Abstract
Background
The PI3K/AKT pathway, extensively studied in cancer, is vital for regulating cell metabolism, differentiation, and proliferation. Pathogenic variants in the PIK3R1 gene, which encodes three regulatory units of class IA PI3Ks, have been found in affected tissue of individuals with vascular lesions. These variants predominantly occur in the iSH2 domain, disrupting inhibitory contacts with the catalytic unit and leading to PI3K activation. Germline variants in this gene are also linked to an immunological condition called Activated PI3K delta syndrome type 2 (APDS2).
Methods
This is a case report and literature review. Clinical data were retrieved from medical records.
Results
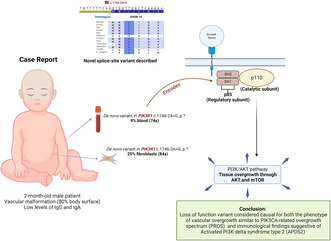
A male patient presented with extensive vascular malformation covering over 90% of his body, along with complete 2–3 toe syndactyly, suggesting a vascular malformation syndrome called PROS. Low levels of IgA and IgG were detected. The patient achieved his developmental milestones and had above‐average weight, height, and head circumference. Exome sequencing of skin and blood DNA revealed a de novo variant in PIK3R1 (c.1746‐2A>G, p.?) in 9% of the patient's blood cells and 25% of cultured fibroblasts. Initially, classified as a variant of uncertain significance, this variant was later confirmed to be the cause.
Conclusions
This is the first intronic SNV in a canonical splice site within iSH2 described, highlighting the importance of iSH2 in the regulation of the PI3K/AKT pathway and its involvement in the development of vascular overgrowth and antibody deficiency.
Keywords: PIK3R1 gene, vascular abnormalities, vascular overgrowth
The PI3K/AKT pathway is important in regulating cell metabolism, differentiation, and proliferation, and is studied in cancer. Variants in the PIK3R1 gene have been associated with vascular lesions and Activated PI3K delta syndrome type 2 (APDS2). This case report describes an infant boy with vascular malformations and syndactyly, along with low levels of IgA and IgG, due to a de novo variant in PIK3R1.
1. INTRODUCTION
The phosphoinositide 3‐kinase (PI3K/AKT) pathway, which involves various downstream effectors such as AKT and mTOR, is persistently activated in numerous vascular malformations and in the context of pathological angiogenesis (Canaud et al., 2021; Le Cras & Boscolo, 2019). It has also been shown to also regulate cell metabolism, differentiation, and proliferation being extensively studied in cancer (Cottrell et al., 2021; Rascio et al., 2021). Class IA PI3Ks are composed of a catalytic subunit (p110α, p110β, or p110δ) and an associated regulatory subunit (p85α, p85β, p50α, p55α, or p55γ) that are obligatory heterodimers (Geering et al., 2007; Siempelkamp et al., 2017). The PIK3R1 (Phosphoinositide‐3‐Kinase Regulatory Subunit 1) gene encodes three of these regulatory units named p85α, p55α, and p50α. The regulatory subunits play a dual role of stabilizing and inhibiting the catalytic units in their basal state, while also facilitating their association with activated receptor tyrosine kinases to enable their subsequent activation (Cottrell et al., 2021; Lemmon & Schlessinger, 2010).
The catalytic subunits differ in tissue expression: p110α and p110β are ubiquitously expressed and p110δ is primarily expressed in immune cells (Dornan et al., 2017). Somatic variants that cause the overactivation of p110α lead to the phenotype of PIK3CA‐related overgrowth syndromes (PROS) such as CLOVES (congenital lipomatous overgrowth, vascular malformations, and epidermal nevi‐ OMIM 612918), for example (Mirzaa et al., 1993). Pathogenic germline variants in PIK3R1 are described associated with the phenotype of Agammaglobulinemia 7 (OMIM 615214), Immunodeficiency 36 (OMIM 616005), and SHORT syndrome (OMIM 269880). Loss of function germline variants in PIKR31 are also associated with a primary antibody deficiency named Activated PI3K delta syndrome type 2 (APDS2) (Szczawińska‐Popłonyk et al., 2022).
Recently affected tissue from 17 individuals with vascular lesions demonstrated that somatic variants in PIK3R1 were responsible for a similar phenotype seen in PROS including vascular abnormalities, mild bone and soft tissue overgrowth, and mild dysmorphic features (Cottrell et al., 2021). Another case in the literature described a male patient presenting with overgrowth, and lymphedema carrying a somatic variant in PIK3R1 (Schönewolf‐Greulich et al., 2022). Interestingly, these somatic variants are primarily concentrated in the iSH2 domain, which would hinder the inhibitory contacts to p110, ultimately resulting in the activation of PI3K.
In this case report, we describe a case of an infant presenting with vascular overgrowth caused by a somatic novel variant in PIK3R1 presenting also with features of APDS2. This is the first intronic SNV in a canonical splice site within iSH2 described, which further corroborates the importance of iSH2 in the regulation of the PI3K/AKT) pathway.
2. MATERIALS AND METHODS
This is a case report and literature review. Data on clinical history, genetic analysis information, and sociodemographic characteristics were retrieved from medical records. This patient and their parents were consented to IRB 19‐003389; details of the research were discussed with the participants, and all questions were answered prior to obtaining research consent.
2.1. Case report
A 2‐month‐old male patient was brought for a second opinion on his extensive vascular malformation and suspicion of vascular malformation syndrome. He has one healthy older brother, and family history was noncontributory. After an uncomplicated pregnancy and delivery, he was born at 40 weeks and 4 days of gestation. The first physical examination reported a diffuse capillary malformation involving approximately 80%–90% of the body surface area (Figure 1a–c). Also noted was complete syndactyly of toes 2–3 bilaterally. His birth weight was 4.28 kg (96th centile—DC Growth Charts). He was evaluated in the newborn nursery by Neurology with a normal head and spinal sacral ultrasound. Segmental overgrowth was not observed. However, the longitudinal growth curve showed symmetrical accelerated growth in height, weight, and OFC with crossing growth centile. The patient also had a normal newborn bloodspot and hearing screen. Ophthalmology examination was also normal.
FIGURE 1.
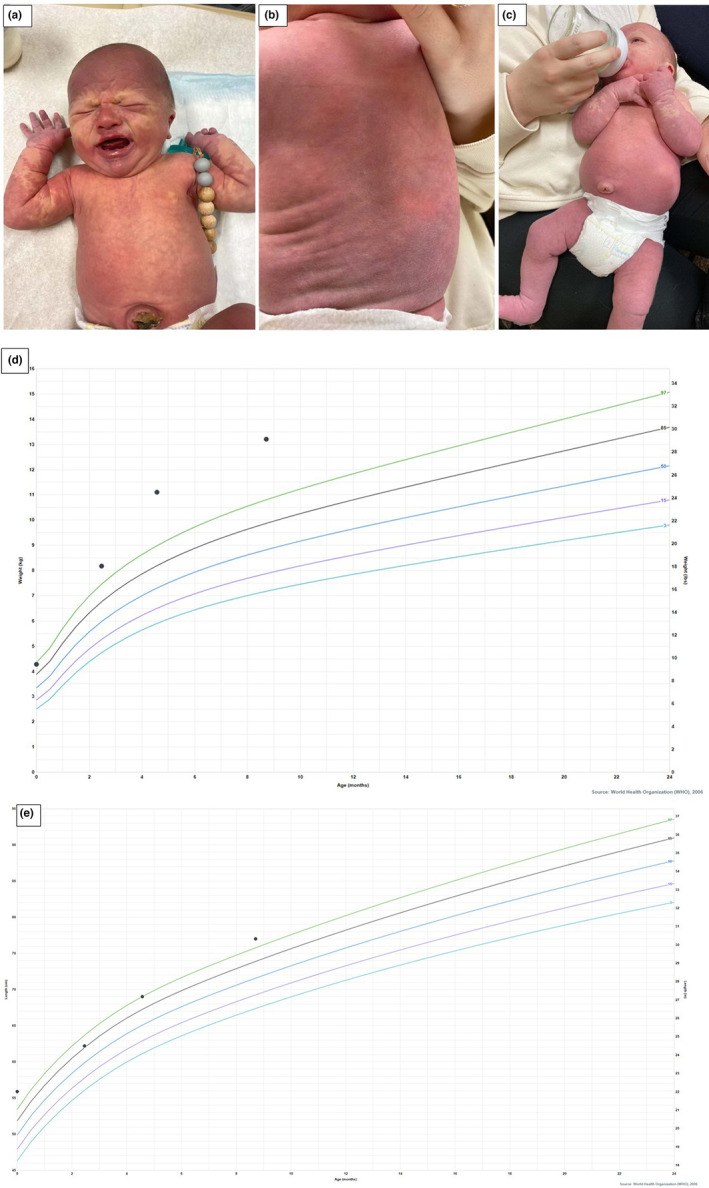
Vascular abnormalities and growth chats of the proband. (a–c) Diffuse capillary malformation involving approximately 80%–90% of the body surface area. (d) Weight‐for‐age percentiles (boys, birth to 2 years of age—World Health Organization, 2006); (e) Length‐for‐age Percentiles (boys, birth to 2 years of age—World Health Organization, 2006).
His weight, height, and head circumference were all above the 97th centile as shown in his growth curves (Figure 1d,e). His developmental history was unremarkable. At 4 months, he was able to pull to sit without head lag, bears weight on legs, follows 180°, reaches, regards hands, holds object briefly, rolls over, smiles, babbles, and laughs, with all neurodevelopmental milestones being met on time.
Brain MRI at the age of 5‐months reported mild thinning of the corpus callosum with mild enlargement of the ventricles with patchy nonspecific T2 signal prolongation within the cerebral white matter. There was no evidence for a capillary pial vascular malformation suggestive of Sturge–Weber syndrome.
Due to the lack of a clear diagnosis, the family elected to pursue exome sequencing (ES) and mitochondrial (mtDNA) testing when the patient was around 9 months old. The testing was performed on DNA derived from the proband's cultured fibroblasts from a 3 mm biopsy punch size from his right lower paraspinal region, and from DNA derived from a blood sample from both parents and the proband. Exome sequencing was performed at GeneDx. The ES identified a de novo variant in PIK3R1 c.1746‐2A>G, p.? (NM_181523.2) present in only 9% of the proband's blood cells (74 sequencing reads) and in 25% of the cultured fibroblasts (84 sequencing reads). This variant was classified as a variant of uncertain significance in the original report, being posteriorly determined as causative after research review.
During the patient's most recent appointment at 12 months of age, it was reported that he had two ear infections occurring 4 weeks apart from each other, both of which resolved with two rounds of oral antibiotic therapy. Additionally, he had a bilateral eye infection that required treatment with antibiotics. Laboratory workup revealed low levels of IgG (208 mg/dL; RR = 246–904 mg/dL), IgA (8 mg/dL; RR = 27–66 mg/dL), but normal IgM (68 mg/dL; RR = 40–143 mg/dL). The patient also had an absolute neutrophil count of 1.25 K/UL (RR = 1.19–7.21 ×10(9)/L). T and B cell subsets were all in the normal range.
3. DISCUSSION
The widespread use of NGS technology has allowed new discoveries of disease‐associated somatic variants in affected tissues due to the increased sensitivity of this technique in detecting low‐level mosaicism in comparison to Sanger sequencing, for example (Canaud et al., 2021; Le Cras & Boscolo, 2019). The variant classification for mosaic variants are, however, not contemplated by the published guidelines of the American College of Medical Genetics and Genomics (ACMG) making the interpretation of the pathogenic role of these variants challenging (Richards et al., 2015). Luckily, the PI3K‐AKT growth signaling pathway is already well recognized as dysregulated in cancer, vascular malformations and overgrowth tissue which allows a better understanding of the mechanism of pathogenicity for the different variants in genes associated in this pathway (Canaud et al., 2021; Chang et al., 2016; Cottrell et al., 2021).
To the best of our knowledge, this is the first time that a somatic SNV in a splice site has been described as associated with this phenotype of vascular malformation. The p85α subunit, which is encoded by PIK3R1, comprises the N‐terminal SH3 domain, a domain that is analogous to the Rho GTPase‐activating protein domain (RhoGAP), and three SH2 domains (nSH2, iSH2, and cSH2) (Chen et al., 2018). The somatic variants associated with a similar phenotype as our proband are interestingly located in both the nSH2 and iSH2 domains (Figure 2a). Previous studies have demonstrated that both the N‐terminal SH3 domain and the RhoGAP‐like domain are located at the interfaces between the p110ɑ and p85ɑ subunits. The N‐terminal adapter‐binding domain (ABD) and protein kinase C homology‐2 (C2) domains of p110ɑ not only bind to the iSH2 domain but also interact with the PI4K kinase domain in p110ɑ (Heurtier et al., 2017). Certain functional variants in cancer such as p.Arg503Trp and Asn564Asp in the nSH2 and iSH2 domains have been demonstrated to activate the canonical PI3K pathway by interfering with the inhibitory contact of the C2 domain in p110α (Chen et al., 2018).
FIGURE 2.
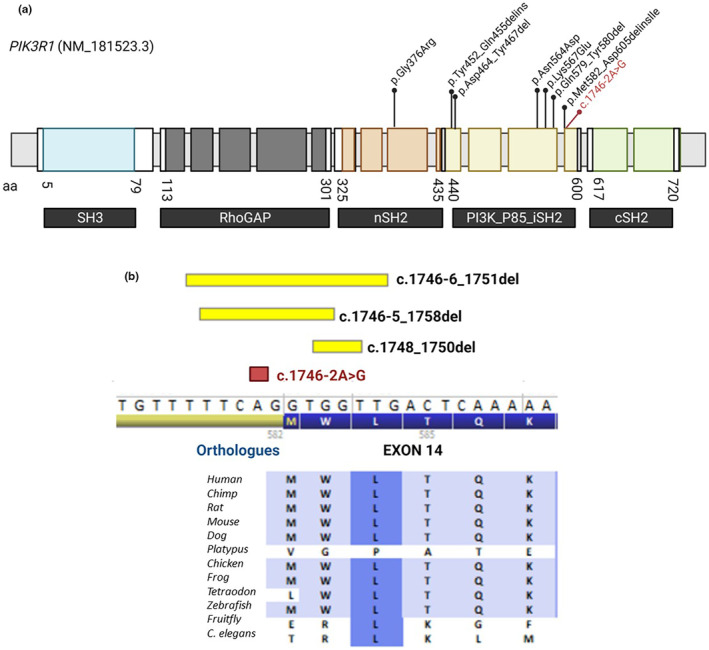
Domains of PIK3R1 and somatic variants in patients with vascular/overgrowth syndrome. (a) The variant found in the proband is depicted in red, with amino acid numbers delimiting the different domains. (b) Variants affecting the Met582 codon modified from Alamut. Yellow variants were already described by Cottrell et al. (2021), in red proband's variant.
In the cases described by Cottrell et al, there were three patients with variants affecting the same intron 13/exon 14 junction as our proband with good clinical overlap (Figure 2b). These variants were named p.(Met582_Asp605delinsIle) based on established nomenclature conventions because of the predicted effect on methionine codon 582 (Cottrell et al., 2021). They were also described as M582_splice in cancer studies that demonstrated, with RNA functional studies, in‐frame exon 14 skipping (Chang et al., 2016). This is corroborated by the new SpliceVault tool which identifies exon 14 skipping as the most common mis‐splicing event in the exon‐intron junction based on the detection of unannotated splice junctions across publicly available RNA‐Seq samples (Dawes et al., 2023). Additionally, these variants are located in a recurrently mutated residue in tumor samples, making them a somatic mutational hotspot (Chang et al., 2016).
Because of the nonstandardized annotation of the variant nomenclature for mosaic variants we have used the criteria applied by Cottrell et al. in classifying our variant. We believe that the following criteria can be applied: PS2 due to the confirmed mosaic and confirmed de novo, PS_Cancer for hotspot, as defined by the author for being a variant well‐represented in cancer and considered to occur in a statistically significant hotspot or region (cancerhotspots.org) within PIK3R1, PS3 for the RNA functional study and PS4_Mod for the same exon skipping effect being seeing it at least other three individuals (Cottrell et al., 2021).
The distinct tissue expression profiles of the different p110 catalytic subunits suggest that loss of function PIK3R1 variants may affect various cell types differently (Dornan et al., 2017). In the case of APDS2, the phenotype usually results from a splice variant that leads to p85α with a central deletion (Δ434–475) (Coulter & Cant, 2018). Given that p85 regulatory subunits are known to inhibit all class IA catalytic subunits, variants associated with APDS2 are anticipated to activate not only p110α but also p110β and p110δ. Previous studies have shown that pathogenic variants in p85α lead to a higher basal activation of the delta subunit of p110 compared to the alpha subunit (>300‐fold × ∼2‐fold) (Dornan et al., 2017). More recently, a study on mouse models and patients' immune cells compared the immune cell profiling of APDS2 and APDS1 (caused by heterozygous gain‐of‐function variants in PIK3CD). The study found that APDS2 patients had lower percentages of B cells and reduced frequencies of more mature subsets, such as naïve and memory B cells, compared to healthy individuals. Interestingly, transitional B cells with PIK3R1 loss of function secreted fourfold less IgM and failed to produce detectable levels of the switched isotypes IgG and IgA (Nguyen et al., 2023). Our proband has only 9% of the pathogenic PIK3R1 variant detected in blood, and additional studies are needed to determine whether this variant is causing their low immunoglobulin levels. The management of individuals with APDS has been quite diverse, spanning from mere monitoring to the administration of selective PI3Kδ inhibitors and hematopoietic stem cell transplant (HSCT) during childhood (Coulter & Cant, 2018).
Currently, there has been a single paper published about a treatment attempt in a patient with a somatic variant in PIK3R1 with Alpelisib (VIJOICE®) (Schönewolf‐Greulich et al., 2022). This drug has been granted accelerated approval by FDA in 2022 for pediatric (2 years of age and older) and adult patients with PROS who need systemic therapy (Center for Drug Evaluation and Research, 2022). Because of the theoretical disease mechanism (LOF variants in p85a × gain of function variants in p100a in PROS) the patient described by Schönewolf‐Greulich et al. (2022) started treatment at 10 months of age. He demonstrated normalization of his growth curve and improvement of the affected vascular and lymphatic tissue after 1 year of treatment with no side effects reported (Schönewolf‐Greulich et al., 2022). Interestingly, no symptoms APDS2 was described for this patient. For PROS, a low dose of Alpelisib was also shown to have striking effects in a cohort of 19 patients with several PROS‐related life‐threatening complications (Mirzaa et al., 1993; Venot et al., 2018). There is a lack of knowledge on the different levels of activation of the somatic mosaic variants in PIK3R1 and PIK3CA in the PI3K pathway. Understanding the degree of activation of p110a resulting from each variant could be a crucial factor in comprehending the safety and appropriate dosage of this therapy. Epigenetic modifications influence several critical signal pathways and therapeutic targeting of this pathway has been showing promising results. Interestingly, PTEN (associated with Multiple Hamartoma Syndrome, OMIM 158350, with overlapping phenotype of tissue overgrowth) was also reported to regulate the PI3K pathway (Tangye et al., 2023). It was already demonstrated that over methylation of PTEN promoter inhibits PTEN expression in tamoxifen‐resistant breast carcinoma cells and activates AKT and 5‐Azacytidine (5‐Aza) methylation of the PTEN promoter (Hussain et al., 2022). Understanding tissue‐specific gene expression and epigenetic regulators in each tissue may enable more effective targeted therapies in the future.
4. CONCLUSION
While somatic LOF variants in PIK3R1, mainly in the nSH2 and iSH2 domains, are associated with a phenotype resembling that of PROS, germline LOF variants can be associated with an immune phenotype named APDS2. In this case report, we describe an individual who presented with both vascular overgrowth similar to PROS and immunological findings suggestive of APDS2, with a mosaic intronic single nucleotide variant (SNV) in a canonical splice site. This case highlights the importance of this gene in the vascular overgrowth syndrome and additionally exemplifies the importance of a complete assessment of other immune complications related to this gene so that appropriate therapeutic options can be pursued. Assessment for immune function in patients with PROS should be considered as this case illustrates that there can be clinical overlap between PROS and APDS2.
AUTHOR CONTRIBUTIONS
Matheus V. M. B. Wilke, Lisa Schimmenti collected, interpreted, and analyzed the data of patient. Lisa Schimmenti, Madeline Q. R. Lopour, and Megha M. Tollefson were directly involved in the care of the patients. Matheus V. M. B. Wilke, Lisa Schimmenti, and Eric W. Klee helped to design this case report. All authors contributed to the study, approved the final version, and agreed to be accountable for the work. All authors were granted complete access to the manuscript and all the data obtained during the study.
FUNDING INFORMATION
There was no funding for the work associated with this publication. None of the authors have been paid by any agency or pharmaceutical company to write this article.
CONFLICT OF INTEREST STATEMENT
None.
ETHICS STATEMENT
The patient and their parents were consented to IRB 19‐003389; details of the research were discussed with the participants, and all questions were answered prior to obtaining research consent.
INFORMED CONSENT STATEMENT
The patient's parental (or legal guardian) provided informed and written consent to present and report their cases.
ACKNOWLEDGMENTS
Not applicable.
Wilke, M. V. M. B. , Schimmenti, L. , Lopour, M. Q. R. , Tollefson, M. M. , & Klee, E. W. (2023). A somatic splice‐site variant in PIK3R1 in a patient with vascular overgrowth and low immunoglobulin levels: A case report. Molecular Genetics & Genomic Medicine, 11, e2271. 10.1002/mgg3.2271
DATA AVAILABILITY STATEMENT
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.
REFERENCES
- Canaud, G. , Hammill, A. M. , Adams, D. , Vikkula, M. , & Keppler‐Noreuil, K. M. (2021). A review of mechanisms of disease across PIK3CA‐related disorders with vascular manifestations. Orphanet Journal of Rare Diseases, 16(1), 306. 10.1186/s13023-021-01929-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Center for Drug Evaluation and Research . (2022). FDA approves alpelisib for PIK3CA‐related overgrowth spectrum. FDA. Retrieved February 9, 2023, from https://www.fda.gov/drugs/resources‐information‐approved‐drugs/fda‐approves‐alpelisib‐pik3ca‐related‐overgrowth‐spectrum [Google Scholar]
- Chang, M. T. , Asthana, S. , Gao, S. P. , Lee, B. H. , Chapman, J. S. , Kandoth, C. , Gao, J. J. , Socci, N. D. , Solit, D. B. , Olshen, A. B. , Schultz, N. , & Taylor, B. S. (2016). Identifying recurrent mutations in cancer reveals widespread lineage diversity and mutational specificity. Nature Biotechnology, 34(2), 155–163. 10.1038/nbt.3391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, L. , Yang, L. , Yao, L. , Kuang, X. Y. , Zuo, W. J. , Li, S. , Qiao, F. , Liu, Y. R. , Cao, Z. G. , Zhou, S. L. , Zhou, X. Y. , Yang, W. T. , Shi, J. X. , Huang, W. , Hu, X. , & Shao, Z. M. (2018). Characterization of PIK3CA and PIK3R1 somatic mutations in Chinese breast cancer patients. Nature Communications, 9, 1357. 10.1038/s41467-018-03867-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cottrell, C. E. , Bender, N. R. , Zimmermann, M. T. , Heusel, J. W. , Corliss, M. , Evenson, M. J. , Magrini, V. , Corsmeier, D. J. , Avenarius, M. , Dudley, J. N. , Johnston, J. J. , Lindhurst, M. J. , Vigh‐Conrad, K. , Davies, O. M. T. , Coughlin, C. C. , Frieden, I. J. , Tollefson, M. , Zaenglein, A. L. , Ciliberto, H. , … Drolet, B. A. (2021). Somatic PIK3R1 variation as a cause of vascular malformations and overgrowth. Genetics in Medicine, 23(10), 1882–1888. 10.1038/s41436-021-01211-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulter, T. I. , & Cant, A. J. (2018). The treatment of activated PI3Kδ syndrome. Frontiers in Immunology, 9, 2043. 10.3389/fimmu.2018.02043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawes, R. , Bournazos, A. M. , Bryen, S. J. , Bommireddipalli, S. , Marchant, R. G. , Joshi, H. , & Cooper, S. T. (2023). SpliceVault predicts the precise nature of variant‐associated mis‐splicing. Nature Genetics, 55(2), 324–332. 10.1038/s41588-022-01293-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dornan, G. L. , Siempelkamp, B. D. , Jenkins, M. L. , Vadas, O. , Lucas, C. L. , & Burke, J. E. (2017). Conformational disruption of PI3Kδ regulation by immunodeficiency mutations in PIK3CD and PIK3R1. Proceedings of the National Academy of Sciences of the United States of America, 114(8), 1982–1987. 10.1073/pnas.1617244114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geering, B. , Cutillas, P. R. , Nock, G. , Gharbi, S. I. , & Vanhaesebroeck, B. (2007). Class IA phosphoinositide 3‐kinases are obligate p85‐p110 heterodimers. Proceedings of the National Academy of Sciences of the United States of America, 104(19), 7809–7814. 10.1073/pnas.0700373104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heurtier, L. , Lamrini, H. , Chentout, L. , Deau, M. C. , Bouafia, A. , Rosain, J. , Plaza, J. M. , Parisot, M. , Dumont, B. , Turpin, D. , Merlin, E. , Moshous, D. , Aladjidi, N. , Neven, B. , Picard, C. , Cavazzana, M. , Fischer, A. , Durandy, A. , Stephan, J. L. , & Kracker, S. (2017). Mutations in the adaptor‐binding domain and associated linker region of p110δ cause activated PI3K‐δ syndrome 1 (APDS1). Haematologica, 102(7), e278–e281. 10.3324/haematol.2017.167601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain, A. , Tebyaniyan, H. , & Khayatan, D. (2022). The role of epigenetic in dental and oral regenerative medicine by different types of dental stem cells: A comprehensive overview. Stem Cells International, 2022, 1–15. 10.1155/2022/5304860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Cras, T. D. , & Boscolo, E. (2019). Cellular and molecular mechanisms of PIK3CA‐related vascular anomalies. Vascular Biology, 1(1), H33–H40. 10.1530/VB-19-0016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemmon, M. A. , & Schlessinger, J. (2010). Cell signaling by receptor‐tyrosine kinases. Cell, 141(7), 1117–1134. 10.1016/j.cell.2010.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mirzaa, G. , Graham, J. M. , & Keppler‐Noreuil, K. (1993). PIK3CA‐related overgrowth spectrum. In Adam M. P., Everman D. B., Mirzaa G. M., Pagon R. A., Wallace S. E., Bean L. J. H., Gripp K. W., & Amemiya A. (Eds.), GeneReviews®. University of Washington; Retrieved February 9, 2023, from http://www.ncbi.nlm.nih.gov/books/NBK153722/ [Google Scholar]
- Nguyen, T. , Lau, A. , Bier, J. , Cooke, K. C. , Lenthall, H. , Ruiz‐Diaz, S. , Avery, D. T. , Brigden, H. , Zahra, D. , Sewell, W. A. , Droney, L. , Okada, S. , Asano, T. , Abolhassani, H. , Chavoshzadeh, Z. , Abraham, R. S. , Rajapakse, N. , Klee, E. W. , Church, J. A. , … Deenick, E. K. (2023). Human PIK3R1 mutations disrupt lymphocyte differentiation to cause activated PI3Kδ syndrome 2. The Journal of Experimental Medicine, 220(6), e20221020. 10.1084/jem.20221020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rascio, F. , Spadaccino, F. , Rocchetti, M. T. , Castellano, G. , Stallone, G. , Netti, G. S. , & Ranieri, E. (2021). The pathogenic role of PI3K/AKT pathway in cancer onset and drug resistance: An updated review. Cancers, 13(16), 3949. 10.3390/cancers13163949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards, S. , Aziz, N. , Bale, S. , Bick, D. , das, S. , Gastier‐Foster, J. , Grody, W. W. , Hegde, M. , Lyon, E. , Spector, E. , Voelkerding, K. , Rehm, H. L. , & ACMG Laboratory Quality Assurance Committee . (2015). Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genetics in Medicine, 17(5), 405–424. 10.1038/gim.2015.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schönewolf‐Greulich, B. , Karstensen, H. G. , Hjortshøj, T. D. , Jørgensen, F. S. , Harder, K. M. , Frevert, S. , Hove, H. , & Diness, B. R. (2022). Early diagnosis enabling precision medicine treatment in a young boy with PIK3R1‐related overgrowth. European Journal of Medical Genetics, 65(10), 104590. 10.1016/j.ejmg.2022.104590 [DOI] [PubMed] [Google Scholar]
- Siempelkamp, B. D. , Rathinaswamy, M. K. , Jenkins, M. L. , & Burke, J. E. (2017). Molecular mechanism of activation of class IA phosphoinositide 3‐kinases (PI3Ks) by membrane‐localized HRas. The Journal of Biological Chemistry, 292(29), 12256–12266. 10.1074/jbc.M117.789263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczawińska‐Popłonyk, A. , Bernat‐Sitarz, K. , Schwartzmann, E. , Piechota, M. , & Badura‐Stronka, M. (2022). Clinical and immunological assessment of APDS2 with features of the SHORT syndrome related to a novel mutation in PIK3R1 with reduced penetrance. Allergologia et Immunopathologia, 50(4), 1–9. 10.15586/aei.v50i4.510 [DOI] [PubMed] [Google Scholar]
- Tangye, S. G. , Nguyen, T. , Deenick, E. K. , Bryant, V. L. , & Ma, C. S. (2023). Inborn errors of human B cell development, differentiation, and function. The Journal of Experimental Medicine, 220(7), e20221105. 10.1084/jem.20221105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venot, Q. , Blanc, T. , Rabia, S. H. , Berteloot, L. , Ladraa, S. , Duong, J. P. , Blanc, E. , Johnson, S. C. , Hoguin, C. , Boccara, O. , Sarnacki, S. , Boddaert, N. , Pannier, S. , Martinez, F. , Magassa, S. , Yamaguchi, J. , Knebelmann, B. , Merville, P. , Grenier, N. , … Canaud, G. (2018). Targeted therapy in patients with PIK3CA‐related overgrowth syndrome. Nature, 558(7711), 540–546. 10.1038/s41586-018-0217-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. (2006). WHO child growth standards: Length/height‐for‐age, weight‐for‐age, weight‐for‐length, weight‐for‐height and body mass index‐for‐age: Methods and development. Retrieved August 23, 2023, from https://www.who.int/publications‐detail‐redirect/924154693X [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors confirm that the data supporting the findings of this study are available within the article [and/or] its supplementary materials.


