Abstract
A 2,3-dihydroxybiphenyl 1,2-dioxygenase from the naphthalenesulfonate-degrading bacterium Sphingomonas sp. strain BN6 oxidized 3-chlorocatechol to a yellow product with a strongly pH-dependent absorption maximum at 378 nm. A titration curve suggested (de)protonation of an ionizable group with a pKa of 4.4. The product was isolated, purified, and converted, by treatment with diazomethane, to a dimethyl derivative and, by incubation with ammonium chloride, to a picolinic acid derivative. Mass spectra and 1H and 13C nuclear magnetic resonance (NMR) data for these two derivatives prove a 3-chloro-2-hydroxymuconic semialdehyde structure for the metabolite, resulting from distal (1,6) cleavage of 3-chlorocatechol. 3-Methylcatechol and 2,3-dihydroxybiphenyl are oxidized by this enzyme, in contrast, via proximal (2,3) cleavage.
Aerobic bacterial degradation of aromatic substrates very often proceeds via intermediate formation of vicinal dihydroxy arenes such as substituted 1,2-dihydroxybenzenes (catechols) or 1,2-dihydroxynaphthalenes. Further down the catabolic pathway, substituted catechols are oxidized by cleavage of the aromatic ring either between the two hydroxy groups (intradiol or ortho cleavage) or at a bond adjacent to the two hydroxy groups (extradiol or meta cleavage). As a rule, which mode of ring fission predominates depends on the other substituents on the aromatic ring. Chlorocatechols generally are mineralized via the ortho-cleavage pathway. Degradation of methylcatechols, in contrast, generally proceeds via meta cleavage (12, 13). 1,2-Dioxygenation (ortho cleavage) of methyl-substituted catechols often results in the formation of dead-end metabolites such as methylmuconolactones (30, 33). Conversion of chlorocatechols by extradiol dioxygenases (meta cleavage), on the other hand, frequently leads to ill-defined compounds which do not allow for productive degradation (16). In the proximal (2,3) extradiol cleavage of 3-chlorocatechol by certain extradiol dioxygenases, for instance, the enzymes are rapidly inactivated, presumably by chelation of the catalytically active ferrous ion or suicide inhibition by an acylchloride (6, 22).
Degradation of polycyclic chlorinated arenes, e.g., polychlorinated biphenyls or naphthalenes, usually requires extradiol cleavage of one of the aromatic rings followed by intradiol cleavage of the chlorinated catechol formed intermediately in this process. There is a continuous search, therefore, for a productive extradiol cleavage of chlorinated catechols, or at least for extradiol dioxygenases which are not inactivated in the course of the degradative process (3, 8). Some evidence has recently been presented for productive extradiol cleavage of chlorocatechols. Pseudomonas putida GJ31, which grows on both toluene and chlorobenzene, effects a proximal cleavage of 3-chlorocatechol and instant hydrolysis of the resulting acylchloride yielding 2-hydroxymuconic acid (compound VI [Fig. 1]). The enzyme involved in this process appears not to be subject to suicide inactivation (21, 25). There is also some evidence for productive degradation of 4-chlorophenol, 3-methyl-4-chlorocatechol, or 2,4-dichlorophenol by extradiol degradative pathways (18, 19, 23).
FIG. 1.
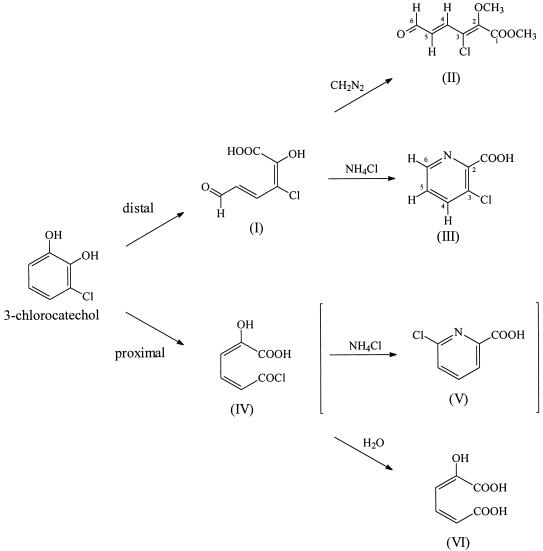
Possible directions for extradiol ring cleavage of 3-chlorocatechol and conversion of the ring fission products. Compounds: I, 3-chloro-2-hydroxymuconic semialdehyde; II, 3-chloro-2-methoxymuconic semialdehyde methyl ester; III, 3-chloropicolinic acid; IV, 2-hydroxymuconic acid chloride; V, 6-chloropicolinic acid; VI, 2-hydroxymuconic acid.
In the course of our studies on the naphthalenesulfonate-degrading strain Sphingomonas sp. strain BN6, we have recently cloned the genes for two extradiol dioxygenases (14, 15). One of the encoded enzymes is the smallest extradiol dioxygenase characterized so far. It oxidizes 3-chlorocatechol to a metabolite which, on the basis of its characteristic yellow color, was tentatively assigned as the product of distal (1,6) extradiol oxidation of 3-chlorocatechol. We have now isolated and structurally characterized this metabolite and attempted to assess the possibility of a further conversion of the ring cleavage product which could represent the first step of a novel metabolic pathway for the degradation of 3-chlorocatechol.
MATERIALS AND METHODS
Bacterial strains.
The 2,3-dihydroxybiphenyl 1,2-dioxygenase (BphC) was produced by using Escherichia coli JM108(pGHS118). Plasmid pGHS118 contained bphC under the control of the lac promoter. The plasmid was constructed by transferring a 507-bp NdeI-BamHI fragment containing the bphC gene from pGHS117 (15) to plasmid pJOE 2783.1. This plasmid was constructed from the pBR322 derivative pBTac1 (Ampr) (9), in which the tac promoter was replaced by a lac wild-type promoter (1). Catechol 2,3-dioxygenase (XylE) was expressed from the pBR322 derivative pJF150 (Ampr) and 2-hydroxymuconic semialdehyde hydrolase was expressed from E. coli C600(pGP1-2)(pJH102), which contained the xylE and xylF genes, respectively, under the control of the T7 promoter (10, 11). P. putida mt-2 was grown with 3-methylbenzoate (29).
Preparation of cell extracts.
Frozen cells were suspended in 50 mM sodium potassium phosphate buffer (pH 7.5) and disrupted by using a French press (Aminco, Silver Spring, Md.) at 80 MPa. Cell debris was removed by centrifugation (100,000 × g, 45 min, 4°C). In the case of the cell extract of E. coli C600(pGP1-2)(pJH102), the buffer additionally contained 2-mercaptoethanol (5 mM) (10). Protein concentration was determined as described by Bradford (7), with bovine serum albumin as the standard.
Enzyme assays.
The assays contained the ring fission products of catechol, 3-chlorocatechol, or 3-methylcatechol (60 μM each, in a total volume of 1 ml). Enzyme activities were determined at 25°C in 50 mM Na-K phosphate buffer (pH 7.5) by measuring the decrease of absorbance at 375, 378, or 388 nm (10, 32). In certain experiments the cell extracts were treated with NAD+ glycohydrolase (NADase) for 15 min at 30°C. For the assay of the 2-hydroxymuconic semialdehyde dehydrogenase, NAD+ (0.5 mM) was added (32). One unit of enzyme activity was defined as the amount of enzyme that converts 1 μmol of substrate per min, using the extinction coefficients reported previously (15).
HPLC.
Formation of ring cleavage products from 3-chloro- and 3-methylcatechol was monitored by high-pressure liquid chromatography (HPLC) (HPLC Millenium Chromatography Manager 2.0, equipped with the programmable multiwavelength detector model 486 and the HPLC pump 510; Waters Associates, Milford, Mass.). Two different solvent systems were used: water-methanol (88/12 [vol/vol]) plus tetrabutylammonium hydrogen sulfate as ion-pair reagent (PicA; Waters); and water-methanol (70.0/29.7 [vol/vol]) plus 0.3% (vol/vol) H3PO4. For all analyses, a reversed-phase column (150 by 4 mm; Grom-Sil C8; Grom, Herrenberg, Germany) was used, with a flow rate of 0.7 ml min−1, and the detection wavelength was set at 210 or 380 nm.
Spectroscopy.
Mass spectrometry (MS) data were obtained on a Finnigan MAT 95 mass spectrometer. The nuclear magnetic resonance (NMR) spectra were recorded with a Bruker AC250 or a ARX500 spectrometer in CDCl3 solution. Chemical shifts (δ) are given in parts per million relative to tetramethylsilane as the internal standard.
Oxidation of 3-chlorocatechol by resting cells.
E. coli JM108(pGHS118) was grown overnight in 3-liter Erlenmeyer flasks (600 ml of culture volume) in Luria-Bertani broth (LB) plus 100 μg of ampicillin ml−1 to an optical density at 546 nm of about 4. Cells were harvested by centrifugation, washed in Na-K phosphate buffer (100 mM, pH 7.5), and resuspended in 1/10 of the original volume of Na-K phosphate buffer. These resting cells were gently shaken at 27°C, and 3-chlorocatechol (from a 50 mM stock solution in Na-K phosphate buffer) was added at constant intervals. Generally, every 30 min an aliquot of 3-chlorocatechol (0.5 mM each; total, 5 mM) was added. Finally, cells were removed by centrifugation (20 min, 12,000 × g), and the supernatant was kept at 4°C.
Purification of the metabolite.
The supernatant containing the oxidation product was applied to a Q-Sepharose fast-flow column (15 by 1.6 cm), using a fast protein liquid chromatography apparatus (24). The sample was washed with 80 ml of Na-K phosphate buffer (50 mM, pH 7.5), and the metabolite was eluted, at a flow rate of 4 ml min−1, with a linear gradient of the same Na-K phosphate buffer plus 1.2 M NaCl. The yellow ring cleavage product was eluted at an NaCl concentration of about 0.4 M. Fractions (8 ml each) containing the product were collected and combined. The resulting pool (approximately 80 ml) was separated from residual macromolecular contaminations by ultrafiltration (Diaflo PM10 membrane with 10,000-molecular-weight cutoff; Amicon), and the filtrate was used for subsequent chemical derivatization.
Preparation of a picolinic acid derivative.
The purified aqueous solution of the ring cleavage product was incubated with NH4Cl (1.2 M) overnight (4, 26). The reaction mixture was adjusted to pH 2 with 2 N HCl and extracted five times with 15 ml each of ethyl acetate, and the extract was dried with anhydrous Na2SO4. Evaporation (40°C) of the solvent yielded colorless crystals (for NMR and MS data, see Tables 1 and 2).
TABLE 1.
MS data for 3-chloropicolinic acid (compound III [Fig. 1]) and 3-chloro-2-methoxymuconic semialdehyde methyl ester (compound II [Fig. 1])a
| Compound | Ionization method | m/z | Relative intensity | Assignment |
|---|---|---|---|---|
| III | Chemical ionization | 158/160 | 100/32 | [MH]+ |
| 140/142 | 72/24 | [MH − H2O]+ | ||
| 124 | 24 | |||
| EI | 157/159 | 4.7/1.6 | M+· | |
| 113/115 | 100/33 | M+· − CO2 | ||
| 78 | 50 | M+· − CO2, C1· | ||
| II | Chemical ionization | 205/207 | 17.4/5.8 | [MH]+ |
| 173/175 | 100/34 | [MH − HOCH3]+ | ||
| 145/147 | 43/14 | [MH − CH3OH, CO]+ | ||
| EI | 204/206 | 8.5/2.7 | M+· | |
| 175/177 | 10.7/3.4 | M+· − CHO· | ||
| 145/147 | 100/32 | M+· − CH3·, CO2 |
The picolinic acid and the muconic semialdehyde derivative were prepared as described in Materials and Methods.
TABLE 2.
1H NMR and 13C NMR data for 3-chloropicolinic acid (compound III [Fig. 1]) and 3-chloro-2-methoxymuconic semialdehyde methyl ester (compound II [Fig. 1])
| Compound | Position | δ (ppm)
|
J (Hz) | |
|---|---|---|---|---|
| 13C | 1H | |||
| III | COOH | 162.04 | ||
| 2 | 142.67 | |||
| 3 | 134.31 | |||
| 4 | 141.76 | 7.96 | 3J (4-H, 5-H) = 8.2 | |
| 5 | 128.80 | 7.57 | 3J (5-H, 6-H) = 4.6 | |
| 6 | 146.24 | 8.58 | 4J (4-H, 6-H) = 1.4 | |
| II | 1 | 162.04 | ||
| 1-OCH3 | 60.16 | 3.96 | ||
| 2 | 149.09 | |||
| 2-OCH3 | 53.10 | 3.86 | ||
| 3 | 126.49 | |||
| 4 | 143.79 | 7.96 | 3J (4-H, 5-H) = 14.9 | |
| 5 | 133.22 | 6.65 | 3J (5-H, 6-H) = 7.8 | |
| 6 | 192.52 | 9.68 | ||
Derivatization of the metabolite with diazomethane.
The purified aqueous solution of the ring cleavage product was acidified with 2 N HCl to pH 2.0 and extracted three times with ethyl acetate (25 ml each). The extract was treated with diazomethane, dried with anhydrous Na2SO4, and evaporated to dryness. The yellow solid residue was purified by column chromatography on silica gel with hexane-ethylacetate (70/30 [vol/vol]). Fractions containing the methylated ring fission product, with an absorption maximum at λ = 297 nm, were combined and evaporated to dryness, yielding a colorless powder (for NMR and MS data, see Tables 1 and 2).
Determination of the pH dependence of the absorption spectrum of 3-chloro-2-hydroxymuconic semialdehyde.
An aqueous solution of the ring cleavage product, prepared as described above, was diluted (1:200 [vol/vol]) with the following buffers (100 mM each): citric acid-HCl (pH 1 to 2), citric acid-NaOH (pH 3 to 5), Na-K phosphate (pH 5.5 to 7.5), Tris-HCl (pH 8 to 9.5), and glycine-NaOH (pH 10 to 13). Absorbance at λ = 378 nm was determined immediately after mixing and corrected for the absorbance of the buffer solution used.
Chemicals.
3-Chlorocatechol was purchased from TCI (Tokyo, Japan). NAD and NADase were obtained from Sigma (Deisenhofen, Germany). The sources of all other chemicals have been described before (14, 15).
RESULTS
Characterization and purification of the 3-chlorocatechol oxidation product.
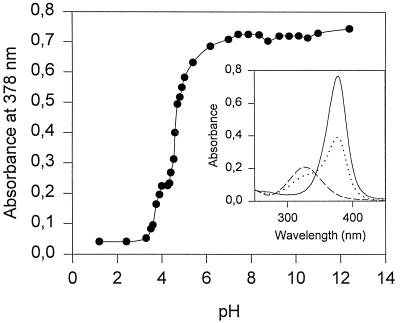
The 2,3-dihydroxybiphenyl 1,2-dioxygenase (BphC) from strain BN6 oxidized 3-chlorocatechol to a product with an absorption maximum (pH 7.5) at λ = 378 nm, which suggested distal extradiol cleavage of 3-chlorocatechol to a chlorinated muconic semialdehyde (15). A plasmid-free strain of E. coli JM108, in contrast, showed no reaction with 3-chlorocatechol. The oxidation product was fairly stable at 4°C: after 40 h in aqueous solution (pH 7.5 or 13.0), loss in absorbance at λ = 378 nm was less than 10%. At pH 2, the half-life was 15 h. The absorption of the ring cleavage product was shown to be strongly pH dependent, the titration curve indicating (de)protonation of an ionizable group with a pKa of 4.4 (Fig. 2). The corresponding pKa values for the extradiol (proximal) cleavage products of catechol and 4-chlorocatechol are 6.7 and 5.2 or 5.8, respectively (27, 37). To obtain the ring cleavage product in sufficient amount for structural analysis, 3-chlorocatechol was converted by resting cells of E. coli JM108(pGHS118); the transformation and the work-up were carried out as described in Materials and Methods.
FIG. 2.
Absorption of the ring cleavage product of 3-chlorocatechol at 378 nm depends on the pH value. The inset shows the absorption spectra at pH 8.0 (——), at pH 4.5 (•••), and at pH 2.0 (–––).
Identification of the ring cleavage product.
The purified ring cleavage product was transformed, by incubation with NH4Cl, into the appropriate picolinic acid derivative. Formation of a chloropicolinic acid is proven by the chemical ionization (CH4-CI) mass spectrum (Table 1), which displays the quasimolecular ion at m/z = 158/160 and fragment ions at m/z = 140/142 and 124, corresponding to the loss of H2O and HCl, respectively. The electron impact (EI) mass spectrum is dominated by a fragment at m/z = 113/115; loss of CO2 from M+· is characteristic for pyridine-2-carboxylic acids. The overall fragmentation pattern closely resembles that reported for 6-chloropyridine-2-carboxylic acid (28).
The 1H NMR spectrum of this derivative (Table 2) shows a set of three aryl protons which, from the coupling pattern, are in adjacent positions of the heterocyclic ring. One resonance must be assigned, due to its pronounced downfield shift (8.58 ppm), to a proton α to the aza nitrogen (17), and the two other aryl hydrogens thence must be in β and γ positions. The 13C NMR spectrum shows, besides the carboxylate resonance, five signals in the aromatic carbon region. Typical for a pyridine, these are differentiated into two resonances at higher field (C-3/5, meta position) and three at low field (C-2/6, ortho position; C-4, para position) where the electron-withdrawing effect of the aza nitrogen is effective (36). The assignments of C-3 and C-5 are straightforward since only one of these two carbons still bears a hydrogen (as demonstrated by the pronounced nuclear Overhauser enhancement effect [NOE]). The downfield shift of C-5 (+5 ppm relative to pyridine) is consistent only with a carboxylic group at C-2. The 134.31-ppm shift for the quaternary carbon C-3 likewise requires the Cl substituent to be in this position (20).
MS and NMR data thus unequivocally prove the 3-chloropicolinic acid structure (compound III [Fig. 1]) for the NH4Cl incubation product and thence the 3-chloro-2-hydroxymuconic semialdehyde (compound I, [Fig. 1]), formed by distal cleavage of 3-chlorocatechol, as the ring cleavage precursor. Proximal cleavage of 3-chlorocatechol (Fig. 1) would result in formation of 2-hydroxymuconic acid (compound VI) (25). Theoretically, the ring cleavage product (compound IV) could be converted by incubation with NH4Cl to 6-chloropicolinic acid (compound V). Both 1H and 13C NMR data for this compound (5, 31) are clearly different from those of the product obtained here.
The purified ring cleavage product was also reacted with diazomethane. The 1H and 13C NMR spectra of the derivative clearly show the presence of two methoxy groups, one of which is incorporated in an ester function (δ = 3.96 and 60.16 ppm, respectively). The proton signal at 9.68 ppm and the corresponding 13C resonance at 192.52 ppm unequivocally establish the presence of an aldehyde functionality linked, as proven by the 1H coupling pattern, to a trans olefinic moiety (3J = 14.9 Hz) (2). The (quasi)molecular ion signals in the CH4-CI and EI mass spectra (Table 1) are consistent with a chlorinated methoxymuconic semialdehyde methyl ester structure (compound II [Fig. 1]). Moreover, the EI fragmentation pattern closely resembles that observed for the cleavage product from 4-chlorocatechol (37). Once again, the spectroscopic data definitively establish the 3-chloro-2-hydroxymuconic semialdehyde structure (compound I [Fig. 1]) for the ring cleavage product and thus prove the distal extradiol cleavage of 3-chlorocatechol.
Proximal cleavage of 3-methylcatechol by BphC.
All extradiol dioxygenases described so far converted the structurally analogous substrate 3-methylcatechol by a proximal ring cleavage mechanism. Oxidation of 3-methylcatechol by BphC was therefore analyzed in detail and compared to cleavage of this compound by the archetypal catechol 2,3-dioxygenase encoded by the TOL plasmid. No differences were observed when the reaction products were analyzed by UV/visible light spectroscopy or HPLC. Furthermore, the respective reaction products (from BphC or catechol 2,3-dioxygenase with 3-methylcatechol as the substrate) were converted by cell extracts from P. putida mt-2 with the same activity. 3-Methylcatechol may hence be concluded to be oxidized by BphC via a proximal ring cleavage mechanism.
Conversion of 3-chloro-2-hydroxymuconic semialdehyde by 2-hydroxymuconic semialdehyde hydrolase.
The ring fission products of both 3- and 4-methylcatechol are further metabolized by the TOL plasmid-encoded enzyme 2-hydroxymuconic semialdehyde hydrolase or 2-hydroxymuconic semialdehyde dehydrogenase (29). Cell extracts from 3-methylbenzoate-grown cells of P. putida mt-2 also converted 3-chloro-2-hydroxymuconic semialdehyde. Experiments with or without NAD+ and NADase indicated the 2-hydroxymuconic semialdehyde hydrolase to be responsible for this reaction (Table 3). The relative activity for the conversion of 3-chloro-2-hydroxymuconic semialdehyde was low, but the reaction rate did not change for more than 30 min. Cell extracts, prepared from P. putida mt-2 grown on succinate, showed only about 5% of the activity found after growth on 3-methylbenzoate. This result proved that an inducible enzyme was responsible for the conversion of 3-chloro-2-hydroxymuconic semialdehyde. The data were confirmed by using the recombinant E. coli strain C600(pGP1-2)(pJH102), expression host for the 2-hydroxymuconic semialdehyde hydrolase from the TOL plasmid. The relative activities observed for the turnover of the ring fission products were almost the same as for the assay with an NADase-treated cell extract of P. putida mt-2.
TABLE 3.
Conversion of 3-chloro-2-hydroxymuconic semialdehyde by cell extracts from P. putida mt-2a
| Substrate (initial concn, 60 μM) | Sp act (μmol min−1 mg of protein−1)
|
||
|---|---|---|---|
| No supple- ments | +NADase | +NAD+ | |
| 2-Hydroxymuconic semialdehyde (ring fission product of catechol) | 0.27 | 0.22 | 0.49 |
| 2-Hydroxy-6-oxo-hepta-2,4-dienoate (ring fission product of 3-methylcatechol) | 2.24 | 2.24 | 2.24 |
| 3-Chloro-2-hydroxymuconic semialdehyde (ring fission product of 3-chlorocatechol) | 0.0007 | 0.0006 | 0.0007 |
Cells were grown with 3-methylbenzoate at 30°C. Experimental details are given in Materials and Methods.
DISCUSSION
The results reported above unequivocally prove distal cleavage of 3-chlorocatechol by BphC from strain BN6. 3-Methylcatechol and 2,3-dihydroxybiphenyl, in contrast, are transformed by this enzyme via proximal cleavage of the aromatic nucleus. The steric demand of the substituent in the 3 position thus appears not to be decisive in directing the regiochemistry of the ring cleavage. The amino acid sequence of BphC shows a very small degree of homology with the main group of extradiol dioxygenases. Nevertheless, the residues involved in chelation of the catalytically active Fe(II) ion are conserved among BphC from strain BN6 and the well-studied catechol 2,3-dioxygenases and 2,3-dihydroxybiphenyl dioxygenases (15). The same catalytic mechanism thus probably operates with all these enzymes.
For the catechol 2,3-dioxygenase from P. putida mt-2, the catalytic cycle is supposed to involve complexation of the catalytically active Fe(II) ion by a monoanionic catecholate as bidentate ligand (35). The different regiochemistry of the ring fission between 3-chloro- and 3-methylcatechol thus may be due to the difference in relative acidity of the two hydroxy groups in the presence of a further methyl or chloro substituent on the aromatic ring. The inductive effect of a chloro substituent should favor formation of a phenolate anion in the ortho position; a methyl group, in contrast, should destabilize an ortho phenolate anion. As ring cleavage by the catechol 2,3-dioxygenase is proposed to proceed via an attack of the activated oxygen species on the (nonhydroxylated) position vicinal to the carbon atom bearing the phenolate anion (35), the higher stability of an ortho- relative to a meta-substituted chlorophenolate anion should favor proximal cleavage of 3-chlorocatechol if the inductive effect argument is valid. The opposite is observed, though, with BphC from strain BN6.
If the different inductive and/or resonance effects of a chloro compared to an alkyl substituent indeed were responsible for the different directions of ring fission between chloro- and methylcatechol, and if the basic biochemistry of ring fission between BphC from strain BN6 and the well-characterized extradiol dioxygenases was in fact conserved, it still remains unclear why most extradiol dioxygenases appear to oxidize chloro- and methylcatechol by a proximal mechanism and suffer suicide inhibition in the attempted conversion of 3-chlorocatechol. The simplest explanation for this may be that in most extradiol dioxygenases, steric hindrance prevents substrate binding in a position in the active site which would allow a distal cleavage of the substrate. This interpretation is supported in some way by modeling experiments with the 2,3-dihydroxybiphenyl dioxygenase from Pseudomonas sp. strain KKS102 (34). These modeling experiments suggested that the enzyme may also accept 4-chloro-2,3-dihydroxybiphenyl as a substrate, but with a slight conformational change in several amino acid side chains.
Evidence for productive conversion of 3-chlorocatechol via proximal extradiol ring cleavage was recently presented (21, 25). The distal cleavage of 3-chlorocatechol seems to be an almost unique feature of BphC from strain BN6. Only a catechol 2,3-dioxygenase from Azotobacter vinelandii 206 has previously been reported to likewise oxidize 3-chlorocatechol to a yellow product and thus presumably performs the same reaction (32). Because BphC oxidized 3,5-dichlorocatechol also by the distal cleavage mechanism (14), this enzyme seems to have the capacity for oxidizing various meta-substituted halogenated catechols by a distal cleavage mechanism without suffering suicide inhibition. Since this enzyme also oxidizes 2,3-dihydroxybiphenyl, it offers a potential for constructing bacteria with the ability to degrade chlorinated biphenyls.
ACKNOWLEDGMENTS
We thank J. Trinkner for performing the MS experiments and D. W. Ribbons (Graz) and H.-J. Knackmuss for advice and helpful discussions.
This work was supported by grant KN 183/5-1 from the Deutsche Forschungsgemeinschaft.
REFERENCES
- 1.Altenbucher, J. Personal communication.
- 2.Andreoni V, Canonica L, Galli E, Gennari C, Treccani V. 2,3-Dihydroxybenzoate pathway in Pseudomonas putida. 1H n.m.r. study on the ring-cleavage site. Biochem J. 1981;194:607–610. doi: 10.1042/bj1940607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arensdorf J J, Focht D D. Formation of chlorocatechol meta cleavage products by a pseudomonad during metabolism of monochlorobiphenyls. Appl Environ Microbiol. 1994;60:2884–2889. doi: 10.1128/aem.60.8.2884-2889.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Asano Y, Yamamoto Y, Yamada H. Catechol 2,3-dioxygenase-catalyzed synthesis of picolinic acids from catechols. Biosci Biotechnol Biochem. 1995;58:2054–2056. [Google Scholar]
- 5.Ashimori A, Ono T, Uchida T, Ohtaki Y, Fukaya C, Watanabe M, Yokoyama K. Novel 1,4-dihydropyridine calcium antagonists. I. Synthesis and hypotensive activity of 4-(substituted pyridyl)-1,4-dihydropyridine derivatives. Chem Pharm Bull. 1990;38:2446–2458. doi: 10.1248/cpb.38.2446. [DOI] [PubMed] [Google Scholar]
- 6.Bartels I, Knackmuss H-J, Reineke W. Suicide inactivation of catechol 2,3-dioxygenase from P. putida mt-2 by 3-halocatechols. Appl Environ Microbiol. 1984;47:500–505. doi: 10.1128/aem.47.3.500-505.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bradford M M. A rapid and sensitive method for the quantitation of protein utilizing the principle of protein dye binding. Anal Biochem. 1976;72:248–252. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 8.Brenner V, Arensdorf J J, Focht D D. Genetic construction of PCB degraders. Biodegradation. 1994;5:359–377. doi: 10.1007/BF00696470. [DOI] [PubMed] [Google Scholar]
- 9.Brosius J, Dull T J, Sleeter D D, Noller H F. Gene organization and primary structure of a ribosomal RNA operon from Escherichia coli. J Mol Biol. 1981;148:107–127. doi: 10.1016/0022-2836(81)90508-8. [DOI] [PubMed] [Google Scholar]
- 10.Diaz E, Timmis K N. Identification of functional residues in a 2-hydroxymuconic semialdehyde hydrolase. J Biol Chem. 1995;270:6403–6411. doi: 10.1074/jbc.270.11.6403. [DOI] [PubMed] [Google Scholar]
- 11.Fischer J. Entwicklung eines regulierbaren Expressionssystems zur effizienten Synthese rekombinanter Proteine in Streptomyces lividans. Ph.D. thesis. Stuttgart, Germany: Universität Stuttgart; 1996. [Google Scholar]
- 12.Gottschalk G, Knackmuss H-J. Bacteria and the biodegradation of chemicals achieved naturally, by combination, or by construction. Angew Chem Int Ed Engl. 1993;32:1398–1408. [Google Scholar]
- 13.Harayama S, Timmis K N. Aerobic biodegradation of aromatic hydrocarbons by bacteria. In: Sigel H, Sigel A, editors. Metal ions in biological systems. Vol. 28. New York, N.Y: Marcel Dekker Inc.; 1992. pp. 99–156. [Google Scholar]
- 14.Heiss G, Müller C, Altenbuchner J, Stolz A. Analysis of a new dimeric extradiol dioxygenase from a naphthalenesulfonate-degrading sphingomonad. Microbiology. 1997;143:1691–1699. doi: 10.1099/00221287-143-5-1691. [DOI] [PubMed] [Google Scholar]
- 15.Heiss G, Stolz A, Kuhm A E, Müller C, Klein J, Altenbuchner J, Knackmuss H-J. Characterization of a 2,3-dihydroxybiphenyl dioxygenase from the naphthalenesulfonate-degrading bacterium strain BN6. J Bacteriol. 1995;177:5865–5871. doi: 10.1128/jb.177.20.5865-5871.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hernandez B S, Arensdorf J J, Focht D D. Catabolic characteristics of biphenyl-utilizing isolates which cometabolize PCBs. Biodegradation. 1995;6:75–82. [Google Scholar]
- 17.Hesse M, Meier H, Zeeh B. Spektroskopische Methoden in der organischen Chemie. Stuttgart, Germany: Thieme Verlag; 1984. p. 152. [Google Scholar]
- 18.Higson F K, Focht D D. Utilization of 3-chloro-2-methylbenzoic acid by Pseudomonas cepacia MB2 through the meta fission pathway. Appl Environ Microbiol. 1992;58:2501–2504. doi: 10.1128/aem.58.8.2501-2504.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hollender J, Dott W, Hopp J. Regulation of chloro- and methylphenol degradation in Comamonas testosteroni JH5. Appl Environ Microbiol. 1994;60:2330–2338. doi: 10.1128/aem.60.7.2330-2338.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kalinowski H-O, Berger S, Braun S. 13C-NMR-Spektroskopie. Stuttgart, Germany: Thieme Verlag; 1984. p. 364. [Google Scholar]
- 21.Kaschabek S R, Kasberg T, Müller D, Mars A E, Janssen D B, Reineke W. Degradation of chloroaromatics: purification and characterization of a novel type of chlorocatechol 2,3-dioxygenase of Pseudomonas putida GJ31. J Bacteriol. 1998;180:296–302. doi: 10.1128/jb.180.2.296-302.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klecka G M, Gibson D T. Inhibition of catechol 2,3-dioxygenase from Pseudomonas putida mt-2 by 3-chlorocatechol. Appl Environ Microbiol. 1981;41:1159–1165. doi: 10.1128/aem.41.5.1159-1165.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koh S-C, McCullar M V, Focht D D. Biodegradation of 2,4-dichlorophenol through a distal meta-fission pathway. Appl Environ Microbiol. 1997;63:2054–2057. doi: 10.1128/aem.63.5.2054-2057.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhm A E, Stolz A, Ngai K-L, Knackmuss H-J. Purification and characterization of a 1,2-dihydroxynaphthalene dioxygenase from a bacterium that degrades naphthalenesulfonic acids. J Bacteriol. 1991;173:3795–3802. doi: 10.1128/jb.173.12.3795-3802.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mars A E, Kasberg T, Kaschabek S R, van Agteren M, Janssen D B, Reineke W. Microbial degradation of chloroaromatics. Use of the meta-cleavage pathway for the mineralization of chlorobenzene. J Bacteriol. 1997;179:4530–4537. doi: 10.1128/jb.179.14.4530-4537.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matthews C, Rossiter J T, Ribbons D W. Production of pyridine synthons by biotransformations of benzene precursors and their cyclization with nitrogen nucleophiles. Biocatal Biotransform. 1995;12:241–254. [Google Scholar]
- 27.Morris C M, Barnsley E A. The cometabolism of 1- and 2-chloronaphthalene by pseudomonads. Can J Microbiol. 1982;28:73–79. [Google Scholar]
- 28.Moser R J, Brown E V. Mass spectra of some 5- and 6-substituted-2-pyridinecarboxylic acids. Nature of fragmentation step for loss of CO2. Org Mass Spectrom. 1970;4:555–561. [Google Scholar]
- 29.Murray K, Duggleby C J, Sala-Trepat J M, Williams P A. The metabolism of benzoate and methylbenzoates via the meta-cleavage pathway by Pseudomonas arvilla mt-2. Eur J Biochem. 1972;28:301–310. doi: 10.1111/j.1432-1033.1972.tb01914.x. [DOI] [PubMed] [Google Scholar]
- 30.Pieper D H, Stadler-Fritzsche K, Knackmuss H-J, Timmis K N. Formation of dimethylmuconolactones from dimethylphenols by Alcaligenes eutrophus JMP 134. Appl Environ Microbiol. 1995;61:2159–2165. doi: 10.1128/aem.61.6.2159-2165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Puszko A. 13C NMR, IR, and UV absorption spectra of 2-halopyridinecarboxylic acids and their N-oxides. Polish J Chem. 1993;67:837–847. [Google Scholar]
- 32.Sala-Trepat J M, Evans W C. The meta cleavage of catechol by Azotobacter species. 4-Oxalocrotonate pathway. Eur J Biochem. 1971;20:400–413. doi: 10.1111/j.1432-1033.1971.tb01406.x. [DOI] [PubMed] [Google Scholar]
- 33.Schmidt S, Cain R B, Rao G V, Kirby G W. Isolation and identification of two novel butenolides as products of dimethylbenzoate metabolism by Rhodococcus rhodochrous N75. FEMS Microbiol Lett. 1994;120:93–98. [Google Scholar]
- 34.Senda T, Sugiyama K, Narita H, Yamamoto T, Kimbara K, Fukuda M, Sato M, Yano K, Mitsui Y. Three-dimensional structures of free form and two substrate complexes of an extradiol ring-cleavage type dioxygenase, the BphC enzyme from Pseudomonas sp. strain KKS102. J Mol Biol. 1996;255:735–752. doi: 10.1006/jmbi.1996.0060. [DOI] [PubMed] [Google Scholar]
- 35.Shu L, Chiou Y-M, Orville A M, Miller M A, Lipscomb J D, Que L., Jr X-ray absorption spectroscopic studies of the Fe(II) active site of catechol 2,3-dioxygenase. Implications for the extradiol cleavage mechanism. Biochemistry. 1995;34:6649–6659. doi: 10.1021/bi00020a010. [DOI] [PubMed] [Google Scholar]
- 36.Simons W W. The Sadtler guide to carbon-13 NMR spectra. Philadelphia, Pa: Sadtler Research Laboratories; 1983. [Google Scholar]
- 37.Wieser M, Eberspächer J, Vogler B, Lingens F. Metabolism of 4-chlorophenol by Azotobacter sp. GP1: structure of the meta cleavage product of 4-chlorocatechol. FEMS Microbiol Lett. 1994;116:73–78. doi: 10.1111/j.1574-6968.1994.tb06678.x. [DOI] [PubMed] [Google Scholar]