first approach and also confirm that septal peptidoglycan biosynthesis occurs in two stages: an early stage that is penicillin insensitive and requires FtsZ but not FtsA, FtsI, or FtsQ and a late penicillin-sensitive stage that requires all of these proteins (16, 46). Studies over the past few years demonstrate that FtsZ assembles into a cytokinetic ring, designated the FtsZ or Z ring, that is required throughout the process of septation and directs the invagination of the septum (1, 2, 4, 8).
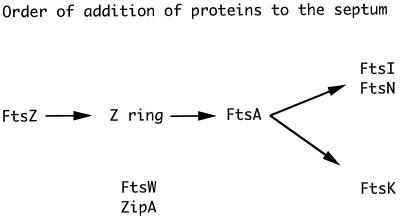
The formation of the Z ring at the future division site is an early event in the cell division cycle (1, 37). The Z ring is postulated to form through the GTP-dependent polymerization of FtsZ (12, 19, 30, 34, 35, 48), which is an ancestral homolog of eukaryotic tubulin (18, 29, 34). About the same time as the Z ring is formed, both ZipA and FtsA are recruited to the division site (5, 21, 32). The recruitment of FtsA, which interacts directly with FtsZ, is dependent on the prior localization of FtsZ (33, 44). However, it is not known if ZipA, which also interacts with FtsZ, precedes or follows FtsZ. ZipA, however, is much less conserved than FtsZ, suggesting that it has a function that has been added to the Z ring rather than having a primary function in localizing the ring (21). In addition to ZipA and FtsA, FtsN has also been localized to the septum (3). It appears at the septum later than FtsZ and FtsA, and its appearance at the septum is dependent on the prior localization of FtsZ and FtsA. In addition, FtsN’s localization depends on functional FtsI and FtsQ. One possibility is that FtsN forms a complex with FtsI and FtsQ that is attached to the Z ring through FtsA and carries out septal peptidoglycan biosynthesis (14, 15).
The order of appearance of division proteins at the septum as determined by immunofluorescence microscopy is fairly consistent with the order of action of these proteins inferred from the terminal phenotype of the corresponding mutants. These latter studies have indicated that FtsZ acts early and FtsA, FtsI, and FtsQ act later (7). More recently, FtsW has been studied and found to act early (28). Consistent with these results, the Z ring has been found to form in ftsA(Ts), ftsI(Ts), ftsQ(Ts), and ftsW(Ts) mutants (1, 27). However, cells depleted for FtsW contained mostly one or no Z rings, whereas FtsN-depleted cells contained multiple Z rings (3, 11). These results suggests that FtsW may have some role in Z ring stability or formation. In cells treated with cephalexin, which specifically blocks FtsI, one or two Z rings were observed after addition of the antibiotic. This result indicated that some Z rings can form in the absence of functional FtsI but that the capacity is limited and not all potential division sites are occupied (37).
From the above results, a reasonable hypothesis is that the Z ring is responsible for septal specific peptidoglycan biosynthesis by recruiting the necessary division proteins to the septum. This recruitment has been shown for FtsA, which has been implicated in FtsI function (42), and FtsN (3, 5). To further test this possibility, we have examined the localization of FtsI and FtsW during the cell division cycle. After this work was submitted, a report was published by Weiss et al. (45), who also observed that FtsI is localized to the septum.
MATERIALS AND METHODS
Bacterial strains and plasmids.
The strains used in this study are E. coli K-12 strains and include W3110 and derivatives of MC4100, including MC4100T(leu::Tn10), MCZ84 [leu::Tn10 ftsZ84(Ts)], MCA12 [leu::Tn10 ftsA12(Ts)], and MC123 [leu::Tn10 fts123(Ts)]; all were described previously (1). Strain JE7947 [ΔftsI::cat recA1 (pHR295)] has the chromosomal ftsI gene disrupted by cat and contains pHR295, which is a mini-F plasmid containing ftsI under lac promoter control (22). DBWC2 (ftsW::cat) has the chromosomal ftsW gene disrupted but carries an intact ftsW gene on pBPW1 under arabinose promoter control (11). pKD162 carries the fusion malG1-33-ftsI37-588 in the expression vector pJF118HE.
Antisera to FtsI and FtsW.
The sequences of FtsI and FtsW were scanned for regions of antigenicity. Peptides corresponding to residues 204 to 220 (PGERIVRKDRYGRVIED) of FtsI and residues 34 to 47 (REKDTDSLIMYDRT) of FtsW were synthesized on a polylysine carrier (MAP-8). The resultant peptides were used for raising peptide-specific antibodies in rabbits (Cocalico, Inc.). The rabbit antisera against FtsI and FtsW were purified through peptide affinity column chromatography. To do this, the FtsI or FtsW peptide was covalently linked to Affi-Gel 10 matrix (Bio-Rad). Each antiserum was then passed over a column containing this matrix with the appropriate linked peptide. The bound antibodies were eluted with glycine-HCl (pH 2.5) and dialyzed against phosphate-buffered saline buffer to give peptide-specific antibodies. Subsequent Western blot analysis with the cell lysates from W3110 containing pKD162 showed that affinity-purified FtsI antibody reacted only with the MalG-FtsI fusion protein which could be neutralized by the FtsI peptide (data not shown). The specificity of the antibodies was also confirmed following depletion experiments with JE7947 (pHR295). Western blotting with purified FtsW antibody also showed that the antibody reacted only with FtsW protein.
Detection of FtsI by Western blotting.
The samples for detecting FtsI in different strains and under different conditions were prepared as follows. JE7947(pHR295) cells (22) were cultured overnight in the LB medium with isopropyl-β-d-thiogalactopyranoside (IPTG; 0.5 mM) and then diluted 50-fold into the same medium. After growth to an optical density at 600 nm (OD600) of 0.4, the cells were washed three times with LB and diluted 1:8 in LB with 0.2% glucose. The culture was sampled at 0, 60, and 180 min. Samples of the wild-type strain MC4100 were taken from both exponentially growing cells and 1 h after furazlocillin addition (the same time point as samples for immunofluorescence). The samples for temperature-sensitive mutants MCZ84 and MCA12 were taken from cultures growing exponentially at 30°C and 30 min after shifting to 42°C. An equivalent amount of OD600 material from each of the samples was loaded onto a sodium dodecyl sulfate–12.5% polyacrylamide gel, electrophoresed, and blotted onto a nitrocellulose membrane. The membrane was probed with FtsI peptide-specific antibody (1:250) prepared as described above. Then the membrane was incubated with horseradish peroxidase-conjugated immunoglobulin G (1:8,000) and developed with enhanced chemiluminescence reagents (Amersham). For detecting FtsW, strain DBWC2 was grown in LB with arabinose as described previously (11). The culture was centrifuged, washed, and resuspended in LB with glucose. Samples were taken at various times and analyzed by Western blotting as described for FtsI except that FtsW-specific antibodies were used with a secondary antibody coupled to alkaline phosphatase.
Growth of cells and immunofluorescent staining.
Cells were grown in LB with antibiotics to select for plasmids and prepared for immunofluorescent staining as described by Addinall et al. (1). In experiments involving temperature-sensitive mutants, cultures growing exponentially at 30°C were shifted to 42°C for 30 min before fixation. For the antibiotic experiments, furazlocillin was used at a concentration of 0.25 μg/ml to inhibit the transpeptidase activity of FtsI. This concentration of the drug had little inhibitory effect on cell growth. Furazlocillin was added to cultures in exponential phase at an OD600 of around 0.08. At various times samples were removed and processed for immunofluorescent staining.
The immunofluorescent staining of cells was performed with either the affinity-purified anti-FtsI antibody or the affinity-purified anti-FtsW antibody as the primary antibody. The antiserum had to be used at a relatively high concentration (1:20 dilution, 30 μg/ml for anti-FtsI and 1:50 dilution for anti-FtsW). This relatively high concentration of antisera may be necessary due to the reported low level of FtsI, about 100 molecules per cell (17, 39). The treatment with lysozyme was carried out for 2, 4, 8, or 16 min, and the immunostaining was done for each time point. The results were consistent for each time point although in different experiments different time points gave clearer results (brighter localized signal relative to the background). Generally the shorter times gave slightly better results. The blocking peptides were added at a concentration of 100 μg/ml. The antibodies against FtsZ, FtsA, FtsK, and FtsN were all described previously (1, 3, 5) and were used at a dilution of 1:250 to 1:1,000. The secondary antibody was conjugated to the fluorophore Cy3 (Jackson Immunoresearch). Observation of stained cells and photography were carried out by using a filter block with a 517 to 552-nm excitation filter and a 590-nm barrier filter (XF34; Omega Optical).
RESULTS
Localization of FtsI (PBP3).
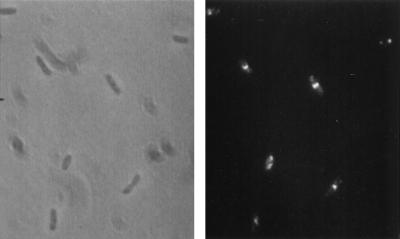
To determine if FtsI was localized to the septum an exponentially growing culture of MC4100 was fixed and processed for immunofluorescence microscopy as described in Materials and Methods. The antibodies to FtsI that were used in this study were affinity purified from an antiserum raised against an internal hydrophilic peptide of FtsI as described in Materials and Methods. The immunofluorescent staining revealed a readily visible band of fluorescence at midcell in approximately 50% of the cells, especially noticeable in cells with a constriction (Fig. 1A). In addition, each cell was faintly stained over the entire cell. To assess the specificity of the immunostaining, we performed a second experiment in which the synthetic peptide was added to block the antibodies. The addition of the blocking peptide prevented the appearance of the bright band of fluorescence otherwise seen at midcell (Fig. 1B), indicating that the bright localized fluorescence was due to the localization of FtsI. The failure of the blocking peptide to inhibit the generalized staining of the cells indicates that this is due to nonspecific staining. We have observed this nonspecific staining with all of the antibodies that we have used in the past, and although it does not obscure the bright localized fluorescence at midcell, it could obscure other, less intense signals.
FIG. 1.
Localization of FtsI in exponentially growing cells. (A) An exponential culture of MC4100 was processed for immunofluorescence microscopy and stained with antibodies to FtsI. The arrows indicate cells that have a clear band of fluorescence at midcell. (B) Cells were stained as for panel A except that the blocking peptide was added. The arrows indicate cells with constrictions that lack the bright band of fluorescence seen in panel A.
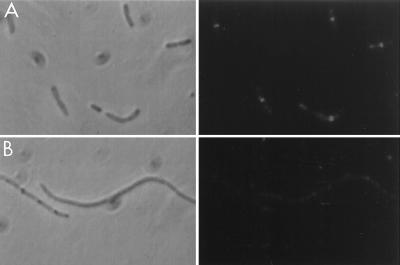
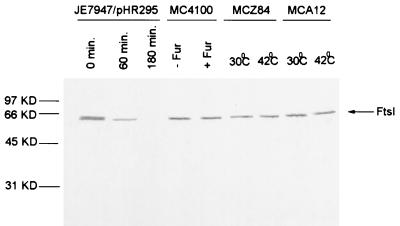
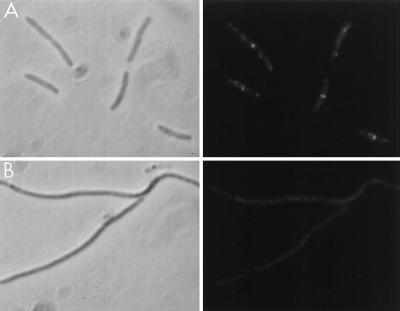
As a further control for specificity of the antibody, we obtained JE7947(pHR295) from H. Hara (22). This strain has the chromosomal ftsI gene replaced by cat with ftsI supplied by the plasmid under lac promoter control. Immunoblot analysis of samples taken at various times after removal of IPTG revealed a single band that disappeared with time (Fig. 2). By 60 min the FtsI band had decreased significantly, and by 180 min it was no longer visible. Microscopic examination of the culture revealed that cells started to filament at 60 min, and by 180 min cells were very filamentous. Immunofluorescence microscopy revealed that FtsI was localized to the septum in cells grown in the presence of IPTG, but no localized signal was detected in the filamentous cells at 180 min after removal of IPTG (Fig. 3).
FIG. 2.
Use of an FtsI depletion strain to examine the specificity of the FtsI peptide antibody. An exponential culture of JE7947(pHR295) growing with IPTG was centrifuged, washed, and resuspended in medium lacking IPTG. Samples were taken 0 (A) and 180 (B) min later and processed for immunofluorescence microscopy using the FtsI peptide-specific antibodies. Left, phase-contrast photomicrograph; right, fluorescence photomicrograph.
FIG. 3.
Western analysis of FtsI levels in strains used for FtsI localization. Cell lysates of the various strains used in this study were analyzed by Western blotting with the anti-FtsI peptide-specific antibodies. JE7947(pHR295) was sampled at 0, 60, and 180 min after removal of IPTG. MC4100 was sampled in the absence and 60 min after the addition of furazlocillin. Samples of the temperature-sensitive mutants MCZ84 and MCA12 were taken from cultures growing at 30°C and 30 min after a shift to 42°C. Samples were adjusted so that an equivalent amount of OD600 material was added in each lane.
Localization of FtsI is dependent on FtsZ and FtsA.
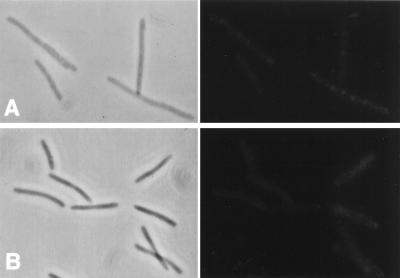
Since FtsI is localized to the septum, it might be recruited by FtsZ or FtsA since they appear at the division site early in the cell division cycle soon after the previous division is completed. To test this possibility, we examined the localization of FtsI in temperature-sensitive ftsZ84(Ts) and ftsA12(Ts) mutants. FtsZ and FtsA are not localized in ftsZ84 filaments, and FtsZ but not FtsA is localized in ftsA12 filaments (1, 5). Localization of FtsI in either of these mutants at the permissive temperature was similar to that for the wild type (data not shown). Figures 4A and B show phase-contrast and immunofluorescent photomicrographs of ftsZ84(Ts) and ftsA12(Ts) filaments, respectively. A diffuse staining of the filaments is visible; however, no observable bands of fluorescence indicative of FtsI localization are apparent. Immunoblot analysis revealed that FtsI was detectable and therefore stable in these filaments at the time of the photomicrographs (30 min after the temperature shift [Fig. 3]).
FIG. 4.
FtsI is not localized in ftsA and ftsZ filaments. Exponential cultures of MCZ84 [ftsZ84(Ts)] (A) and MCA12 [ftsA12(Ts)] (B) growing at 30°C were shifted to 42°C for 30 min. Samples were taken and immunostained for FtsI. Left, phase-contrast micrographs; right, fluorescence photomicrographs. Samples were also analyzed by Western blotting to determine the level of FtsI (Fig. 3).
Localization of FtsZ in FtsI-depleted filaments.
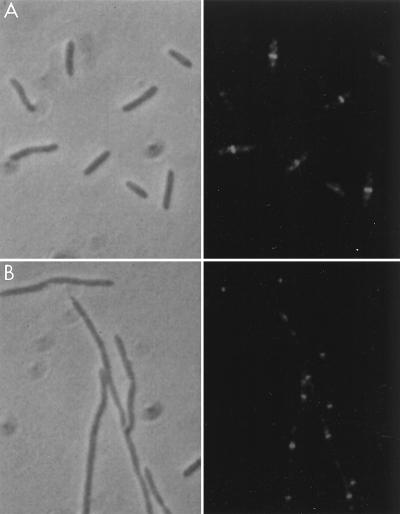
Since FtsI was not localized in the absence of FtsZ and FtsA, it is likely to depend on these proteins, directly or indirectly, for its localization. We therefore examined FtsZ localization in the FtsI-depleted filaments. Cells of JE7947(pHR295) were immunostained 180 min after IPTG removal (Fig. 5). As seen in Fig. 5, Z rings were abundant in these depleted filaments and were present at regular intervals throughout the filaments. These results indicate that FtsI is not required for FtsZ localization.
FIG. 5.
FtsZ localization in FtsI-depleted filaments. An exponential culture of JE7947(pHR295) growing with IPTG was centrifuged, washed, and resuspended in medium lacking IPTG. Cells were processed for immunofluorescence microscopy at 0 (A) and 180 (B) min after IPTG removal. Left, phase-contrast photomicrographs; right, fluorescence photomicrographs.
Localization of Fts proteins in the presence of furazlocillin.
Several antibiotics are known that preferentially target FtsI (PBP3), the product of the ftsI gene. These antibiotics inhibit septation resulting in filamentation similar to that caused by temperature-sensitive mutations in ftsI. One of these, furazlocillin, has been shown to be highly specific for FtsI (9). To test the effect of this antibiotic on the localization of FtsI as well as other Fts proteins, furazlocillin was added at 0.25 μg/ml. At this concentration, filamentation without cell lysis was observed for up to 2 to 3 h after addition of the drug. At 60 min after addition of furazlocillin, cells were processed for immunofluorescence microscopy. Staining these furazlocillin-induced filaments with antibodies to FtsZ revealed that FtsZ was localized in one of two patterns (Fig. 6A). The first had a ring at midcell, and the second had rings at the 1/4 and 3/4 positions with the midcell site vacant. This is the pattern previously reported by Pogliano et al. (37), who used cephalexin, another antibiotic that specifically targets FtsI. Their interpretation of this pattern is that Z rings that have not yet begun to constrict are stable (those observed at midcell) whereas FtsZ rings that are in the process of constriction fall apart upon addition of the antibiotic. In the latter cells, Z rings are able to form at 1/4 and 3/4 of the cell length after addition of cephalexin.
FIG. 6.
Effect of furazlocillin on the localization of Fts proteins. Furazlocillin (0.25 μg/ml) was added to an exponential culture of MC4100. Samples were taken before and 60 min after the addition. The samples were processed for immunostaining or for Western blotting (Fig. 3). The localization results are presented as a pair of photographs, phase contrast (left) and immunofluorescence (right). (A) Stained for FtsZ; (B) stained for FtsA; (C) stained for FtsI; (D) stained for FtsN; (E) stained for FtsK.
Staining the furazlocillin-treated cells for FtsA revealed the same pattern as observed with FtsZ (Fig. 6B), consistent with the previous observations that FtsA localization mimics that of FtsZ. We next examined the localization of FtsI and FtsN. We observed that neither of these proteins is localized in furazlocillin-treated cells (Fig. 6C and D, FtsI and FtsN, respectively) even though FtsZ and FtsA are. We confirmed that FtsI was stable following furazlocillin treatment (Fig. 3). Thus, blocking the transpeptidase activity of FtsI blocks its localization and prevents the localization of FtsN as well. We also examined the localization of FtsK in furazlocillin-treated cells (Fig. 6E). FtsK is an essential cell division protein (6) that is localized to the septum dependent on FtsZ and FtsA (43, 49). The present results indicate that FtsK is recruited to the septum independent of FtsI and FtsN although all three require FtsZ and FtsA. This order of addition of proteins to the septum is summarized in Fig. 7.
FIG. 7.
Model for the assembly of proteins at the septum. In this model, FtsZ polymerizes into the Z ring at the septum. This is accompanied by ZipA and FtsA, both of which interact with FtsZ. Although FtsA has been shown to follow FtsZ, this has not been demonstrated for ZipA and it is possible that ZipA precedes FtsZ. Following these three proteins, FtsN, FtsI, and FtsK are localized to the septum. FtsW is also localized; however, it is not clear at what point it assembles, although genetic studies suggest that it acts early. FtsQ is required for FtsN assembly, but it is not known if it is assembled at the septum. Since it is not clear at which point FtsW and ZipA are recruited to the septum, they are listed below the diagram.
Localization of FtsW.
Previous results have indicated that FtsW is required for cell division and that it is required early (11, 28). To determine if FtsW, which spans the membrane multiple times, is also localized to the septum, we prepared an antiserum to a hydrophilic peptide of FtsW. The antiserum was subsequently affinity purified by using the peptide. The affinity-purified antibodies were used to stain wild-type cells for the localization of FtsW (Fig. 8). The pattern of fluorescence observed was very similar to that observed for FtsI except that we also saw slight staining of the cell poles. The staining of cell poles was in small cells indicating that it may be the new cell pole that had retained some FtsW from the previous division. This staining was not present in all small cells. Many cells had a bright band of fluorescence at midcell. In constricting cells this band was shorter, indicating that FtsW was in a ring that decreased in diameter during septation. Occasionally we observed cells (<1%) in which the pattern of staining was not consistent with localization at midcell. Since these were rare, we assume that it is an artifact associated with the high concentration of antibody or due to variations in the lysis and fixation. The frequency of cells with FtsW localized was quite high (>50%).
FIG. 8.
Localization of FtsW to the septum. An exponential culture of MC4100 was processed for immunofluorescence microscopy and stained with antibodies to FtsW. Left, phase-contrast photomicrograph; right, immunofluorescence photomicrograph. Cells are observed with a band of fluorescence at midcell. In a few cells, a spot of fluorescence is observed at the cell pole; in others (<1%), the staining is not a clear pattern.
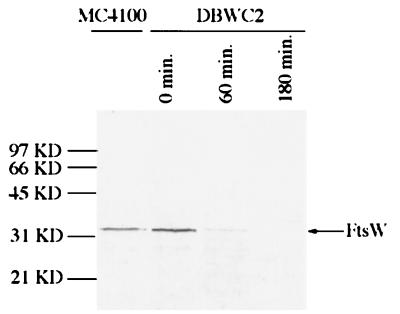
As a control for antibody specificity we used strain DBWC2, in which ftsW is supplied by a plasmid under arabinose promoter control. In the presence of arabinose, FtsW was detected at the septum, as in wild-type cells, whereas filaments obtained from a culture grown without arabinose for 180 min displayed no localization (Fig. 9). Western analysis of cell lysates of DBWC2 at 0, 60, and 180 min after removal of arabinose revealed one band (Fig. 10). At 0 min this band was stronger in intensity than in a control (MC4100), indicating that expression from the plasmid under our growth conditions was in slight excess over the wild type (MC4100 [Fig. 10]). However, the band had decreased significantly in intensity by 60 min and was no longer detectable by 180 min. We also attempted to stain for FtsW in ftsZ84(Ts) filaments. The staining pattern was not clear, as the high background made it difficult to interpret (data not shown). Thus, it is not clear at what step FtsW adds to the septum and what other proteins are required for its localization.
FIG. 9.
Use of an FtsW depletion strain to examine the specificity of the FtsW peptide antibody. DBWC2 growing in the presence of arabinose was centrifuged, washed, and resuspended in medium containing glucose. Samples were taken at 0 (A) and 180 (B) min after resuspension and stained with the FtsW peptide-specific antibodies for fluorescence microscopy. Left, phase-contrast micrographs; right, fluorescence micrographs.
FIG. 10.
Western analysis of an FtsW depletion strain. DBWC2 growing in the presence of arabinose was centrifuged, washed, and resuspended in medium containing glucose. Samples were taken at 0, 60, and 180 min after resuspension and analyzed by Western blotting using FtsW peptide-specific antibodies.
DISCUSSION
Our major findings are that the cell division proteins FtsI and FtsW are localized to the septum during cell division. FtsI and FtsW were localized in dividing cells at midcell, and the fluorescent bands corresponding to localized FtsI or FtsW decreased in diameter as cell division proceeded. These findings are consistent with a model in which FtsI is activated to carry out septum-specific peptidoglycan biosynthesis by recruitment to the septum. FtsW is also localized to the septum, again consistent with the suggestion that FtsI and FtsW act in concert to carry out septal peptidoglycan biosynthesis.
Our results do not address the different requirements for the two stages of septal peptidoglycan biosynthesis, the early penicillin-insensitive stage and a late penicillin-sensitive stage, observed by labeling the peptidoglycan (16, 36, 46). Those studies indicate that FtsZ but not FtsA, FtsI, and FtsQ is required for the early stage and that all are required for the late stage. Although FtsA is recruited to the septum concurrently with FtsZ, it is not required for the early stage. However, consistent with FtsA being required for the late stage, it is required to recruit FtsI, as well as FtsN, to the septum. The components, besides FtsZ, required for the early penicillin-insensitive stage are unknown (36).
We have suggested that one function of the Z ring is to recruit other division proteins to the septum (8, 30). This has been shown to be the case for the cytoplasmic protein FtsA (5, 32) and the cytoplasmic membrane protein FtsN (3), a protein with topology similar to that of FtsI. For FtsA, this is likely to be a direct interaction between FtsA and FtsZ, as the FtsA localization pattern always mimics that of FtsZ and these two proteins have been shown to interact in the yeast two-hybrid system (32, 33, 44). The localization of FtsN to the septum depends on the Z ring but is unlikely to interact directly with FtsZ. First, the N-terminal cytoplasmic tail and transmembrane region of FtsN are not required for localization (3, 15). Furthermore, FtsN localization depends on the prior localization of FtsA and requires functional FtsQ and FtsI. Consistent with this latter result, we observed that FtsN was not localized in the presence of furazlocillin an antibiotic that specifically blocks FtsI function.
The requirements for the localization of FtsI, a bitopic membrane protein with the same topology as FtsN (10), appears similar to that of FtsN. We observed that FtsI localization is dependent on FtsZ and FtsA. In an ftsZ mutant in which FtsZ localization is blocked, no localization of FtsI was observed. In an ftsA mutant where FtsA localization is blocked but FtsZ localization occurs, no FtsI localization was detected. Furthermore, FtsI was not localized in the presence of furazlocillin even though FtsZ and FtsA are localized.
The absence of FtsN localization in the presence of furazlocillin is quite interesting since it suggests that FtsN localization is dependent on a functional FtsI. It is also consistent with our previous observation that FtsN is not localized in an ftsI(Ts) mutant (3). Interestingly, the ftsN gene was isolated as a multicopy suppressor of ftsA12(Ts) but was found to be an even more effective suppressor of ftsI23(Ts) (14). All of these results are consistent with FtsN interacting with FtsI as previously suggested (14).
Previously, we have shown that the periplasmic domain of FtsN is sufficient for localization provided it is exported (3). The results with furazlocillin, which binds to the active site of FtsI located in the periplasm, indicate that the periplasmic domain of FtsI is important for localization and suggest that FtsI may have to be active in order to be localized. In contrast to FtsN, it has been shown that the N-terminal cytoplasmic region and transmembrane region of FtsI are essential for its function (20). Its possible that these regions, as well as the periplasmic domain, are required for FtsI localization to the septum.
Interestingly, a 100-fold overproduction of FtsI does not block septation (13). This result is also an indication that FtsI is locally activated in order to affect peptidoglycan biosynthesis and consistent with our suggestion that FtsI is activated by its recruitment to the septum. In contrast to the lack of effect by overproduction of the wild-type protein, overproduction of a mutant protein in which the active-site serine was replaced by cysteine resulted in inhibition of division (13). One possibility is that this protein competes with the wild type for targeting to the septum. In contrast, our results suggest that FtsI modified with furazlocillin is not capable of localization.
We used two approaches to block FtsI function, either a specific antibiotic or cells depleted of FtsI. Although both approaches blocked cell division, we noted differences in Z-ring formation. Our results with the addition of furazlocillin were the same as reported by Pogliano et al. (37), who used cephalexin. Most filaments contained either one centrally located Z ring or had two Z rings, one at the 1/4 position and another at the 3/4 position. In contrast, filaments arising from the depletion of FtsI had multiple Z rings that were regularly spaced, indicating that most potential division sites were occupied. Results with an ftsI temperature-sensitive mutant gave intermediate numbers of Z rings (1, 37). We are not sure why these different approaches to blocking ftsI function give different results. However, the depletion studies are likely to the least obtrusive, arguing that Z-ring formation does not require ftsI function.
Previous studies indicated two possible products for the ftsW gene, a long form and a short form (27). These would arise from translation beginning at two different in phase initiation codons that are 30 codons apart. Genetic analysis indicated that the short form is sufficient for normal growth and morphology. However, in our Western analysis we detect only one band, and the molecular weight clearly indicates that it is the long form.
Our observation that FtsW is localized to the septum is consistent with its essential role in cell division (11). The rather high frequency of cells observed with FtsW localized is consistent with FtsW acting early as determined from genetic studies (11, 27, 28). Although we observed clear localization of FtsW as indicated by the fluorescence at midcell, it was not possible to determine the dependency on other proteins due to high background. Thus, it is not clear if FtsW colocalizes with FtsZ to the future division site or if it lags behind or possibly even precedes FtsZ.
ACKNOWLEDGMENTS
We thank H. Hara for sending a strain.
This work was supported by NIH grant GM29764.
REFERENCES
- 1.Addinall S G, Bi E, Lutkenhaus J. FtsZ ring formation in fts mutants. J Bacteriol. 1996;178:3877–3884. doi: 10.1128/jb.178.13.3877-3884.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Addinall S G, Cao C, Lutkenhaus J. Temperature shift experiments with an ftsZ84(Ts) strain reveal rapid dynamics of FtsZ localization and indicate that the Z ring is required throughout septation and cannot reoccupy division sites once constriction has initiated. J Bacteriol. 1997;179:4277–4284. doi: 10.1128/jb.179.13.4277-4284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Addinall S G, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in E. coli. Mol Microbiol. 1997;25:303–310. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- 4.Addinall S G, Lutkenhaus J. FtsZ spirals and arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol Microbiol. 1996;22:231–238. doi: 10.1046/j.1365-2958.1996.00100.x. [DOI] [PubMed] [Google Scholar]
- 5.Addinall S G, Lutkenhaus J. FtsA is localized to the septum in an FtsZ-dependent manner. J Bacteriol. 1996;178:7167–7172. doi: 10.1128/jb.178.24.7167-7172.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Begg K J, Dewar S J, Donachie W D. A new Escherichia coli cell division gene, ftsK. J Bacteriol. 1995;177:6211–6222. doi: 10.1128/jb.177.21.6211-6222.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Begg K J, Donachie W D. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J Bacteriol. 1985;163:615–622. doi: 10.1128/jb.163.2.615-622.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- 9.Botta G A, Park J T. Evidence for involvement of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. J Bacteriol. 1981;145:333–340. doi: 10.1128/jb.145.1.333-340.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bowler L D, Spratt B G. Membrane topology of penicillin-binding protein 3 of Escherichia coli. Mol Microbiol. 1989;3:1277–1286. doi: 10.1111/j.1365-2958.1989.tb00278.x. [DOI] [PubMed] [Google Scholar]
- 11.Boyle D S, Khattar M M, Addinall S G, Lutkenhaus J, Donachie W D. ftsW is an essential cell division gene in Escherichia coli. Mol Microbiol. 1997;24:1263–1273. doi: 10.1046/j.1365-2958.1997.4091773.x. [DOI] [PubMed] [Google Scholar]
- 12.Bramhill D, Thompson C M. GTP-dependent polymerization of Escherichia coli FtsZ protein to form tubules. Proc Natl Acad Sci USA. 1994;91:5813–5817. doi: 10.1073/pnas.91.13.5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Broome-Smith J K, Hedge P J, Spratt B G. Production of thiol-penicillin-binding protein 3 of Escherichia coli using a two primer method of site-directed mutagenesis. EMBO J. 1985;4:231–235. doi: 10.1002/j.1460-2075.1985.tb02340.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dai K, Xu Y, Lutkenhaus J. Cloning and characterization of ftsN, an essential cell division gene in Escherichia coli, isolated as a multicopy suppressor of ftsA12(Ts) J Bacteriol. 1993;175:3790–3797. doi: 10.1128/jb.175.12.3790-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dai K, Xu Y, Lutkenhaus J. Topological characterization of the essential Escherichia coli cell division protein FtsN. J Bacteriol. 1996;178:1328–1334. doi: 10.1128/jb.178.5.1328-1334.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.de Pedro M A, Quintela J C, Holtje J-V, Schwarz H. Murein segregation in Escherichia coli. J Bacteriol. 1997;179:2823–2834. doi: 10.1128/jb.179.9.2823-2834.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dougherty T J, Kennedy K, Kessler R E, Pucci M J. Direct quantitation of the number of individual penicillin-binding proteins per cell in Escherichia coli. J Bacteriol. 1996;178:6110–6115. doi: 10.1128/jb.178.21.6110-6115.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erickson H P. FtsZ, a prokaryotic homolog of tubulin? Cell. 1995;80:367–370. doi: 10.1016/0092-8674(95)90486-7. [DOI] [PubMed] [Google Scholar]
- 19.Erickson H P, Taylor D W, Taylor K A, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheet and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA. 1996;93:519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guzman L M, Weiss D S, Beckwith J. Domain-swapping analysis of FtsI, FtsL, and FtsQ, bitopic membrane proteins essential for cell division in Escherichia coli. J Bacteriol. 1997;179:5094–5103. doi: 10.1128/jb.179.16.5094-5103.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hale C A, de Boer P A J. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell. 1997;88:175–185. doi: 10.1016/s0092-8674(00)81838-3. [DOI] [PubMed] [Google Scholar]
- 22.Hara J, Yasuda S, Horiuchi K, Park J T. A promoter for the first nine gene of the Escherichia coli mra cluster of cell division and cell envelope biosynthesis genes, including ftsI and ftsW. J Bacteriol. 1997;179:5802–5811. doi: 10.1128/jb.179.18.5802-5811.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ikeda M, Sato T, Wachi M, Jung H K, Ishino F, Kobayashi Y, Matsuhashi M. Structural similarity among Escherichia coli FtsW and RodA proteins and Bacillus subtilis SpoVE protein, which function in cell division, cell elongation, and spore formation, respectively. J Bacteriol. 1989;171:6375–6378. doi: 10.1128/jb.171.11.6375-6378.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishino F, Matsuhashi M. Peptidoglycan synthetic enzyme activities of highly purified penicillin-binding protein 3 in Escherichia coli: a septum-forming reaction sequence. Biochem Biophys Res Commun. 1981;101:905–911. doi: 10.1016/0006-291x(81)91835-0. [DOI] [PubMed] [Google Scholar]
- 25.Ishino F, Park W, Tomioka S, Tamaki S, Takase I, Kunugita K, Matsuzawa H, Asoh S, Ohta T, Spratt B G, Matsuhashi M. Peptidoglycan synthetic activities in membranes of Escherichia coli caused by overproduction of penicillin-binding protein 2 and RodA protein. J Biol Chem. 1986;261:7024–7031. [PubMed] [Google Scholar]
- 26.James R, Haga A Y, Pardee A B. Inhibition of an early event in the cell division cycle of Escherichia coli by FL1060, an amidinopenicillanic acid. J Bacteriol. 1975;122:1283–1292. doi: 10.1128/jb.122.3.1283-1292.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khattar M M, Addinall S G, Stedul K H, Boyle D S, Lutkenhaus J, Donachie W D. Two polypeptide products of the Escherichia coli cell division gene ftsW and a possible role for FtsW in FtsZ function. J Bacteriol. 1997;179:784–793. doi: 10.1128/jb.179.3.784-793.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Khattar M M, Begg K J, Donachie W D. Identification of FtsW and characterization of a new ftsW division mutant of Escherichia coli. J Bacteriol. 1994;176:7140–7147. doi: 10.1128/jb.176.23.7140-7147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lowe J, Amos L. Crystal structure of the bacterial cell-division protein FtsZ. Nature. 1998;391:203–206. doi: 10.1038/34472. [DOI] [PubMed] [Google Scholar]
- 30.Lutkenhaus J. FtsZ ring in bacterial cytokinesis. Mol Microbiol. 1993;9:404–409. doi: 10.1111/j.1365-2958.1993.tb01701.x. [DOI] [PubMed] [Google Scholar]
- 31.Lutkenhaus J, Addinall S G. Bacterial cell division and the Z ring. Annu Rev Biochem. 1997;66:93–116. doi: 10.1146/annurev.biochem.66.1.93. [DOI] [PubMed] [Google Scholar]
- 32.Ma X, Ehrhardt D W, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ma X, Sun Q, Wang R, Singh G, Jonietz E I, Margolin W. Interactions between heterologous FtsA and FtsZ proteins at the FtsZ ring. J Bacteriol. 1997;179:6788–6797. doi: 10.1128/jb.179.21.6788-6797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mukherjee A, Lutkenhaus J. Guanine nucleotide-dependent assembly of FtsZ into filaments. J Bacteriol. 1994;176:2754–2758. doi: 10.1128/jb.176.9.2754-2758.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mukherjee A, Lutkenhaus J. Dynamic assembly of FtsZ regulated by GTP hydrolysis. EMBO J. 1998;17:462–469. doi: 10.1093/emboj/17.2.462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nanninga N. Cell division and peptidoglycan assembly in Escherichia coli. Mol Microbiol. 1991;5:791–795. doi: 10.1111/j.1365-2958.1991.tb00751.x. [DOI] [PubMed] [Google Scholar]
- 37.Pogliano J, Pogliano K, Weiss D S, Losick R, Beckwith J. Inactivation of FtsI inhibits constriction of the FtsZ cytokinetic ring and delays the assembly of FtsZ rings at potential division sites. Proc Natl Acad Sci USA. 1997;94:559–564. doi: 10.1073/pnas.94.2.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spratt B G. Distinct penicillin-binding proteins involved in the division, elongation and shape of Escherichia coli K-12. Proc Natl Acad Sci USA. 1975;72:2999–3003. doi: 10.1073/pnas.72.8.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Spratt B G. Properties of penicillin-binding proteins of Escherichia coli K12. Eur J Biochem. 1977;72:341–352. doi: 10.1111/j.1432-1033.1977.tb11258.x. [DOI] [PubMed] [Google Scholar]
- 40.Spratt B G. Temperature-sensitive cell division mutants of Escherichia coli with thermolablie penicillin binding proteins. J Bacteriol. 1977;131:293–305. doi: 10.1128/jb.131.1.293-305.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Spratt B G, Boyd A, Stoker N. Defective and plaque-forming lambda transducing bacteriophage carrying penicillin-binding protein-cell shape genes: genetic and physical mapping and identification of gene products from thelip-dacA-rodA-pbpA-leuS region of the Escherichia coli chromosome. J Bacteriol. 1980;143:569–581. doi: 10.1128/jb.143.2.569-581.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tormo A, Ayala J A, de Pedro M A, Vicente M. Interaction of FtsA and PBP3 proteins in the Escherichia coli septum. J Bacteriol. 1986;166:985–992. doi: 10.1128/jb.166.3.985-992.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang, L., and J. Lutkenhaus. Unpublished data.
- 44.Wang X, Huang J, Mukherjee A, Cao C, Lutkenhaus J. Analysis of the interaction of FtsZ with itself, GTP and FtsA. J Bacteriol. 1997;179:5551–5559. doi: 10.1128/jb.179.17.5551-5559.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weiss D S, Pogliano K, Carson M, Guzman L M, Fraipont C, Nguyen-Disteche M, Losick R, Beckwith J. Localization of the Escherichia coli cell division protein Fts1 (PBP3) to the division site and cell pole. Mol Microbiol. 1997;25:671–681. doi: 10.1046/j.1365-2958.1997.5041869.x. [DOI] [PubMed] [Google Scholar]
- 46.Wientjes F B, Nanninga N. Rate and topography of peptidoglycan synthesis during cell division in Escherichia coli: concept of a leading edge. J Bacteriol. 1989;171:3412–3419. doi: 10.1128/jb.171.6.3412-3419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Woldringh C L, Huls P, Pas E, Brakenhoff G J, Nanninga N. Topography of peptidoglycan synthesis during elongation and polar cap formation in a cell division mutant of Escherichia coli MC4100. J Gen Microbiol. 1987;133:575–586. [Google Scholar]
- 48.Yu X-C, Margolin W. Ca2+-mediated GTP-dependent dynamic assembly of bacterial cell division protein FtsZ into asters and polymer networks in vitro. EMBO J. 1997;16:5455–5463. doi: 10.1093/emboj/16.17.5455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yu X-C, Tran A H, Sun Q, Margolin W. Localization of cell division protein FtsK to the Escherichia coli septum and identification of a potential N-terminal targeting domain. J Bacteriol. 1998;180:1296–1304. doi: 10.1128/jb.180.5.1296-1304.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]