Abstract
Tn4652 is a derivative of the toluene degradation transposon Tn4651 that belongs to the Tn3 family of transposons (M. Tsuda and T. Iino, Mol. Gen. Genet. 210:270–276, 1987). We have sequenced the transposase gene tnpA of transposon Tn4652 and mapped its promoter to the right end of the element. The deduced amino acid sequence of tnpA revealed 96.2% identity with the putative transposase of Tn5041. Homology with other Tn3 family transposases was only moderate (about 20 to 24% identity), suggesting that Tn4652 and Tn5041 are distantly related members of the Tn3 family. Functional analysis of the tnpA promoter revealed that it is active in Pseudomonas putida but silent in Escherichia coli, indicating that some P. putida-specific factor is required for the transcription from this promoter. Additionally, tnpA promoter activity was shown to be modulated by integration host factor (IHF). The presence of an IHF-binding site upstream of the tnpA promoter enhanced the promoter activity. The positive role of IHF was also confirmed by the finding that the enhancing effect of IHF was not detected in the P. putida ihfA-deficient strain A8759. Moreover, the Tn4652 terminal sequences had a negative effect on transcription from the tnpA promoter in the ihfA-defective strain. This finding suggests that IHF not only enhances transcription from the tnpA promoter but also alleviates the negative effect of terminal sequences of Tn4652 on the promoter activity. Also, an in vitro binding assay demonstrated that both ends of Tn4652 bind IHF from a cell lysate of E. coli.
Transposons are discrete DNA segments that can move from one genetic location to another. This process does not involve homologous recombination systems of the host but requires a gene product encoded by the moving element itself—transposase. Transposase interacts site specifically with the ends of the transposon, cleaves the DNA at both termini of the element, and carries out the strand transfer reaction (reviewed in references 22 and 34).
Transposition of a mobile element is precisely controlled and depends on the availability of the active transposase. Moreover, in several cases the transposition reaction itself is controlled and modulated by some other transposon-encoded protein(s) and/or host factors (29). One of the host factors participating in the transposition is integration host factor (IHF) (17, 32, 41, 42).
IHF is a sequence-specific sharply DNA bending heterodimeric protein which is involved in a variety of cellular processes including λ site-specific recombination, transposition, replication, and positive and negative control of gene expression (15). IHF has been found to regulate gene expression in a number of gram-negative bacteria (21). IHF genes from diverse bacterial species are well conserved (8, 12). In most cases, the role of IHF is architectural: it facilitates the formation of nucleoprotein complexes through strong bending of DNA. However, activation of transcription from λ pL1 and Mu phage Pe promoters involves direct interaction of IHF with RNA polymerase (20, 44).
Many mobile DNA elements carry IHF-binding sites at one or both termini (14, 18, 25, 32, 46). For γδ (Tn1000), it was shown that binding of IHF to the ends of the transposon facilitates binding of transposase (46). Mostly, IHF affects transposition positively (10, 35, 42). For example, in the well-studied Mu phage transposition, IHF acts positively both by enhancing transcription from the early promoter Pe and favoring the stabile synaptic complex formation that is required in the initial step of transposition (2, 44). However, there are also reports about the negative role of IHF on transposition (17, 41).
According to Kleckner (28), transposable elements from bacteria can be divided into three classes. Class II contains evolutionarily related elements mostly belonging to the Tn3 family of transposons. Tn3 family transposons translocate replicatively and generate 5-bp direct duplications of the target DNA (40). Members of the Tn3 family exhibit similar inverted repeats 35 to 48 bp in length and similar transposases. Comparison of the Tn3 family transposases showed their clustering into three subgroups (26). Tn3 and Tn21 subgroups associate transposons from gram-negative bacteria, while transposons from gram-positive bacteria belong to the third subgroup. Transposases of IS1071 and recently characterized mercury resistance transposon Tn5041 are more diverse and cannot be included to any of these three subgroups (26).
Pseudomonas putida PaW85 carries in its chromosome transposon Tn4652, a 17-kb derivative of the 56-kb toluene degradation transposon Tn4651 coding for xyl genes (43). Tsuda and Iino (43) have shown that Tn4652 belongs to the Tn3 family of transposons, as determined from its transposition properties. Genetic analysis on Tn4652 localized the putative transposase gene to a 3.0-kb segment at the end of the right arm of the element (43). However, regulation of the Tn4652 transposase gene as well as the mechanism of transposition reactions of Tn4652 have remained unexplored.
This study aims to elucidate the regulation of the Tn4652 transposase gene. We sequenced the Tn4652 transposase gene tnpA and localized the promoter of the gene to the right end of the element. Analysis of the deduced amino acid sequence of the tnpA gene revealed highest homology (96.2% identity) with the transposase of Tn5041. Study of the regulation of the tnpA promoter from Tn4652 demonstrated that (i) the promoter was active in P. putida but silent in Escherichia coli and (ii) the IHF-binding site at positions −73 to −85 relative to the transcription start point affected transcription from the tnpA promoter in P. putida positively. Gel mobility shift experiments with cell lysates of E. coli and P. putida were carried out to examine binding of IHF to the ends of Tn4652 in vitro.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The bacterial strains and plasmids used in this study are described in Table 1. Construction of the new broad-host-range promoter-probe vector pKTlacZ is depicted in Fig. 1. Bacteria were grown on LB medium (33). Antibiotics were added at the indicated final concentrations: for E. coli, ampicillin at 100 μg/ml and tetracycline at 15 μg/ml; for P. putida, carbenicillin at 1,500 μg/ml and streptomycin at 500 μg/ml. P. putida was incubated at 30°C, and E. coli was incubated at 37°C. Early-stationary-phase cultures were used for enzyme assays. E. coli was transformed with plasmid DNA as described by Hanahan (23). P. putida was electrotransformed by using the protocol of Sharma and Schimke (39).
TABLE 1.
Bacterial strains and plasmids used
| Strain or plasmid | Genotype or construction | Source or reference |
|---|---|---|
| E. coli | ||
| HB101 | subE44 subF58 hsdS3(rB−mB−) recA13 ara-14 proA2 lacY1 galK2 rpsL20 xyl-5 mtl-1 | 6 |
| WM2015 | subE thi Δ(lac-pro) | 30 |
| WM2017 | WM2015 himA::TcrhimD::Cmr | 30 |
| P. putida | ||
| PaW85 | Tn4652 | 4 |
| PRS2000 | Tn4652-free | 45 |
| KT2442 | Tn4652 xylRS Pu-lacZ Rifr Smr | 8 |
| A8759 | KT2442 Tn4652 ihfA::Kmr Pu-lacZ | 8 |
| Plasmids | ||
| pBluescript KS | Cloning vector (Apr) | Stratagene |
| pEST1332 | Plasmid pAYC32 carrying promoterless pheBA operon | 27 |
| p1332S/C | PCR-generated 122-bp Tn4652 right-end fragment (primers Osac and Ocla) cloned into pEST1332 | This work (Fig. 2) |
| p1332IHF/C | PCR-generated 83-bp Tn4652 right-end fragment (primers Oihf and Ocla) cloned into pEST1332 | This work (Fig. 2) |
| p1332D/C | SacI-DraI deletant of p1332S/C | This work (Fig. 2) |
| p1332S/N | NheI-ClaI deletant of p1332S/C | This work (Fig. 2) |
| pKT240 | Cloning vector (Apr Kmr) | 3 |
| pKRZ-1 | Cloning vector (Apr Kmr) | 37 |
| pKTlacZ | Promoter probe vector containing lacZ gene from pKRZ-1 cloned into pKT240 | This work (Fig. 1) |
| pKTlacZS/C | PCR-generated 122-bp Tn4652 right-end fragment (primers Osac and Ocla) cloned into pKTlacZ | This work (Fig. 2) |
| pKTlacZIHF/C | PCR-generated 83-bp Tn4652 right-end fragment (primers Oihf and Ocla) cloned into pKTlacZ | This work (Fig. 2) |
| pKTlacZD/C | SacI-DraI deletant of pKTlacZS/C | This work (Fig. 2) |
| pHNβα | Plasmid carrying E. coli IHF genes ihfA and ihfB | 31 |
| pUC18 | Cloning vector (Apr) | 48 |
| pUCPu130 | 129-bp DpnI fragment of Pu promoter region of xyl genes in TOL plasmid cloned into pUC18 | This work |
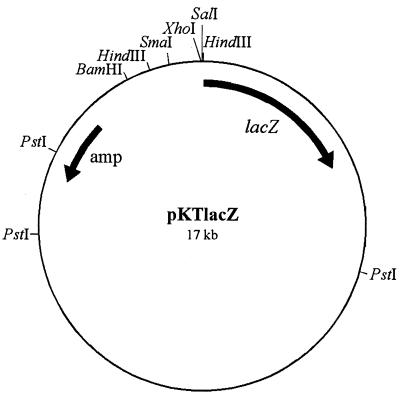
FIG. 1.
Map of the broad-host-range promoter probe vector pKTlacZ. An about 5-kb HindIII-PstI fragment carrying the lacZ gene originates from plasmid pKRZ-1 (37). After this fragment was cloned into pBluescriptSK(+) it was recut with XhoI and SmaI and subcloned into pKT240 opened with XhoI and Ecl136II. Suitable cloning sites are BamHI, HindIII (two sites), SmaI, XhoI, and SalI.
DNA manipulations and mRNA mapping.
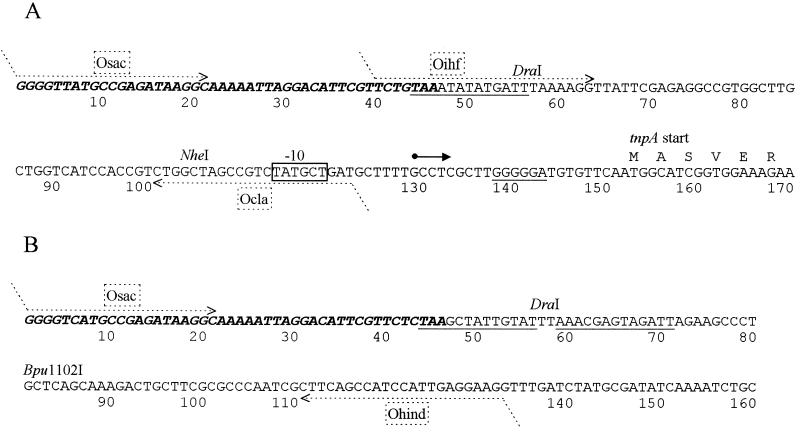
DNA sequencing was performed with a Sequenase version 2.0 DNA sequencing kit (Amersham). Subclones of the tnpA promoter region (Table 1) were obtained by cloning PCR products. The following oligonucleotides, containing suitable restriction sites (SacI and ClaI; boldfaced) and complementary to nucleotides (nt) 1 to 21, 40 to 63, and 101 to 122 relative to the right end of the Tn4652, were used in cloning: Osac (5′-CGTGAGCTCGGGGTTATGCCGAGATAAGGC-3′), Oihf (5′-CGTGAGCTCTGTAAATATATGATTTAAAAGG-3′), and Ocla (5′-CGTATCGATCAGCATAGACGGCTAGCCAG-3′). Locations of these oligonucleotides are shown in Fig. 2A.
FIG. 2.
Sequence analysis of the right end (A) and left end (B) of Tn4652. The 48-bp inverted repeats are in boldface italics. Potential IHF-binding sites resembling the E. coli IHF-binding consensus sequence WATCAANNNNTTR and ribosome-binding site of the tnpA gene are underlined. The transcription start of tnpA is indicated by the solid arrow, and the putative −10 hexamer of the promoter is boxed. The deduced amino acid sequence of the tnpA gene is presented starting from the second ATG. The first six amino acids are shown. Locations of primers used in PCR for cloning of the tnpA promoter and for generating DNA fragments for the gel mobility shift assay are indicated by dotted-line arrows. 5′ ends of the oligonucleotides not complementary to the termini of Tn4652 are indicated by sloping dotted lines. Primers Osac, Oihf, and Ocla contain restriction site SacI or ClaI for cloning of the tnpA promoter.
A reverse transcriptase reaction was carried out to identify the 5′ end of mRNA initiated from the tnpA promoter by a procedure described previously by our group (36). Total RNA (20 μg), purified from P. putida PaW85, P. putida PRS2000, and E. coli HB101 cells as described by Blomberg et al. (5), was used as the template. Oligonucleotide 5′-GTATGCTTGGCAGTCGT-3′, complementary to nt −120 to −136 relative to the start codon of the reporter gene pheB, was used in the primer extension analysis.
Enzyme assays.
The catechol 1,2-dioxygenase (C12O) assay was carried out as described by Hegeman (24). The β-Galactosidase (β-Gal) assay was performed as specified by Miller (33). Protein concentration in cell lysates was measured by the Bradford method (7).
Gel mobility shift assay.
Cell lysates used in gel shift assays were prepared from 30-ml early-stationary-phase cultures. The cells were pelleted and sonicated in 1× binding buffer (25 mM Tris-HCl [pH 7.5], 0.05 mM EDTA, 5 mM dithiothreitol, 25 mM NaCl, 50 mM KCl, 5% glycerol). Protein concentration in cleared lysates was 15 to 20 mg/ml; 1 to 3 μl of undiluted lysate or lysate diluted in 1× binding buffer was used in gel shift assays.
The following DNA fragments were used in gel shift binding assays: (i) a 108-bp DNA restriction fragment containing the right end of transposon Tn4652 up to the NheI restriction site (Fig. 2A); (ii) a 140-bp DNA restriction fragment containing the left end of the transposon up to the Bpu1102I restriction site (Fig. 2B); and (iii) a 140-bp DNA restriction fragment containing a 129-bp DpnI segment of the Pu promoter region cloned into pUC18 (Table 1). These DNA fragments were end labeled with [α-32P]dCTP, using the Klenow fragment of DNA polymerase I, and subsequently purified through an polyacrylamide gel. The binding reaction was carried out in a volume of 20 μl. About 1 ng (1,000 cpm) of DNA probe was incubated at 20°C for 20 min with different cell lysates in 1× binding buffer containing 1 μg of bovine serum albumin and 5 μg of salmon sperm DNA. The following specific nonlabeled competitor DNAs containing IHF-binding sites were generated by PCR: (i) a 122-bp fragment of the right end of Tn4652, amplified by using primers Osac and Ocla (Fig. 2A); (ii) a 132-bp fragment of the left end of Tn4652, amplified by using primers Osac and Ohind (5′-CGTAAGCTTCCTCAATGGATGGCTGAAG-3′ [Fig. 2B]); and (iii) a 250-bp DNA fragment including a 129-bp DpnI segment of the Pu promoter region cloned into pUC18 (Table 1), amplified by using pUC18 reverse and forward primers. When the specific competitor DNA was used, the cell lysate was added last to the binding reaction. After incubation, the reaction mixture was loaded on a 1-h-prerun 5% nondenaturing polyacrylamide gel. Electrophoresis was carried out at room temperature in 0.5× Tris-borate-EDTA buffer at 10 V/cm for 2 h. The gels were dried and autoradiographed or exposed to a phosphorimager screen.
Nucleotide sequences accession numbers.
The 3,348-bp sequence of the right arm of Tn4652 has been assigned accession no. X83686 in the EMBL database. The accession number of the 604-bp-long sequence of the left end of Tn4652 is X83687.
RESULTS
Sequence of the Tn4652 transposase shows highest homology with the putative transposase of Tn5041.
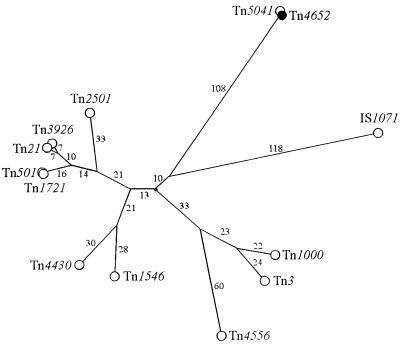
Genetic analysis has localized the transposase gene of Tn4652 to the right arm of the transposon (43). A 3.2-kb DraI-HindIII fragment from Tn4652 DNA, known to contain the transposase gene tnpA, was subcloned into the pBluescript KS(+) vector. Sequencing of the DNA fragment revealed a single 3,012-bp open reading frame (ORF) directed inward from the right end of the transposon. The ORF has two potential ATG start codons, separated by 6 bp (Fig. 2A). Since the potential ribosome-binding site overlaps the first ATG, initiation of translation of tnpA from the second ATG is more likely. The predicted protein, starting from the second ATG, is 1,001 amino acids long, with a calculated molecular mass of 114 kDa. Sequence comparison with the translated sequences of genes in the EMBL database by using the FASTA and BLAST programs revealed a high degree of homology of the Tn4652 tnpA with the putative transposase of the mercury resistance transposon Tn5041 (96.2% identity). Homology with other transposases of Tn3 family transposons (Tn501, Tn1721, Tn1546, Tn21, Tn4430, Tn3926, Tn2501, Tn3, Tn4556, Tn1000, and IS1071) was much lower (about 20 to 24% identity and 30 to 36% similarity). In most of the Tn3 family transposons, the 3′ ends of the transposase genes terminate within one of the terminal repeats of the element (40). Contrary to that, the direction of the tnpA gene of Tn4652 is opposite, starting from the right end of the transposon. Multiple alignment of Tn3 family transposase sequences homologous to Tn4652 transposase was performed via the CBRG server (http://cbrg.inf.ethz.ch/) by using the Darwin program. Alignment revealed stronger conservation in C termini of these proteins (data not shown). The phylogenetic tree of the entire protein sequences demonstrated that the Tn4652 transposase is quite distantly related to other members of the Tn3 family and might constitute a new Tn3 family subgroup together with Tn5041 (Fig. 3).
FIG. 3.
Unrooted phylogenetic tree of the Tn3 family transposase proteins. Multiple alignment of transposase sequences and construction of the phylogenetic tree were carried out via the CBRG server as described in the text. PAM distances are indicated at branches of the tree. DNA accession numbers and hosts (in parentheses): Tn2501 (E. coli), Y00502; Tn3926 (E. coli), X14236; Tn21 (E. coli), X04891; Tn501 (P. aeruginosa), X03406; Tn1721 (E. coli), X61367; Tn4430 (Bacillus thuringiensis), X07651; Tn1546 (Enterococcus faecium), M97297; Tn4556 (Streptomyces fradiae), M29297; Tn3 (E. coli), V00613; Tn1000 (E. coli), X60200; IS1071 (Alcaligenes sp. strain BR60), M65135; Tn5041 (Pseudomonas sp.), X98999; Tn4652 (P. putida), X83686.
Mapping of the tnpA promoter.
The ORF of tnpA gene starts at 152 bp from the right end of transposon Tn4652. To map the tnpA promoter, we constructed plasmid p1332S/C by cloning the 122-bp DNA segment covering the right end of the transposon upstream of the promoterless pheBA operon in plasmid pEST1332 (Table 1 and Fig. 2A). In addition, plasmids p1332D/C and p1332S/N, containing Tn4652 right-end DNA from nt 58 to 122 and from nt 1 to 104, respectively, were constructed (Table 1 and Fig. 2A). E. coli HB101 and P. putida PaW85 and PRS2000 were transformed with these plasmids. As many transposase promoters are downregulated by transposon-encoded repressor proteins (29), the transposon Tn4652-free P. putida strain PRS2000 was used as a reference strain to distinguish potential effects of chromosomally encoded transposon protein(s) on the promoter activity in strain PaW85.
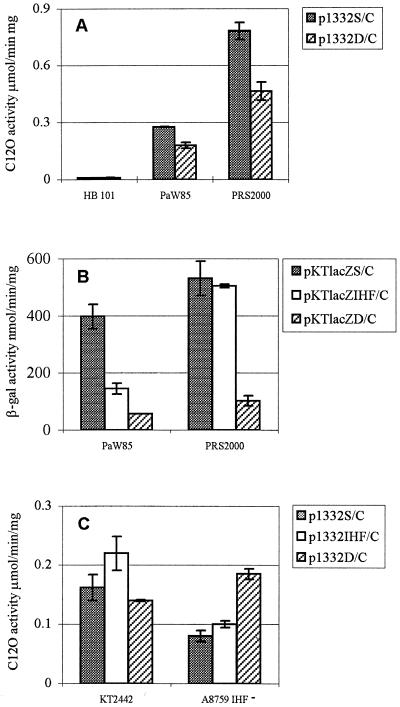
The C12O assay was carried out to study expression of the reporter gene pheB in the plasmids constructed. P. putida PaW85 and PRS2000 harboring plasmids p1332S/C and p1332D/C revealed promoter activity (Fig. 4A), but no C12O activity was detected in bacteria carrying plasmid p1332S/N, indicating that the tnpA promoter was disrupted in this construct (data not shown). Data in Fig. 4A show that C12O activities measured in P. putida PRS2000 were more than twofold higher than those measured in P. putida PaW85. Additionally, bacteria harboring p1332S/C revealed about twofold-higher enzyme activities than bacteria containing p1332D/C. None of the promoter constructs studied revealed activity in E. coli (Fig. 4A).
FIG. 4.
C12O (A and C) and β-Gal (B) activities measured in E. coli HB101 and different P. putida strains carrying different tnpA promoter constructs. P. putida PaW85 carries in the chromosome a copy of Tn4652, and strain PRS2000 is Tn4652 free. P. putida A8759 is an ihfA-deficient derivative of strain KT2442. Bacterial strains and tnpA gene promoter constructs are listed in Table 1. Data (means ± standard deviations) of at least five independent experiments are presented. For plasmid pEST1332, the basal level of expression of C12O is less than 0.01 μmol/min/mg; for pKTlacZ, the level of expression of β-Gal is less than 2 nmol/min/mg.
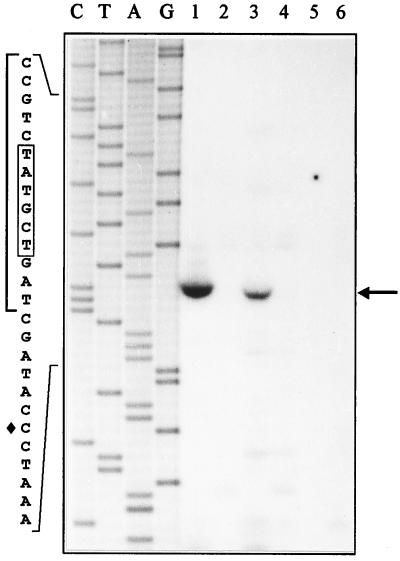
To map the 5′ end of the mRNA initiated from the tnpA promoter, primer extension analysis was carried out. Total RNA extracted from E. coli HB101, P. putida PaW85, and P. putida PRS2000 carrying plasmid p1332S/C, which exhibited tnpA promoter activity, or pEST1332 as a negative control was used as a template for the reverse transcriptase reaction. The results are presented in Fig. 5. Consistent with enzyme assays, no specific transcript was initiated from the tnpA promoter in E. coli (Fig. 5, lane 5). Easily detectable primer extension products could be established by using total RNA extracts both from cells of P. putida PRS2000(p1332S/C) and PaW85(p1332S/C) (Fig. 5, lanes 1 and 3). Primer extension assay localized the putative transcription start point 23 bp upstream of the tnpA gene start codon. The sequence TATGCT, resembling the ς70-recognized promoter consensus TATAAT, was found 10 bp upstream of the transcription start point. However, the −35 region of the promoter was not homologous with ς70-recognized consensus hexamer TTGACA.
FIG. 5.
Mapping of the 5′ end of mRNA initiated from the tnpA promoter. The primer extension product is indicated by the arrow. Lanes 1 to 6 present primer extension reactions carried out with total RNA prepared from P. putida PRS2000 (lanes 1 and 2), P. putida PaW85 (lanes 3 and 4), and E. coli HB101 (lanes 5 and 6) carrying tnpA promoter-containing plasmid p1332S/C (lanes 1, 3, and 5) or pEST1332 (lanes 2, 4, and 6) as a negative control. Lanes C, T, A, and G show DNA sequencing reactions of plasmid p1332S/C; 26 nt of this sequence is presented at the left, and the transcription start point of the tnpA gene is marked by a diamond. DNA originated from the right end of Tn4652 in p1332S/C is indicated by the vertical bold line, and the −10 region of the tnpA promoter is boxed.
The IHF-binding site affects positively transcription from the tnpA promoter.
Results presented in Fig. 4A suggest that the region from bp 1 to 56 bp of the right end of Tn4652 has a positive effect on the transcription from the tnpA promoter (compare p1332S/C and p1332D/C). Sequence analysis of the transposon right end revealed a potential IHF-binding site flanking the DraI site in p1332S/C (Fig. 2A). To test the effect of the presence of an IHF-binding site upstream of the tnpA promoter on expression of the reporter gene, the enzyme assay using the widely used β-Gal reporter system was performed. For that purpose, we constructed plasmids pKTlacZS/C, pKTlacZIHF/C, and pKTlacZD/C by cloning different DNA fragments from the tnpA promoter region (bp 1 to 122, 39 to 122, and 58 to 122, respectively) upstream of the β-Gal gene lacZ in the broad-host-range vector pKTlacZ (Table 1, Fig. 1, and Fig. 2). Plasmid pKTlacZIHF/C, which contains an IHF site adjacent to the DraI site, lacks the last 39 bp from the transposon end. Results of the β-Gal assay presented in Fig. 4B confirmed previous data obtained with the C12O reporter system (Fig. 4A): the presence of the Tn4652 terminal sequences (bp 1 to 57) upstream of the tnpA promoter enhances transcription from the promoter. Moreover, while in the C12O reporter system the positive effect was nearly twofold, the β-Gal system exhibited five- to sixfold enhancement. In Tn4652-free P. putida PRS2000, the presence of an IHF-binding site upstream of the DraI site was sufficient to complement the positive effect of the transposon right end to the tnpA promoter activity (Fig. 4B; compare pKTlacZIHF/C with pKTlacZD/C and pKTlacZS/C). However, in Tn4652-containing P. putida PaW85, the positive effect of an IHF-binding site in plasmid p1332IHF/C was lower than in p1332S/C (Fig. 4B).
Analogously to the C12O reporter, no promoter activity was detected if the β-Gal reporter was used in E. coli (data not shown).
tnpA promoter activity in ihfA-deficient P. putida A8759.
To elucidate the role of IHF in the tnpA promoter activity, a C12O assay using P. putida KT2442 and in its ihfA-deficient derivative P. putida A8759 was carried out. Usage of the β-Gal reporter system was excluded since both of these strains carry a copy of the lacZ gene under the control of the Pu promoter in the chromosome (Table 1). In addition to plasmids p1332S/C and p1332D/C characterized before, plasmid p1332IHF/C was constructed analogously to pKTlacZIHF/C (Table 1, Fig. 1, and Fig. 2). Figure 4C shows that enzyme activities in ihfA-deficient P. putida A8759 harboring either p1332S/C or p1332IHF/C were about twofold lower than in bacteria carrying plasmid p1332D/C. Thus, the DNA region containing the IHF-binding site had no enhancing effect on the tnpA promoter activity in the ihfA-deficient P. putida strain. In contrast, an obvious negative effect of terminal sequences of Tn4652 on transcription from the tnpA promoter could be seen in the ihfA-deficient P. putida strain A8759.
E. coli IHF specifically binds to both ends of Tn4652.
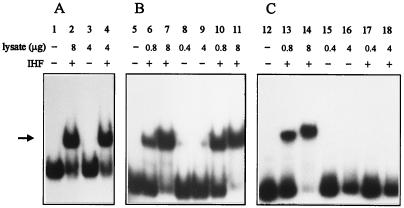
Sequence analysis of the left terminus of Tn4652 revealed two potential IHF-binding sites from bp 44 to 56 and from bp 59 to 71 (Fig. 2B). To test the possibility that IHF can bind to both ends of the transposon, a gel mobility shift assay was carried out. For binding reactions, crude lysates prepared from both E. coli and P. putida PaW85 cells were used. Figure 6 demonstrates that IHF from E. coli specifically retards DNA fragments containing either the left end (Fig. 6A) or right end (Fig. 6B) of the transposon. No probe retardation was detected when cell extract from E. coli WM2017 defective for IHF was used (Fig. 6, lanes 3, 8, and 9). Complementation of this IHF-negative strain with plasmid pHNβα carrying ihfA and ihfB restored the shift (Fig. 6, lanes 4, 10, and 11). However, we could not detect any specific shift with cell lysate from P. putida PaW85 either with the right end or with the left end of the transposon (data not shown). Additionally, a gel shift assay with the DNA fragment of the Pu promoter region known to contain an IHF-binding site (1, 13) was carried out as a control to test whether this site binds IHF from cell lysate of P. putida. However, although the DNA segment of the Pu promoter region specifically bound IHF from E. coli (Fig. 6C, lanes 13 and 14), no probe retardation was detected in the cell lysate from P. putida (Fig. 6C, lanes 17 and 18).
FIG. 6.
Gel shift assay of in vitro binding of IHF from cell lysates of E. coli and P. putida PaW85 to the left end of Tn4652 (A), to the right end of Tn4652 (B), and to the DNA fragment containing the Pu promoter region (C). Cell lysates used were from E. coli WM2015 (lanes 2, 6, 7, 13, and 14), E. coli WM2017 defective in the ihfA and ihfB genes (lanes 3, 8, 9, 15, and 16), E. coli WM2017 complemented with plasmid pHNβα (lanes 4, 10, and 11), and P. putida PaW85 (lanes 17 and 18). No cell lysate was added to reaction mixtures in lanes 1, 5, and 12. The specific IHF-DNA complex is indicated by the arrow.
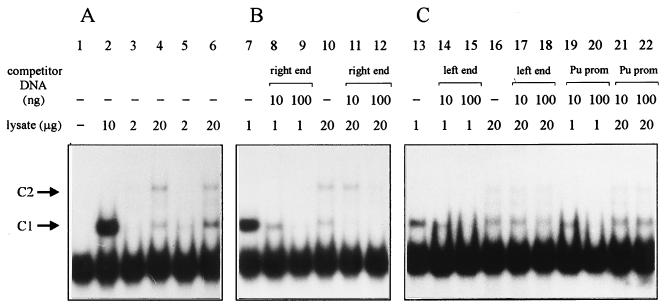
Recently, it has been reported that the IHF content of Pseudomonas aeruginosa is about 30 times lower than that in E. coli (12). To test whether the amount of IHF was too low to detect the shift (up to ∼4 μg of total cell protein per reaction was used), we repeated the gel mobility shift assay with more concentrated P. putida cell lysates. Indeed, 20 μg of total protein from P. putida PaW85 retarded the Tn4652 right-end DNA probe and revealed the presence of two distinct complexes, C1 and C2 (Fig. 7A, lanes 3 and 4). C1 moved as fast as the complex containing E. coli IHF (Fig. 7A, lane 2), which suggested that C1 could represent P. putida IHF bound to a probe. However, two complexes were also seen if lysate from the P. putida ihfA-defective strain A8759 was used in the gel shift assay (Fig. 7A, lanes 5 and 6). To test whether these complexes were specific for the right end of Tn4652, competition experiments with nonlabeled DNA probes were carried out. Addition to the binding reaction of the right-end DNA as a competitor suppressed the formation of C1 effectively, while suppression of C2 needed more competitor DNA (Fig. 7B, lanes 11 and 12). In contrast, DNA fragments of the left end of Tn4652 and from the Pu promoter (which were shown to bind IHF from E. coli) did not compete out either C1 or C2 (Fig. 7C, lanes 17, 18, 21, and 22). Both of these competitor DNAs successfully suppressed complex formation of the E. coli IHF with the tnpA promoter region (Fig. 7C, lanes 14, 15, 19, and 20).
FIG. 7.
(A) Gel shift assays demonstrating specific binding of some unknown factor(s) of P. putida to the right end of Tn4652; (B) competition with nonlabeled right-end DNA; (C) competition with DNA fragments containing either the left end of Tn4652 or the Pu promoter region. The two complexes (C1 and C2) formed are indicated by arrows; C1 in lanes 2, 7, 8, 13, 14, and 19 represents binding of IHF from E. coli HB101 cell lysate to the DNA probe. Cell lysates used were from E. coli HB101 (lanes 2, 7 to 9, 13 to 15, 19, and 20), P. putida PaW85 (lanes 3, 4, 10 to 12, 16 to 18, 21, and 22), and P. putida A8759 defective in the ihfA gene (lanes 5 and 6). No cell lysate was added to the reaction mixture in lane 1. In some experiments, a weak band between C1 and C2 was detected when P. putida crude lysate was used.
DISCUSSION
Many transposons require bacterial host proteins for transposition. IHF is known to participate in transposition of several transposons (32, 41, 42, 47), and it also modulates transposase expression in some cases (44). The experiments presented in this report show that transcription from the Tn4652 transposase promoter is positively affected by IHF.
We found that both ends of Tn4652 contain sequences similar to the IHF-binding consensus sequence (Fig. 2). The putative IHF-binding site at the right end of the transposon is located at positions from −73 to −85 relative to the transcription start point of the tnpA gene. Transposase promoter constructs carrying sequences of the right end of Tn4652 including an IHF-binding site revealed enhanced activity of the reporter gene pheB or lacZ in P. putida in comparison with the constructs lacking the IHF site upstream of the tnpA promoter (Fig. 4A and B). Enzyme assay using the P. putida ihfA-defective strain A8759 confirmed that IHF was involved in stimulation of transcription from the tnpA promoter. No positive effect of the IHF-binding site on promoter activity was detected in this strain (Fig. 4C). In contrast, the right end of the transposon had a negative effect on tnpA promoter activity when IHF was absent: both constructs p1332S/C and p1332IHF/C containing the IHF site exhibited even lower enzyme activity than p1332D/C in the ihfA-defective strain A8759 (Fig. 4C). This finding indicates that the IHF site, if not occupied by IHF protein, can suppress the tnpA promoter activity. It is known that IHF is involved in activation of the Pe promoter of bacteriophage Mu by a dual mechanism. IHF stimulates transcription from the Pe promoter directly and also indirectly via alleviation of the H-NS-mediated repression (44). Analogously, we suggest that binding of IHF to the right end of Tn4652 enhances transcription from the tnpA promoter not only directly but also indirectly by competing with some unknown negatively acting factor for the binding site.
Enzyme assay demonstrated that transcription from the tnpA promoter was higher in the Tn4652-free P. putida strain PRS2000 than that in strains PaW85 and KT2442, which contain a copy of Tn4652 in the chromosome (Fig. 4A and C). We propose that the chromosomally located copy of Tn4652 may code for functions affecting the tnpA promoter activity in P. putida PaW85 and KT2442. Since terminal sequences of transposons are presumed to bind transposase, it is possible that transcription from the tnpA promoter is modulated by the transposase of Tn4652, too.
Enzyme assay revealed that the Tn4652 tnpA gene promoter is silent in E. coli (Fig. 4A). Comparison of promoter specificities of RNA polymerases from E. coli and Pseudomonas spp. revealed that they transcribe similarly well different promoters of both species (16, 19). Considering these experiments, we do not believe that the difference between the E. coli and P. putida polymerases causes the silence of the tnpA promoter in E. coli. The possibility that activation of the transcription from the promoter needs some Tn4652-encoded factor could be also eliminated because the promoter is functional in Tn4652-free P. putida strain PRS2000. Thus, the presence of some host factor specific to P. putida is required for the promoter function. We propose two alternative explanations for the silence of the tnpA promoter in E. coli. First, transcription initiation from the tnpA promoter needs an activator protein that is missing in E. coli. Many ς70-dependent promoters lacking a well-conserved −35 region are known to be subjected to activation by the regulatory proteins (9). Correspondingly, the −35 region of the tnpA promoter revealed no homology with the ς70-recognized −35 consensus hexamer although the −10 region TATGCT of the tnpA promoter was considerably homologous with the ς70-recognized −10 hexamer consensus sequence TATAAT. On the other hand, the tnpA promoter might not be necessarily recognized by ς70. Therefore, an alternative sigma factor, absent in E. coli, might be required for promoter activation. This possibility is illustrated by the fact that alternative sigma factors of pseudomonads, not complemented in E. coli, are essential for the expression of several iron-regulated promoters of Pseudomonas strains (11, 38).
Up to now, there had been no reports about in vitro binding experiments with P. putida IHF. Using the gel mobility shift assay, we demonstrated that both ends of Tn4652 can bind IHF from cell lysate of E. coli (Fig. 6A and B). However, we could not detect an IHF-caused shift under the same conditions when the cell extract of P. putida PaW85 was used. We have also carried out gel shift experiments with lysate of an E. coli IHF-defective mutant complemented with plasmids carrying cloned IHF genes of P. putida. However, we did not detect IHF-caused retardation of a DNA fragment containing the right end of the transposon or the Pu promoter region of the TOL plasmid as a control (data not shown). This indicates that the properties of P. putida and E. coli IHF are different, and the experimental conditions used in in vitro binding assay were not optimal for the binding of P. putida IHF. However, in vivo experiments with hybrid IHF protein containing P. putida and E. coli subunits have showed that the hybrid protein efficiently functioned as a regulator of the pL promoter in E. coli (8), which suggests that in vivo binding properties of the hybrid IHF protein may be similar to those of E. coli IHF. Nevertheless, it would be interesting to compare the properties of IHF purified from P. putida with that from E. coli.
Gel mobility shift experiments with the transposon right-end DNA probe and crude lysate of P. putida PaW85 confirmed formation of two specific complexes (C1 and C2). Probably neither of them corresponded to IHF bound to the probe (Fig. 7), because these complexes were also detected by using cell lysate from P. putida ihfA-defective strain A8759 (Fig. 7A). Also, neither the Tn4652 left-end nor the Pu promoter-region DNA containing an IHF site suppressed formation of these complexes. Therefore, we consider that complexes detected by using the right end of the transposon represent some other protein(s) bound to the probe. Since C1 and C2 were formed with cell lysate from Tn4652-free P. putida PRS2000 as well (data not shown), we suggest that some P. putida host protein(s) participates in these complexes. Although the identity of the protein(s) is not established, it is tempting to speculate that complexes C1 and C2 contain the repressor protein which acts negatively on transcription from the tnpA promoter in a P. putida ihfA-deficient strain (Fig. 4C). Still, we cannot exclude the possibility that an activator, essential for the activity of the tnpA promoter in P. putida, was bound to the right end of Tn4652 in the gel shift assay. However, further experiments are needed to test these possibilities.
Our results demonstrate that IHF from E. coli binds specifically to both ends of Tn4652, just adjacent to the terminal inverted repeats that are presumed to bind the transposase. Other mobile elements are also known to contain IHF-binding sites at one or both ends (18, 25, 32, 46). It is known that γδ transposase of γδ (Tn1000) transposon and IHF bind cooperatively to both ends of the element (46). Additionally, IHF is required in in vitro reactions of IS10 transposition (35). Therefore, we suggest that besides activation of the tnpA promoter, IHF may participate in Tn4652 transposition also either by modulating the binding of transposase to the ends of the transposon or by influencing formation of nucleoprotein complexes needed in subsequent transposition reactions.
ACKNOWLEDGMENTS
We thank V. de Lorenzo for kindly providing P. putida KT2442 and A8759, W. Messer for E. coli WM2015 and WM2017, J. F. Gardner for plasmid pHNβα, and A. M. Chakrabarty for plasmid pKRZ-1. We also thank T. Alamäe, V. Kõiv, and A. Tamm for critically reading the manuscript and for helpful discussions. We are grateful to A. Abroi for discussions and for obliging help in the computer analysis.
This work was supported by grant 2323 from the Estonian Science Foundation, grant LCO000 from the International Science Foundation, and grant LKH100 from the Joint Program of the Government of Estonia and the International Science Foundation.
REFERENCES
- 1.Abril M A, Buck M, Ramos J L. Activation of the Pseudomonas TOL plasmid upper pathway operon. Identification of binding sites for the positive regulator XylR and for integration host factor protein. J Biol Chem. 1991;266:15832–15838. [PubMed] [Google Scholar]
- 2.Allison R G, Chaconas G. Role of the A protein-binding sites in the in vitro transposition of Mu DNA. J Biol Chem. 1992;267:19963–19970. [PubMed] [Google Scholar]
- 3.Bagdasarian M M, Amann E, Lurz R, Rueckert B, Bagdasarian M. Activity of the hybrid trp-lac(tac) promoter of Escherichia coli in Pseudomonas putida. Construction of broad-host-range controlled-expression vectors. Gene. 1983;26:273–282. doi: 10.1016/0378-1119(83)90197-x. [DOI] [PubMed] [Google Scholar]
- 4.Bayley S A, Duggleby C J, Worsey M J, Williams P A, Hardy K G, Broda P. Two modes of loss of the TOL function from Pseudomonas putida mt-2. Mol Gen Genet. 1977;154:203–204. doi: 10.1007/BF00330838. [DOI] [PubMed] [Google Scholar]
- 5.Blomberg P, Wagner E G, Nordström K. Control of replication of plasmid R1: the duplex between the antisence RNA, CopA, and its target, CopT, is processed specifically in vivo and in vitro by RNase III. EMBO J. 1990;9:2331–2340. doi: 10.1002/j.1460-2075.1990.tb07405.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyer H W, Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969;41:459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Calb R, Davidovitch A, Koby S, Giladi H, Goldenberg D, Margalit H, Holtel A, Timmis K, Sanchez-Romero J M, de Lorenzo V, Oppenheim A B. Structure and function of the Pseudomonas putida integration host factor. J Bacteriol. 1996;178:6319–6326. doi: 10.1128/jb.178.21.6319-6326.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Collado-Vides J, Magasanik B, Gralla J D. Control site location and transcriptional regulation in Escherichia coli. Microbiol Rev. 1991;55:371–394. doi: 10.1128/mr.55.3.371-394.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Craigie R, Arndt-Jovin D J, Mizuuchi K. A defined system for the DNA strand-transfer reaction at the initiation of bacteriophage Mu transposition: protein and DNA substrate requirements. Proc Natl Acad Sci USA. 1985;82:7570–7574. doi: 10.1073/pnas.82.22.7570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cunliffe H E, Merriman T R, Lamont I L. Cloning and characterisation of pvdS, a gene required for pyoverdine synthesis in Pseudomonas aeruginosa: PvdS is probably an alternative sigma factor. J Bacteriol. 1995;177:2744–2750. doi: 10.1128/jb.177.10.2744-2750.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delic-Attree I, Toussaint B, Froger A, Willison J C, Vignais P M. Isolation of an IHF-deficient mutant of a Pseudomonas aeruginosa mucoid isolate and evaluation of the role of IHF in algD gene expression. Microbiology. 1996;142:2785–2793. doi: 10.1099/13500872-142-10-2785. [DOI] [PubMed] [Google Scholar]
- 13.de Lorenzo V, Herrero M, Metzke M, Timmis K N. An upstream XylR- and IHF-induced nucleoprotein complex regulates the sigma 54-dependent Pu promoter of TOL plasmid. EMBO J. 1991;10:1159–1167. doi: 10.1002/j.1460-2075.1991.tb08056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Meirsman C, van Soom C, Verreth C, van Gool A, van der Leyden J. Nucleotide sequence analysis of IS427 and its target sites in Agrobacterium tumefaciens T37. Plasmid. 1990;24:227–234. doi: 10.1016/0147-619x(90)90006-x. [DOI] [PubMed] [Google Scholar]
- 15.Friedman D I. Integration host factor: a protein for all reasons. Cell. 1988;55:545–554. doi: 10.1016/0092-8674(88)90213-9. [DOI] [PubMed] [Google Scholar]
- 16.Fujita M, Anemura A. Purification and characterization of a DNA-dependent RNA polymerase from Pseudomonas putida. Biosci Biotechnol Biochem. 1992;56:1797–1800. doi: 10.1271/bbb.56.1797. [DOI] [PubMed] [Google Scholar]
- 17.Gama M-J, Toussaint A, Higgins N P. Stabilization of bacteriophage Mu repressor-operator complexes by the Escherichia coli integration host factor protein. Mol Microbiol. 1992;6:1715–1722. doi: 10.1111/j.1365-2958.1992.tb00896.x. [DOI] [PubMed] [Google Scholar]
- 18.Gamas P, Chandler M G, Prentki P, Galas D J. Escherichia coli integration host factor binds specifically to the ends of the insertion sequence IS1 and to its major insertion hot-spot in pBR322. J Mol Biol. 1987;195:261–272. doi: 10.1016/0022-2836(87)90648-6. [DOI] [PubMed] [Google Scholar]
- 19.Gao J, Gussin G. RNA polymerases from Pseudomonas aeruginosa and Pseudomonas syringae respond to Escherichia coli activator proteins. J Bacteriol. 1991;173:394–397. doi: 10.1128/jb.173.1.394-397.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Giladi H, Igarashi K, Ishihama A, Oppenheim A B. Stimulation of the Phage (lambda) pL promoter by integration host factor requires the carboxy terminus of the α-subunit of RNA polymerase. J Mol Biol. 1992;227:985–990. doi: 10.1016/0022-2836(92)90514-k. [DOI] [PubMed] [Google Scholar]
- 21.Goosen N, van de Putte P. The regulation of transcription initiation by integration host factor. Mol Microbiol. 1995;16:1–7. doi: 10.1111/j.1365-2958.1995.tb02386.x. [DOI] [PubMed] [Google Scholar]
- 22.Hallet B, Sherratt D J. Transposition and site-specific recombination: adapting DNA cut-and-paste mechanisms to a variety of genetic rearrangements. FEMS Microbiol Rev. 1997;21:157–178. doi: 10.1111/j.1574-6976.1997.tb00349.x. [DOI] [PubMed] [Google Scholar]
- 23.Hanahan D. Studies on the transformation of E. coli with plasmids. J Mol Biol. 1983;166:577–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 24.Hegeman G D. Synthesis of the enzymes of the mandelate pathway by Pseudomonas putida. I. Synthesis of the enzymes by wild type. J Bacteriol. 1966;91:1140–1154. doi: 10.1128/jb.91.3.1140-1154.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huisman O, Errada P R, Signon L, Kleckner N. Mutational analysis of IS10’s outside end. EMBO J. 1989;8:2101–2109. doi: 10.1002/j.1460-2075.1989.tb03619.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kholodii G Y, Yurieva O V, Gorlenko ZhM, Mindlin S Z, Bass I A, Lomovskaya O L, Kopteva A V, Nikiforov V G. Tn5041: a chimeric mercury resistance transposon closely related to the toluene degradative transposon Tn4651. Microbiology. 1997;143:2549–2556. doi: 10.1099/00221287-143-8-2549. [DOI] [PubMed] [Google Scholar]
- 27.Kivisaar M, Hõrak R, Kasak L, Heinaru A, Habicht J. Selection of independent plasmids determining phenol degradation in Pseudomonas putida and the cloning and expression of genes encoding phenol monooxygenase and catechol 1,2-dioxygenase. Plasmid. 1990;24:25–36. doi: 10.1016/0147-619x(90)90022-5. [DOI] [PubMed] [Google Scholar]
- 28.Kleckner N. Transposable elements in prokaryotes. Annu Rev Genet. 1981;15:341–404. doi: 10.1146/annurev.ge.15.120181.002013. [DOI] [PubMed] [Google Scholar]
- 29.Kleckner N. Regulation of transposition in bacteria. Annu Rev Cell Biol. 1990;6:297–327. doi: 10.1146/annurev.cb.06.110190.001501. [DOI] [PubMed] [Google Scholar]
- 30.Langer U, Richter S, Roth A, Weigel C, Messer W. A comprehensive set of DnaA-box mutations in the replication origin, oriC, of Escherichia coli. Mol Microbiol. 1996;21:301–311. doi: 10.1046/j.1365-2958.1996.6481362.x. [DOI] [PubMed] [Google Scholar]
- 31.Lee E C, Hales L M, Gumport R I, Gardner J F. The isolation and characterization of mutants of the integration host factor (IHF) of Escherichia coli with altered, expanded DNA-binding specificities. EMBO J. 1992;11:305–313. doi: 10.1002/j.1460-2075.1992.tb05053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Makris J C, Nordmann P L, Reznikoff W S. Integration host factor plays a role in IS50 and Tn5 transposition. J Bacteriol. 1990;172:1368–1373. doi: 10.1128/jb.172.3.1368-1373.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. [Google Scholar]
- 34.Mizuuchi K. Transpositional recombination: mechanistic insights from studies of Mu and other elements. Annu Rev Biochem. 1992;61:1011–1051. doi: 10.1146/annurev.bi.61.070192.005051. [DOI] [PubMed] [Google Scholar]
- 35.Morisato D, Kleckner N. Tn10 transposition and circle formation in vitro. Cell. 1987;51:101–111. doi: 10.1016/0092-8674(87)90014-6. [DOI] [PubMed] [Google Scholar]
- 36.Nurk A, Tamm A, Hõrak R, Kivisaar M. In vivo generated fusion promoters in Pseudomonas putida. Gene. 1993;127:23–29. doi: 10.1016/0378-1119(93)90612-7. [DOI] [PubMed] [Google Scholar]
- 37.Rothmel R K, Shinabarger D L, Parsek M R, Aldrich T L, Chakrabarty A M. Functional analysis of the Pseudomonas putida regulatory protein CatR: transcriptional studies and determination of the CatR DNA-binding site by hydroxyl-radical-footprinting. J Bacteriol. 1991;173:4717–4724. doi: 10.1128/jb.173.15.4717-4724.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sexton R, Gill P R, Callanan M J, O’Sullivan D J, Dowling D N, O’Gara F. Iron-responsive gene expression in Pseudomonas fluorescens M114: cloning and characterization of a transcription-activating factor, PbrA. Mol Microbiol. 1995;15:297–306. doi: 10.1111/j.1365-2958.1995.tb02244.x. [DOI] [PubMed] [Google Scholar]
- 39.Sharma R C, Schimke R T. Preparation of electro-competent E. coli using salt-free growth medium. BioTechniques. 1996;20:42–44. doi: 10.2144/96201bm08. [DOI] [PubMed] [Google Scholar]
- 40.Sherratt D. Tn3 and related transposable elements: site-specific recombination and transposition. In: Berg D E, Howe M M, editors. Mobile DNA. Washington, D.C: American Society for Microbiology; 1989. pp. 163–184. [Google Scholar]
- 41.Signon L, Kleckner N. Negative and positive regulation of Tn10/IS10-promoted recombination by IHF: two distinguishable processes inhibit transposition off of multicopy plasmid replicons and activate chromosomal events that favour evolution of new transposons. Genes Dev. 1995;9:1123–1136. doi: 10.1101/gad.9.9.1123. [DOI] [PubMed] [Google Scholar]
- 42.Surette M G, Lavoie B D, Chaconas G. Action at a distance in Mu DNA transposition: an enhancer-like element is the site of activation of supercoiling relief activity by integration host factor (IHF) EMBO J. 1989;8:3483–3489. doi: 10.1002/j.1460-2075.1989.tb08513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsuda M, Iino T. Genetic analysis of a transposon carrying toluene degrading genes on a TOL plasmid pWWO. Mol Gen Genet. 1987;210:270–276. doi: 10.1007/BF00325693. [DOI] [PubMed] [Google Scholar]
- 44.van Ulsen P, Hillebrand M, Zulianello L, van de Putte P, Goosen N. Integration host factor alleviates the H-NS-mediated repression of the early promoter of bacteriophage Mu. Mol Microbiol. 1996;21:567–578. doi: 10.1111/j.1365-2958.1996.tb02565.x. [DOI] [PubMed] [Google Scholar]
- 45.Wheelis M L, Ornston L N. Genetic control of enzyme induction in β-ketoadipate pathway of Pseudomonas putida: deletion mapping of cat mutations. J Bacteriol. 1972;109:790–795. doi: 10.1128/jb.109.2.790-795.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wiater L A, Grindley N D F. γδ transposase and integration host factor bind cooperatively at both ends of γδ. EMBO J. 1988;7:1907–1911. doi: 10.1002/j.1460-2075.1988.tb03024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wiater L A, Grindley N D. Integration host factor increases the transpositional immunity conferred by gamma delta ends. J Bacteriol. 1990;172:4951–4958. doi: 10.1128/jb.172.9.4951-4958.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yanisch-Perron C, Vieira J, Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]