Abstract
Background
Nucleic acid‐based assays provide an opportunity to screen for genetically encoded diseases like spinal muscular atrophy (SMA), before the onset of symptoms. Nowadays, such assays could be easily utilized as high‐throughputs in SMA to detect a homozygous deletion of exon 7 of the survival motor neuron 1 gene (SMN1) that is responsible for >95% of SMA patients.
Methods
We developed a new line method (NLM) as a direct real time PCR test procedure without nucleic acid extraction in dried blood spots (DBS) to screen for homozygous deletion of exon 7 of the SMN1 gene. Performance of this setup was evaluated on 580 DBS newborn samples and air dried 50 DBS from whole blood including 20 samples for homozygous deletion of the SMN1 gene detected earlier with MLPA.
Results
We found all 580 newborn DBS samples as wild type. DBS prepared from 50 whole blood samples also including 20 affected people were correctly identified as homozygous deletions and 30 wild types of exon 7 of SMN1 as before with MLPA. When the MLPA method was taken as the gold standard, the sensitivity and specificity of the NLM test were found 100% for the detection of SMN1 exon 7 homozygous deletion.
Conclusion
In the NLM, the total test duration has been reduced to less than 75 min without requiring any extra process such as DNA extraction step and sample plate preparation after the punching step. Thereby, newborn SMA screening with the NLM has gained an environmentally friendly feature with not requiring additional tedious steps.
Keywords: DBS, direct test, newborn screening, SMA
1. INTRODUCTION
SMA is one of the most common autosomal‐recessive diseases, with an incidence of ~1 in 6000–10,000 live births and a carrier frequency of 1 in 35–117, depending on ethnicity (Hendrickson et al., 2009; Luo et al., 2014). The disease is characterized by the progressive degeneration and loss of anterior horn cells in the spinal cord and brainstem nuclei causing symmetric muscle weakness and atrophy (Meldrum et al., 2007). SMA is separated into different phenotypes, according to the age of onset and the motor function status. Generally, SMA type I is the most severe and common type, which affectsabout 50% of patients. Life duration is generally considered to be <2 years because of the rapid, progressive muscle weakness and respiratory failure (Czibere et al., 2020, Finkel et al., 2018; Kolb & Kissel, 2015; Mercuri, Darras et al., 2018). Although therapeutic approachings have been developed recently available, the success of these therapeutic interventions strongly depends on early starting due to the substantial loss of motor neurons that occurs at 3 months of age (Czibere et al., 2020; Finkel et al., 2018; Kolb & Kissel, 2015; Mercuri, Finkel et al., 2018; Saffari et al., 2019). By luck, nowadays, a genetic test provides identification of the patients before the onset of symptoms and offers treatment options.
There are many challenges for genetic testing of SMA caused mostly by SMN1 (MIM *600354) exon 7 homozygous deletion in newborn DBS samples, such as DNA extraction, selection of an appropriate assay, and the sample recording and tracking. Therefore, to overcome these challenges easily, we developed a simple and uncomplicated test for the detection of SMN1 exon 7 homozygous deletion in a rapid direct real time PCR application as NLM. In the procedure, first step is 1.2 mm DBS punch has been directly sent to the real time test plate with DBS puncher equipment. As the second step there is only need to add DBA mix and real time PCR assay could be directly started. Thereby, the sample plate after the punching step did not require extra time consuming procedures such as DNA extraction, sample recording and tracking.
2. MATERIALS AND METHODS
2.1. Ethics and samples
The research was started after obtaining the approval from the ethics committee (Acibadem University ATADEK‐2019‐16/4) and informed consent for all parents. In this study, 580 DBS (Whatman 903) samples used in this study were gathered from the newborn who applied to Istanbul Acıbadem Hospital. Additional 50 air dried blood spots (I.W. Tremont Co.) were prepared with the bloods that were provided from a previously published study (Cavdarli et al., 2020). The results obtained with MLPA (MRC Holland SALSA MLPA P060) were not shared with the technical team carrying out the wet lab research.
2.2. Punch sizes and DNA isolation
Punch sizes of 1.2 mm and 3.2 mm diameters were taken from DBS samples (BSD600 Ascent Juniour and DBS Card Punch Analytical SS) into the white 0.2 mL real time PCR test plates. DNA isolation, purity and concentration were not performed as they were not needed for the direct real time PCR. DNA isolations were only done with 3.2 mm DBS punches by using the SNP DBS DNA Extraction Kit (21S‐03‐50, SNP Biotechnology) for confirmation tests.
2.3. Internal control or reference gene
In real‐time PCR studies, a housekeeping gene should be used as an internal control to analyze the reaction and amplification efficiency. We used FII‐Prothrombin Gene (MIM* 176930) as an internal control or reference gene like published before (Cavdarli et al., 2020).
2.4. Primer‐Probe design
Primers and probes are designed for point mutation (c.840C>T) and deletion in exon 7 of the SMN1 gene and for the reference gene “prothrombin”. The probe used for SMN1 was labeled with FAM dye‐stain, while the probe used for the reference gene was marked with TEXAS RED dye‐stain. The primer and probe sequences are listed in Table 1.
TABLE 1.
Primer and probe sequences.
| Ref Forward | GGTTCCCAATAAAAGTGACTCTCAG |
| Ref Reverse | CCAGGTGGTGGATTCTTAAGTCTTC |
| Ref Prob | 5′ Tex Red‐AATGCTCCCAGTGCTATTCATGGGCAG‐3′ BHQ‐2 |
| SMN1 Forward | AACTTCCTTTATTTTCCTTACAGGGTTAC |
| SMN1 Reverse | CTTTCATAATGCTGGCAGACTTACTC |
| SMN1 Prob | 5′ FAM‐TCAAAAAGAAGGAAGGTGCTCACATTCC‐3′ BHQ‐1 |
2.5. Direct real time PCR and DBS mix
In the study, all primers and probes were optimized to operate multiplex reactions in a single tube. The final volume of the reaction mix was 22 μL, which included 1X DBS mix (SNP Biotechnology), 5 pmol of SMN1 P1 and P2, 2.5 pmol SMN1 Probe—FAM, 11 pmol of Internal Control P1 and P2, 2.5 pmol Internal Control probe—TEXAS RED, and 1.0 Unit AB+ Taq DNA Polymerase (Nanohelix). The direct real time PCR in Bio‐Rad CFX 96 (Bio‐Rad) was performed without DNA extraction in the real time PCR plate containing DBS mix and DBS punches. Triple repeats were done for each sample. The real time PCR program was consisting of 35 cycles including the initial DNA extraction step (5 min at 95°C), denaturation (10 s at 95°C) and annealing/extension/reading (1 min at 60°C).
2.6. Principle of the 5′Nuclease method
The test principle is based on the 5′‐3′ exonuclease activity of Taq DNA polymerase. The probe has a reporter dye at the 5′ end and a quencher dye at the 3′ end. The quencher dye suppresses the reporter dye irradiation and at the same time prevents the probe from acting as a primer. During PCR, the probe between the reporter and the quencher is broken apart with enzyme activity, and radiation is produced when suppression is eliminated. This process only takes place on probes hybridized over the target region. As the amount of amplification increases, the luminescence increases linearly with the release of the reporter dye, and this increase is detected simultaneously by the device.
3. RESULTS
3.1. Newborn DBS samples and punch sizes
In this study, 580 DBS (Whatman 903) samples used were detected as wild type for SMN1 Exon 7. The best results were obtained from DBS samples with 1.2 mm punch sizes (Figure 1a). Intended results were not always obtained from the 3.2 mm punch size due to some amplification errors (Figure 1b).
FIGURE 1.
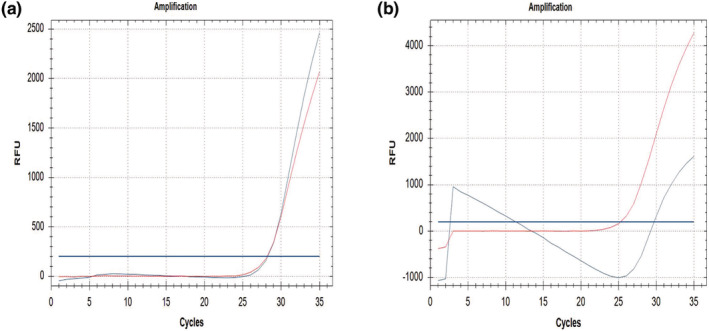
(a) Wild type sample, 1.2 mm punch Whatman 903 DBS. (b) Amplification error, 3.2 mm punch Whatman 903 DBS. SMN1 Exon 7 labelled with FAM‐blue colour and reference gene labelled with TEXAS RED‐red colour.
3.2. Air dried DBS samples and punch sizes
Punch size results of 50 air dried blood spots (I.W. Tremont Co.) for SMN1 Exon 7 yielded similar results as newborn DBS samples. That is, the best results were always obtained from air dried DBS samples with 1.2 mm punch size (Figure 2a). Expected results could not be orderly obtained from the 3.2 mm punch sizes and some amplification errors appeared (Figure 2b). Also, 20 air dried blood spot samples previously known were detected as SMN1 Exon 7 homozygous deletion (Figure 3) and the rest of air dried blood spot samples were typed correctly as wild type for SMN1 Exon 7.
FIGURE 2.
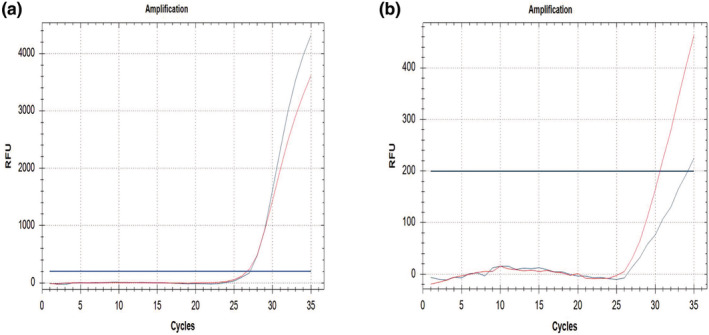
(a) Wild type sample, 1.2 mm punch I.W. Tremont Co. DBS. (b) Unacceptable result, 3.2 mm punch I.W. Tremont Co. DBS. SMN1 Exon 7 labelled with FAM‐blue colour and reference gene labelled with TEXAS RED‐red colour.
FIGURE 3.
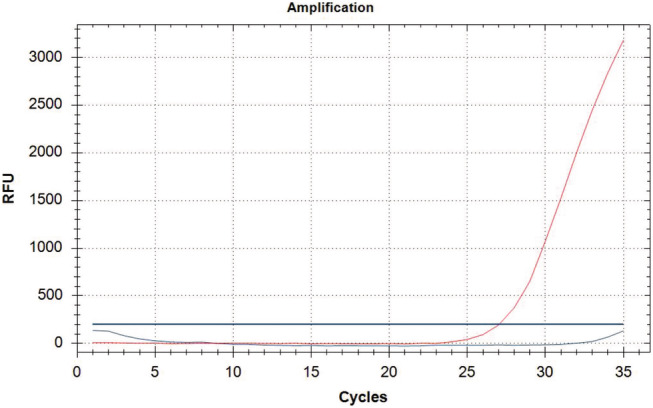
1.2 mm I.W. Tremont Co.– SMN1 Exon 7 Homozygous deletion. SMN1 Exon 7 labelled with FAM‐blue colour and reference gene labelled with TEXAS RED‐red colour.
3.3. Direct real‐time PCR
When real time PCR were repeated three times with the samples containing DBS mix and DBS punches with 1.2 mm size, we constantly obtained the similar Ct results for target and reference genes (Table 2). Preferable amplification plots were also seen in I.W. Tremont Co. 1.2 mm punch size samples (Figure 4).
TABLE 2.
Reproducibility study with Whatman 903 and I.W. Tremont Co. DBS papers.
| Whatman 903 | I.W. Tremont Co. | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Repeats | FAM | Standard deviation σ | TEXAS RED | Standard deviation σ | Repeats | FAM | Standard deviation σ | TEXAS RED | Standard deviation σ | ||||||||
| 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | 1 | 2 | 3 | ||||||
| Samples | Samples | ||||||||||||||||
| 1 | 29.36 | 30.86 | 28.41 | 1.23 | 29.53 | 30.80 | 28.77 | 1.02 | 13 | 26.41 | 26.15 | 26.20 | 0.14 | 26.32 | 26.21 | 26.18 | 0.07 |
| 2 | 29.30 | 28.90 | 29.35 | 0.25 | 29.23 | 29.09 | 29.22 | 0.08 | 14 | 26.38 | 26.74 | 26.68 | 0.19 | 26.43 | 26.68 | 26.59 | 0.13 |
| 3 | 28.85 | 27.89 | 28.44 | 0.48 | 29.68 | 28.53 | 28.36 | 0.72 | 15 | 26.19 | 26.48 | 25.28 | 0.63 | 26.31 | 26.61 | 26.10 | 0.26 |
| 4 | 27.91 | 28.26 | 27.24 | 0.52 | 28.09 | 28.17 | 27.62 | 0.30 | 16 | 26.25 | 26.40 | 24.28 | 1.18 | 26.25 | 26.63 | 25.83 | 0.40 |
| 5 | 29.26 | 29.07 | 27.93 | 0.72 | 29.34 | 29.31 | 28.25 | 0.62 | 17 | 28.43 | 28.21 | 29.04 | 0.43 | 27.78 | 27.98 | 28.43 | 0.33 |
| 6 | 28.33 | 30.24 | 28.15 | 1.16 | 28.37 | 30.02 | 28.46 | 0.93 | 18 | 29.78 | 28.20 | 28.12 | 0.94 | 29.14 | 28.29 | 28.06 | 0.57 |
| 7 | 27.49 | 27.39 | 26.96 | 0.28 | 27.68 | 27.60 | 27.23 | 0.24 | 19 | 27.14 | 27.19 | 27.10 | 0.05 | 27.14 | 27.24 | 27.21 | 0.05 |
| 8 | 29.49 | 28.12 | 27.59 | 0.98 | 29.22 | 28.36 | 27.94 | 0.65 | 20 | 27.55 | 27.91 | 27.43 | 0.25 | 27.51 | 27.60 | 27.48 | 0.06 |
| 9 | 28.93 | 28.79 | 28.65 | 0.14 | 28.42 | 28.76 | 28.57 | 0.17 | 21 | 28.00 | 26.61 | 26.39 | 0.87 | 27.47 | 26.75 | 26.52 | 0.49 |
| 10 | 29.88 | 29.36 | 29.26 | 0.33 | 29.70 | 29.16 | 29.10 | 0.33 | 22 | 25.67 | 25.81 | 25.88 | 0.10 | 26.04 | 26.26 | 26.16 | 0.11 |
| 11 | 28.82 | 29.30 | 28.79 | 0.29 | 27.69 | 28.26 | 27.79 | 0.30 | 23 | 29.08 | 28.61 | 28.97 | 0.25 | 28.02 | 27.78 | 27.84 | 0.12 |
| 12 | 29.81 | 30.19 | 29.03 | 0.59 | 29.68 | 29.53 | 29.04 | 0.33 | 24 a | — | — | — | 26.19 | 27.27 | 26.23 | 0.61 | |
Note: Threshold Values; FAM and TEXAS RED‐200. Bold indicates Standard Deviaton value of each sample.
Sample is SMN1 Exon 7 Homozygous deleted.
FIGURE 4.
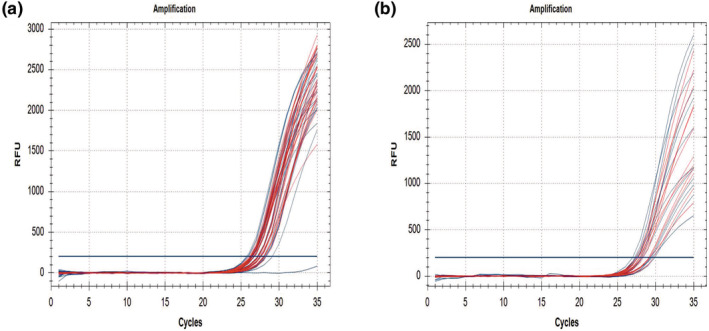
The appereance of amplification plots (a) I.W. Tremont Co. DBS; one sample is homozygous deleted, (b) Whatman 903 DBS. SMN1 Exon 7 labelled with FAM‐blue colour and reference gene labelled with TEXAS RED‐red colour.
3.4. Confirmation test
The results of 50 air dried DBS samples were found to be fully compatible in 1.2 mm punch size samples with previous MLPA data. MLPA results with 50 DBS samples taken randomly from 580 newborn DBS Samples were found to be fully compatible (Table 3).
TABLE 3.
Evaluation of Direct SMA Screening test versus MLPA.
| Value | Calculation | Wild type | Homozygous deletion | Results |
|---|---|---|---|---|
| Accuracy | (TP + TN)/(TP + TN + FP + FN) | (80 + 20)/(80 + 20 + 0 + 0) | (20 + 80)/(20 + 80 + 0 + 0) | 1 |
| Sensitivity | TP/(TP + FN) | 80/(80 + 0) | 20/(20 + 0) | 1 |
| Specificity | TN/(TN + FP) | 20/(20 + 0) | 80/(80 + 0) | 1 |
| False positive rate | FP/(TN + FP) | 0/(20 + 0) | 0/(80 + 0) | 0 |
| False negative rate | FN/(TP + FN) | 0/(80 + 0) | 0/(20 + 0) | 0 |
| Efficiency | (TP + TN)/(TP + TN + FP + FN) | (80 + 20)/(80 + 20 + 0 + 0) | (20 + 80)/(20 + 80 + 0 + 0) | 1 |
Abbreviations: FN, false negative; FP, false positive; TN, true negative; TP, true positive.
4. DISCUSSION
SMA is the second leading autosomal recessively inherited disease in humans after cystic fibrosis (Czibere et al., 2020; Phan et al., 2015). Therefore, many countries have recently introduced newborn SMA screening programs to detect the SMN1 exon 7 deletion, homozygous status. In this study, we basically developed the NLM with the DBS mix using reduced punch size to minimize its inhibitory effect such as DBS paper, hemoglobin, immunoglobulin, and so forth on the direct real time PCR test. In the new direct test system, we achieved the expected results with 1.2 mm punch size (Figures 1 and 2). We ensured that NLM can be applied as direct test to large number of samples in the shortest time by real‐time PCR method. Moreover, NLM has a significant advantage to minimize the contamination risk due to not including many working steps such as pipetting and centrifugation in the laboratory where thousand tests per a day carried out. Thereby, many countries can effortlessly use this user‐friendly direct real time PCR test system and reduce a severe financial burden on their national budgets because the NLM can be run without many special reagents, kits and trained personnel. In the procedure, you only need to apply two sequential steps; first to place a DBS punch into real time PCR plate well with a DBS puncher, and second, to add DBS mix into each well and then directly head for real time PCR running step. Due to without any extra work and effort for nucleic acid extraction from DBS, it will seriously diminish time‐consuming steps, lab errors, cost and additional laborious problems such as sample recording and tracking. At the end of the run time, approximately 73 minutes for CFX‐96 Real Time PCR equipment (Bio‐Rad, USA), the results can be obtained manually or by custom designed software; IntRa‐Q (SNP Biotechnology/Turkey) for CFX‐96 Real Time PCR equipment. IntRa‐Q can only facilitate and prop to procure the results from CFX‐96 Real Time PCR equipment easily. The NLM does not depend on the software to work properly. That is, it does not use the IntRa‐Q software to reach the results. In brief, this new SMA screening system with direct real time PCR tests demanding no extraction can also be used everywhere to assay with any real time PCR equipment.
In this study, we found all 580 newborn DBS samples as wild type in the direct real time PCR test. On the other hand, we correctly detected all 20 SMN1 exon 7 homozygous deletion in DBS samples prepared with patients’ blood (Figure 3). Internal control of all samples was positive and their Ct values were below 32. Intended results were obtained with direct real time PCR tests by repeating three times in DBS punches including 1.2 mm samples size. The results of two kinds of DBS paper showed minor differences. We got nearly the same ct values in three repeats with I.W. Tremont Co. DBS paper whereas the ct values showed slight difference with Whatman 903 DBS paper (Table 2). Although the standard deviation results of Whatman 903 and I.W. Tremont Co. DBS papers were found close to each other, the I.W. Tremont Co. DBS papers has been more attractive in appearance of amplification plots and the amount of fluorescence values (Figure 4a). The amplification plots of Whatman 903 DBS had a little dispersed appearance (Figure 4b). According to the data, we can claim that newborn DBS samples can be run once to reach final results.
All results of 50 DBS samples randomly selected from 580 DBS samples and 50 whole blood‐impregnated DBS samples were compatible with MLPA data. When the MLPA method was taken as the gold standard, the sensitivity and specificity of the NLM test were found 100% for the detection of SMN1 exon 7 homozygous deletion. These results show that the positive and negative predictive values of the newly developed extraction free test system were found 100% (Table 3). From all reasons above, the method was considered as successful and validated.
Newborn SMA screening with the NLM that we have developed has an environmentally friendly feature without requiring DNA extraction step. The DNA extraction process needs various harmful chemical substances, extra energy consumption and countless plastic tubes and tips usage. Therefore, there is no need for DNA extraction procedure in the NLM, which promotes a sustainable environment and waste management, which has gained great importance all over the world in recent years.
In the literature, it is seen that methods requiring consecutive two‐step PCR (Shinohara et al., 2019) or post‐PCR processes such as melting curve/agarose gel electrophoresis (Takeuchi et al., 2019; Wijaya, Nishio, Niba, Okamoto, et al., 2021; Wijaya, Nishio, Niba, Shiroshita, et al., 2021) are applied in direct PCR studies for SMA newborn screening. These methods have a low potential for application in SMA newborn screening programs due to involving high risk of contamination and operational challenges. However, the single‐step and post‐PCR‐free NLM we have described in this study have a high application potential for newborn SMA screening programs. Especially, the use of I.W. Tremont Co. DBS paper, which we recommend made it possible to observe standard sigmoidal Real Time PCR plots in the TaqMan® Real Time PCR assay.
To date, we have found that in all countries having a newborn screening program for SMA, a homozygous deletion of Exon 7 of SMN1 is detected, due to some ethical worries. In fact, when the analysis for the SMA carrier status in newborn SMA screening programs is carried out, extra information for the baby as carrier status may also be estimated. If carrier status of a baby for SMA was detected, the mother/father of the baby would be warned and also tested for their carrier status. Thus, parents who were found to be carriers may be provided genetic counseling for planning their next pregnancies. In fact, we noticed that the direct real time PCR method used in this study can also be available quantitatively for newborn SMA carrier screening in the near future.
In conclusion, the SMA screening program can be a very useful program for a country's health and economy in the long run.
AUTHOR CONTRIBUTIONS
Ayhan Kubar designed the work, performed interpretation of results, drafted and revised the manuscript. Sehime Gulsum Temel, Serdar Beken, Gizem Onder, Ozden Hatırnaz, Ayse Korkmaz, Yasemin Alanay, Ugur Ozbek and Sebnem Ozemri Sag contributed specimen collection, clinical follow‐up and evaluation. Mahmut Cerkez Ergoren performed statistics analysis. Elif Kubar, Candan Zeynep Sonmezalp and Ozlem Doğan performed laboratory work, preparing manuscript to submission and redaction. All authors read and approved the final manuscript.
FUNDING INFORMATION
No financial assistance was received in support of this study.
CONFLICT OF INTEREST STATEMENT
The author(s) declare(s) that there is no conflict of interest regarding the publication of this paper.
ETHICS STATEMENT
Ethics approval statement of Acibadem University ATADEK‐2019‐16/4.
ACKNOWLEDGMENTS
We thank all the newborns, patients and their family members who participated in this study, and we thank the SNP Biotechnology R&D Ltd. team for their environmental friendly approach and work.
Kubar, A. , Temel, S. G. , Beken, S. , Onder, G. , Hatirnaz, O. , Korkmaz, A. , Alanay, Y. , Ozbek, U. , Sag, S. O. , Ergoren, M. C. , Kubar, E. , Sonmezalp, C. Z. , & Doğan, O. (2023). A new line method; A direct test in spinal muscular atrophy screening for DBS. Molecular Genetics & Genomic Medicine, 11, e2270. 10.1002/mgg3.2270
DATA AVAILABILITY STATEMENT
The data used to support the findings of this study are included within the article and additionally it can be provided from the corresponding author upon request.
REFERENCES
- Cavdarli, B. , Ozturk, F. N. , Guntekin Ergun, S. , Ergun, M. A. , Dogan, O. , & Percin, E. M. (2020). Intelligent ratio: A new method for carrier and newborn screening in spinal muscular atrophy. Genetic Testing and Molecular Biomarkers, 24, 9, 569–577. [DOI] [PubMed] [Google Scholar]
- Czibere, L. , Burggraf, S. , Fleige, T. , Glück, B. , Keitel, L. M. , Landt, O. , Durner, J. , Röschinger, W. , Hohenfellner, K. , Wirth, B. , Müller‐Felber, W. , Vill, K. , & Becker, M. (2020). High‐throughput genetic newborn screening for spinal muscular atrophy by rapid nucleic acid extraction from dried blood spots and 384‐well qPCR. European Journal of Human Genetics, 28, 23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finkel, R. S. , Mercuri, E. , Meyer, O. H. , Simonds, A. K. , Schroth, M. K. , Graham, R. J. , Kirschner, J. , Iannaccone, S. T. , Crawford, T. O. , Woods, S. , Muntoni, F. , Wirth, B. , Montes, J. , Main, M. , Mazzone, E. S. , Vitale, M. , Snyder, B. , Quijano‐Roy, S. , Bertini, E. , … SMA Care group . (2018). Diagnosis and management of spinal muscular atrophy: Part 2: Pulmonary and acute care; Medications, supplements and immunizations; Other organ systems; And ethics. Neuromuscular Disorders, 28, 197–207. [DOI] [PubMed] [Google Scholar]
- Hendrickson, B. C. , Donohoe, C. , Akmaev, V. R. , Sugarman, E. A. , Labrousse, P. , Boguslavskiy, L. , Flynn, K. , Rohlfs, E. M. , Walker, A. , Allitto, B. , Sears, C. , & Scholl, T. (2009). Differences in SMN1 allele frequencies among ethnic groups within North America. Journal of Medical Genetics, 46, 641–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolb, S. J. , & Kissel, J. T. (2015). Spinal muscular atrophy. Neurologic Clinics, 33(4), 831–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, M. , Liu, L. , Peter, I. , Zhu, J. , Scott, S. A. , Zhao, G. , Eversley, C. , Kornreich, R. , Desnick, R. J. , & Edelman, L. (2014). An Ashkenazi Jewish SMN1 haplotype specific to duplication alleles improves pan‐ethnic carrier screening for spinal muscular atrophy. Genetics in Medicine, 16(2), 149–156. [DOI] [PubMed] [Google Scholar]
- Meldrum, C. , Scott, C. , & Swoboda, K. J. (2007). Spinal muscular atrophy genetic counseling access and genetic knowledge: parents' perspectives. Journal of Child Neurology, 22, 1019–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercuri, E. , Darras, B. T. , Chiriboga, C. A. , Day, J. W. , Campbell, C. , Connolly, A. M. , Iannaccone, S. T. , Kirschner, J. , Kuntz, N. L. , Saito, K. , Shieh, P. B. , Tulinius, M. , Mazzone, E. S. , Montes, J. , Bishop, K. M. , Yang, Q. , Foster, R. , Gheuens, S. , Bennett, C. F. , … CHERISH Study Group . (2018). Nusinersen versus sham control in later‐onset spinal muscular atrophy. The New England Journal of Medicine, 378, 625–635. [DOI] [PubMed] [Google Scholar]
- Mercuri, E. , Finkel, R. S. , Muntoni, F. , Wirth, B. , Montes, J. , Main, M. , Mazzone, E. S. , Vitale, M. , Snyder, B. , Quijano‐Roy, S. , Bertini, E. , Davis, R. H. , Meyer, O. H. , Simonds, A. K. , Schroth, M. K. , Graham, R. J. , Kirschner, J. , Iannaccone, S. T. , Crawford, T. O. , … Szlagatys‐Sidorkiewicz, A. (2018). Diagnosis and management of spinal muscular atrophy: Part 1: Recommendations for diagnosis, rehabilitation, orthopedic and nutritional care. Neuromuscular Disorders, 28, 103–115. [DOI] [PubMed] [Google Scholar]
- Phan, H. C. , Taylor, J. L. , Hannon, H. , & Howell, R. (2015). Newborn screening for spinal muscular atrophy: Anticipating an imminent need. Seminars in Perinatology, 39, 217–229. [DOI] [PubMed] [Google Scholar]
- Saffari, A. , Kölker, S. , Hoffmann, G. F. , Weiler, M. , & Ziegler, A. (2019). Novel challenges in spinal muscular atrophy—How to screen and whom to treat? Annals of Clinical and Translational Neurology, 6, 197–205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara, M. , Niba, E. T. , Wijaya, Y. O. , Takayama, I. , Mitsuishi, C. , & Kumasaka, S. (2019). A novel system for spinal muscular atrophy screening in newborns: Japanese pilot study. International Journal of Neonatal Screening, 5, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi, A. , Tode, C. , Nishino, M. , Wijaya, Y. O. , Niba, E. T. , & Awano, H. (2019). Newborn screening for spinal muscular atrophy: DNA preparation from dried blood spot and DNA polymerase selection in PCR. The Kobe Journal of Medical Sciences, 65(3), 95–99. [PMC free article] [PubMed] [Google Scholar]
- Wijaya, Y. O. , Nishio, H. , Niba, E. T. , Okamoto, K. , Shintaku, H. , & Takeshima, Y. (2021). Detection of spinal muscular atrophy patients using dried saliva spots. Genes, 12, 1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wijaya, Y. O. , Nishio, H. , Niba, E. T. , Shiroshita, T. , Kato, M. , & Bouike, Y. (2021). Dried blood spot screening system for spinal muscular atrophy with allele‐specific polymerase chain reaction and melting peak analysis. Genetic Testing and Molecular Biomarkers, 25(4), 293–301. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are included within the article and additionally it can be provided from the corresponding author upon request.


